dmrt1 Is Responsible for Androgen-Induced Masculinization in Nile Tilapia
Abstract
1. Introduction
2. Materials and Methods
2.1. Nile tilapia Culture and Genetic Sex Identification
2.2. Nile tilapia Mutant Lines
2.3. Steroid Treatment
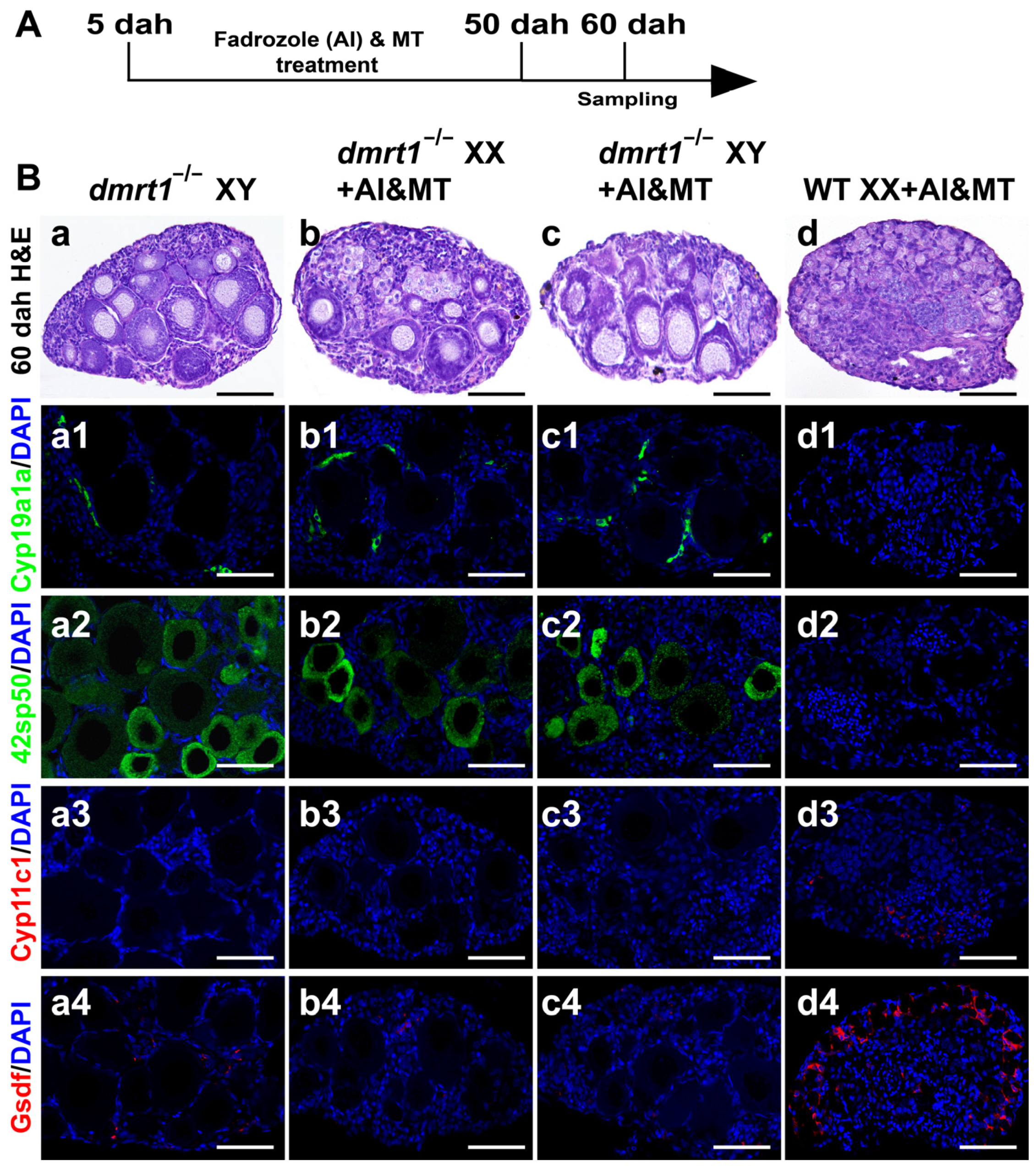
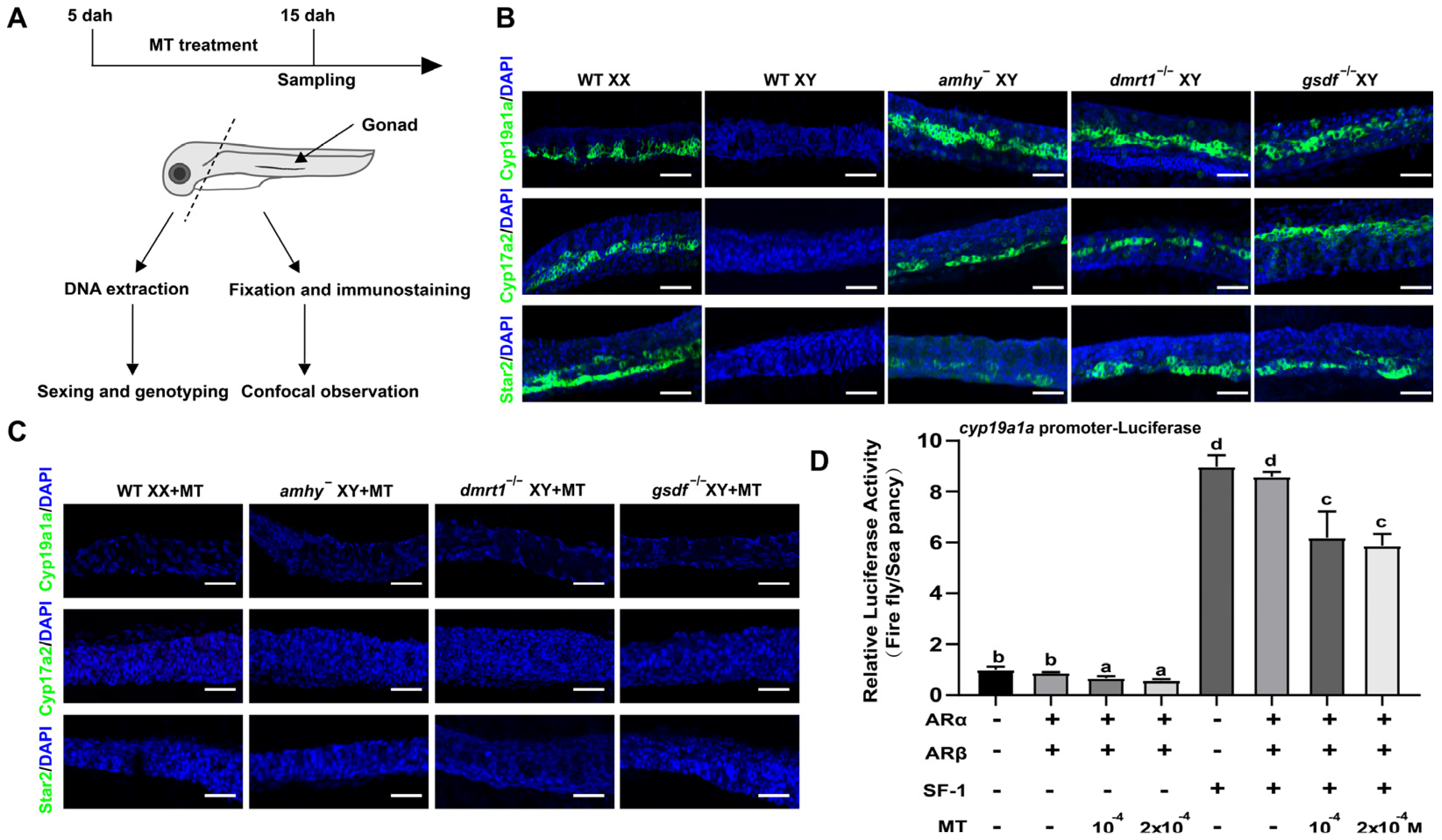
2.4. Histological Observation and Immunofluorescence
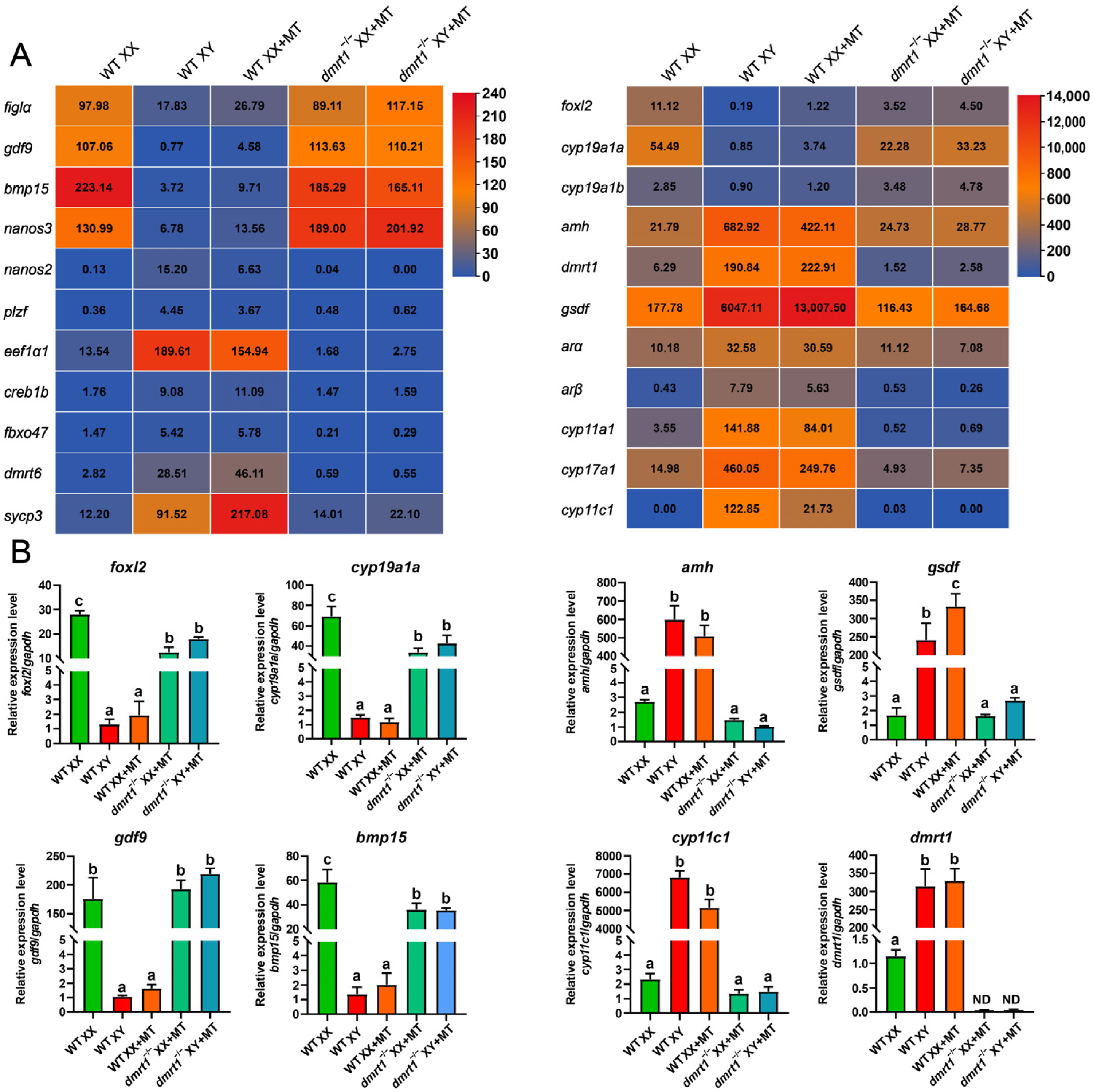
2.5. RNA Isolation, Transcriptome Sequencing, and Real-Time PCR Verification
2.6. Whole-Mount Gonad Immunofluorescence
2.7. Luciferase Assay
2.8. EdU Incorporation and Germ Cell Proliferation Analysis
2.9. Data Analysis
3. Results
3.1. amhy and gsdf Mutants But Not dmrt1 Mutants Were Masculinized by MT Treatment
3.2. Gonads of dmrt1 Mutants Retained Female Identity after MT Treatment
3.3. Co-Treatment with MT and AI Failed to Induce Sex Reversal in dmrt1 Mutants
3.4. MT Treatment Inhibited Early Steroidogenesis Independent of Male Pathway Genes
3.5. Dmrt1 Is Required for MT-Induced Inhibition of Germ Cell Proliferation
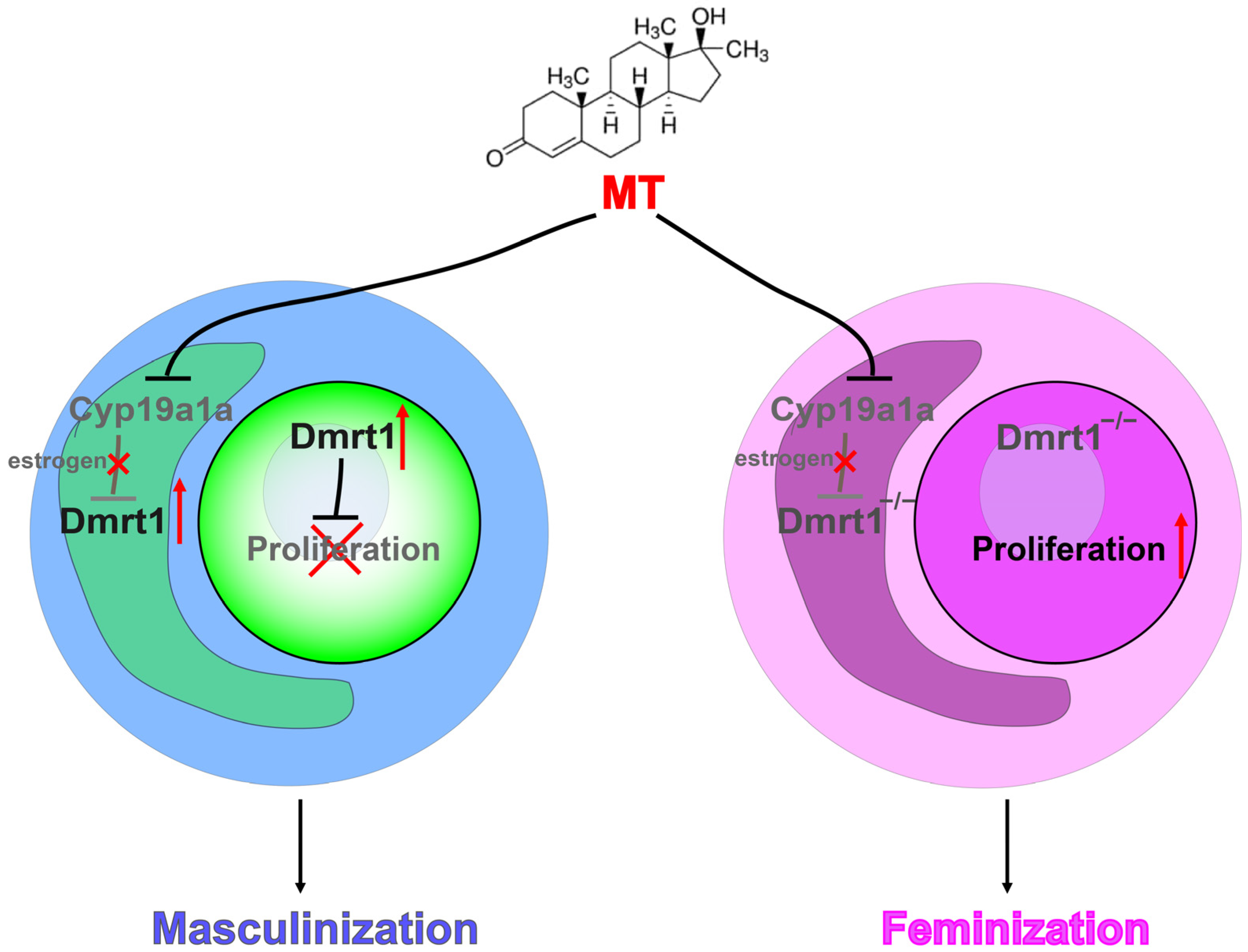
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Capel, B. Vertebrate sex determination: Evolutionary plasticity of a fundamental switch. Nat. Rev. Genet. 2017, 18, 675–689. [Google Scholar] [CrossRef] [PubMed]
- Nagahama, Y.; Chakraborty, T.; Paul-Prasanth, B.; Ohta, K.; Nakamura, M. Sex determination, gonadal sex differentiation, and plasticity in vertebrate species. Physiol. Rev. 2021, 101, 1237–1308. [Google Scholar] [CrossRef] [PubMed]
- Devlin, R.H.; Nagahama, Y. Sex determination and sex differentiation in fish: An overview of genetic, physiological, and environmental influences. Aquaculture 2002, 208, 191–364. [Google Scholar]
- Yamamoto, T. Artificial induction of functional sex-reversal in genotypic females of the medaka (Oryzias latipes). J. Exp. Zool. 1958, 137, 227–263. [Google Scholar] [CrossRef] [PubMed]
- Owusu-Frimpong, M.; Nijjhar, B. Induced sex reversal in Tilapia nilotica (Cichlidae) with methyl testosterone. Hydrobiologia 1981, 78, 157–160. [Google Scholar] [CrossRef]
- Komen, J.; Lodder, P.A.; Huskens, F.; Richter, C.J.; Huisman, E.A. Effects of oral administration of 17α-methyltestosterone and 17β-estradiol on gonadal development in common carp, Cyprinus carpio L. Aquaculture 1989, 78, 349–363. [Google Scholar] [CrossRef]
- Feist, G.; Yeoh, C.G.; Fitzpatrick, M.S.; Schreck, C.B. The production of functional sex-reversed male rainbow trout with 17α-methyltestosterone and 11 β-hydroxyandrostenedione. Aquaculture 1995, 131, 145–152. [Google Scholar] [CrossRef]
- Orn, S.; Holbech, H.; Madsen, T.H.; Norrgren, L.; Petersen, G.I. Gonad development and vitellogenin production in zebrafish (Danio rerio) exposed to ethinylestradiol and methyltestosterone. Aquat. Toxicol. 2003, 65, 397–411. [Google Scholar] [CrossRef]
- Kanamori, A.; Yamamura, A.; Koshiba, S.; Lee, J.S.; Orlando, E.F.; Hori, H. Methyltestosterone efficiently induces male development in the self-fertilizing hermaphrodite fish, Kryptolebias marmoratus. Genesis 2006, 44, 495–503. [Google Scholar] [CrossRef]
- Xu, D.; Yang, F.; Chen, R.; Lou, B.; Zhan, W.; Hayashida, T.; Takeuchi, Y. Production of neo-males from gynogenetic yellow drum through 17α-methyltestosterone immersion and subsequent application for the establishment of all-female populations. Aquaculture 2018, 489, 154–161. [Google Scholar] [CrossRef]
- Liu, S.; Xu, P.; Liu, X.; Guo, D.; Chen, X.; Bi, S.; Lai, H.; Wang, G.; Zhao, X.; Su, Y.; et al. Production of neo-male mandarin fish Siniperca chuatsi by masculinization with orally administered 17α-methyltestosterone. Aquaculture 2021, 530, 735904. [Google Scholar] [CrossRef]
- Li, N.; Oakes, J.A.; Storbeck, K.H.; Cunliffe, V.T.; Krone, N.P. The P450 side-chain cleavage enzyme Cyp11a2 facilitates steroidogenesis in zebrafish. J. Endocrinol. 2020, 244, 309–321. [Google Scholar] [CrossRef]
- Zhai, G.; Shu, T.; Xia, Y.; Lu, Y.; Shang, G.; Jin, X.; He, J.; Nie, P.; Yin, Z. Characterization of sexual trait development in cyp17a1-deficient zebrafish. Endocrinology 2018, 159, 3549–3562. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Tang, H.; Liu, Y.; Chen, Y.; Li, G.; Liu, X.; Lin, H. Targeted disruption of aromatase reveals dual functions of cyp19a1a during sex differentiation in zebrafish. Endocrinology 2017, 158, 3030–3041. [Google Scholar] [CrossRef] [PubMed]
- Bhandari, R.K.; Nakamura, M.; Kobayashi, T.; Nagahama, Y. Suppression of steroidogenic enzyme expression during androgen-induced sex reversal in Nile tilapia (Oreochromis niloticus). Gen. Comp. Endocrinol. 2007, 150, 521, Erratum in Gen. Comp. Endocrinol. 2006, 145, 20–24. [Google Scholar] [CrossRef]
- Wang, J.Y.; Ma, Y.X.; Hu, Q.M.; Peng, F.; Zhou, M.; Ji, X.S.; Zhao, Y. All-male Nile tilapia larvae production using high-temperature and low dose of MT combination treatment. Aquaculture 2022, 546, 737311. [Google Scholar] [CrossRef]
- Horie, Y.; Myosho, T.; Sato, T.; Sakaizumi, M.; Hamaguchi, S.; Kobayashi, T. Androgen induces gonadal soma-derived factor, Gsdf, in XX gonads correlated to sex-reversal but not Dmrt1 directly, in the teleost fish, northern medaka (Oryzias sakaizumii). Mol. Cell. Endocrinol. 2016, 436, 141–149. [Google Scholar] [CrossRef]
- Lee, S.L.J.; Horsfield, J.A.; Black, M.A.; Rutherford, K.; Fisher, A.; Gemmell, N.J. Histological and transcriptomic effects of 17α-methyltestosterone on zebrafish gonad development. BMC Genom. 2017, 18, 557. [Google Scholar] [CrossRef]
- Wang, D.S.; Zhou, L.Y.; Kobayashi, T.; Matsuda, M.; Shibata, Y.; Sakai, F.; Nagahama, Y. Doublesex- and Mab-3-related transcription factor-1 repression of aromatase transcription, a possible mechanism favoring the male pathway in tilapia. Endocrinology 2010, 151, 1331–1340. [Google Scholar] [CrossRef]
- El-Zaeem, S.Y.; El-Hanafy, A.; El-Dahhar, A.A.; Elmaghraby, A.M.; Hendy, A.M. A New Investigation to Discriminate Sexes in Alive Nile Tilapia (Oreochromis niloticus) Using Cyp19a1a and Dmrt1 Gene Expression in Tail Fin Tissues. Mar. Biotechnol. 2024, 26, 423–431. [Google Scholar] [CrossRef]
- Papoulias, D.M.; Noltie, D.B.; Tillitt, D.E. Effects of methyl testosterone exposure on sexual differentiation in medaka, Oryzias latipes. Mar. Environ. Res. 2000, 50, 181–184. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T.; Nagahama, Y. Molecular aspects of gonadal differentiation in a teleost fish, the Nile tilapia. Sex. Dev. 2009, 3, 108–117. [Google Scholar] [CrossRef] [PubMed]
- Ijiri, S.; Kaneko, H.; Kobayashi, T.; Wang, D.S.; Sakai, F.; Paul-Prasanth, B.; Nakamura, M.; Nagahama, Y. Sexual dimorphic expression of genes in gonads during early differentiation of a teleost fish, the Nile tilapia Oreochromis niloticus. Biol. Reprod. 2008, 78, 333–341. [Google Scholar] [CrossRef]
- Li, M.; Sun, Y.; Zhao, J.; Shi, H.; Zeng, S.; Ye, K.; Jiang, D.; Zhou, L.; Sun, L.; Tao, W.; et al. A Tandem Duplicate of Anti-Müllerian Hormone with a Missense SNP on the Y Chromosome Is Essential for Male Sex Determination in Nile Tilapia, Oreochromis niloticus. PLoS Genet. 2015, 11, e1005678. [Google Scholar] [CrossRef]
- Jiang, D.N.; Yang, H.H.; Li, M.H.; Shi, H.J.; Zhang, X.B.; Wang, D.S. gsdf is a downstream gene of dmrt1 that functions in the male sex determination pathway of the Nile tilapia. Mol. Reprod. Dev. 2016, 83, 497–508. [Google Scholar] [CrossRef]
- Dai, S.; Qi, S.; Wei, X.; Liu, X.; Li, Y.; Zhou, X.; Xiao, H.; Lu, B.; Wang, D.; Li, M. Germline sexual fate is determined by the antagonistic action of dmrt1 and foxl3/foxl2 in tilapia. Development 2021, 148, dev199380. [Google Scholar] [CrossRef]
- Zhang, X.; Li, M.; Ma, H.; Liu, X.; Shi, H.; Li, M.; Wang, D. Mutation of foxl2 or cyp19a1a Results in Female to Male Sex Reversal in XX Nile Tilapia. Endocrinology 2017, 158, 2634–2647. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.L.; Jiang, D.N.; Zeng, S.; Hu, C.J.; Ye, K.; Yang, C.; Yang, S.J.; Li, M.H.; Wang, D.S. Screening and characterization of sex-linked DNA markers and marker-assisted selection in the Nile tilapia (Oreochromis niloticus). Aquaculture 2014, 433, 19–27. [Google Scholar] [CrossRef]
- Yu, X.; Wu, L.; Xie, L.; Yang, S.; Charkraborty, T.; Shi, H.; Wang, D.; Zhou, L. Characterization of two paralogous StAR genes in a teleost, Nile tilapia (Oreochromis niloticus). Mol. Cell. Endocrinol. 2014, 392, 152–162. [Google Scholar] [CrossRef]
- Yang, L.; Zhang, X.; Liu, S.; Zhao, C.; Miao, Y.; Jin, L.; Wang, D.; Zhou, L. Cyp17a1 is Required for Female Sex Determination and Male Fertility by Regulating Sex Steroid Biosynthesis in Fish. Endocrinology 2021, 162, bqab205. [Google Scholar] [CrossRef]
- Yang, L.; Wu, Y.; Su, Y.; Zhang, X.; Chakraborty, T.; Wang, D.; Zhou, L. Cyp17a2 is involved in testicular development and fertility in male Nile tilapia, Oreochromis niloticus. Front. Endocrinol. 2022, 13, 1074921. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.S.; Kobayashi, T.; Zhou, L.Y.; Paul-Prasanth, B.; Ijiri, S.; Sakai, F.; Okubo, K.; Morohashi, K.; Nagahama, Y. Foxl2 up-regulates aromatase gene transcription in a female-specific manner by binding to the promoter as well as interacting with ad4 binding protein/steroidogenic factor 1. Mol. Endocrinol. 2007, 21, 712–725. [Google Scholar] [CrossRef] [PubMed]
- Mor, G.; Eliza, M.; Song, J.; Wiita, B.; Chen, S.; Naftolin, F. 17alpha-methyl testosterone is a competitive inhibitor of aromatase activity in Jar choriocarcinoma cells and macrophage-like THP-1 cells in culture. J. Steroid Biochem. Mol. Biol. 2001, 79, 239–246. [Google Scholar] [CrossRef]
- Nakamura, M.; Bhandari, R.K.; Higa, M. The role estrogens play in sex differentiation and sex changes of fish. Fish Physiol. Biochem. 2003, 28, 113–117. [Google Scholar] [CrossRef]
- Tao, W.; Yuan, J.; Zhou, L.; Sun, L.; Sun, Y.; Yang, S.; Li, M.; Zeng, S.; Huang, B.; Wang, D. Characterization of gonadal transcriptomes from Nile tilapia (Oreochromis niloticus) reveals differentially expressed genes. PLoS ONE 2013, 8, e63604. [Google Scholar] [CrossRef]
- Castañeda-Cortés, D.C.; Zhang, J.; Boan, A.F.; Langlois, V.S.; Fernandino, J.I. High temperature stress response is not sexually dimorphic at the whole-body level and is dependent on androgens to induce sex reversal. Gen. Comp. Endocrinol. 2020, 299, 113605. [Google Scholar] [CrossRef]
- Papaconstantinou, A.D.; Umbreit, T.H.; Goering, P.L.; Brown, K.M. Effects of 17 alpha-methyltestosterone on uterine morphology and heat shock protein expression are mediated through estrogen and androgen receptors. J. Steroid Biochem. Mol. Biol. 2002, 82, 305–314. [Google Scholar] [CrossRef]
- Feldkoren, B.I.; Andersson, S. Anabolic-androgenic steroid interaction with rat androgen receptor in vivo and in vitro: A comparative study. J. Steroid Biochem. Mol. Biol. 2005, 94, 481–487. [Google Scholar] [CrossRef]
- Shi, Y.; Liu, X.; Zhang, H.; Zhang, Y.; Lu, D.; Lin, H. Molecular identification of an androgen receptor and its changes in mRNA levels during 17α-methyltestosterone-induced sex reversal in the orange-spotted grouper Epinephelus coioides. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2012, 163, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Qi, S.; Dai, S.; Zhou, X.; Wei, X.; Chen, P.; He, Y.; Kocher, T.D.; Wang, D.; Li, M. Dmrt1 is the only male pathway gene tested indispensable for sex determination and functional testis development in tilapia. PLoS Genet. 2024, 20, e1011210. [Google Scholar] [CrossRef] [PubMed]
- Mustapha, U.F.; Huang, Y.; Huang, Y.Q.; Assan, D.; Shi, H.J.; Jiang, M.Y.; Deng, S.P.; Li, G.L.; Jiang, D.N. Gonadal development and molecular analysis revealed the critical window for sex differentiation, and E2 reversibility of XY-male spotted scat, Scatophagus argus. Aquaculture 2021, 544, 737147. [Google Scholar] [CrossRef]
- Ioannidis, J.; Taylor, G.; Zhao, D.; Liu, L.; Idoko-Akoh, A.; Gong, D.; Lovell-Badge, R.; Guioli, S.; McGrew, M.J.; Clinton, M. Primary sex determination in birds depends on DMRT1 dosage, but gonadal sex does not determine adult secondary sex characteristics. Proc. Natl. Acad. Sci. USA 2021, 118, e2020909118. [Google Scholar] [CrossRef]
- Ge, C.; Ye, J.; Zhang, H.; Zhang, Y.; Sun, W.; Sang, Y.; Capel, B.; Qian, G. Dmrt1 induces the male pathway in a turtle species with temperature-dependent sex determination. Development 2017, 144, 2222–2233. [Google Scholar]
- Zarkower, D.; Murphy, M.W. DMRT1: An Ancient Sexual Regulator Required for Human Gonadogenesis. Sex. Dev. 2022, 16, 112–125. [Google Scholar] [CrossRef]
- Nakamura, M.; Kobayashi, T.; Chang, X.T.; Nagahama, Y. Gonadal sex differentiation in teleost fish. J. Exp. Zool. 1998, 281, 362–372. [Google Scholar] [CrossRef]
- Crowder, C.M.; Lassiter, C.S.; Gorelick, D.A. Nuclear Androgen Receptor Regulates Testes Organization and Oocyte Maturation in Zebrafish. Endocrinology 2018, 159, 980–993. [Google Scholar] [CrossRef]
- Herpin, A.; Schindler, D.; Kraiss, A.; Hornung, U.; Winkler, C.; Schartl, M. Inhibition of primordial germ cell proliferation by the medaka male determining gene Dmrt1bY. BMC Dev. Biol. 2007, 7, 99. [Google Scholar] [CrossRef]
- Adolfi, M.C.; Fischer, P.; Herpin, A.; Regensburger, M.; Kikuchi, M.; Tanaka, M.; Schartl, M. Increase of cortisol levels after temperature stress activates dmrt1a causing female-to-male sex reversal and reduced germ cell number in medaka. Mol. Reprod. Dev. 2019, 86, 1405–1417. [Google Scholar] [CrossRef]
- Sakae, Y.; Oikawa, A.; Sugiura, Y.; Mita, M.; Nakamura, S.; Nishimura, T.; Suematsu, M.; Tanaka, M. Starvation causes female-to-male sex reversal through lipid metabolism in the teleost fish, medaka (Olyzias latipes). Biol. Open 2020, 9, bio050054. [Google Scholar] [CrossRef] [PubMed]
- Krentz, A.D.; Murphy, M.W.; Kim, S.; Cook, M.S.; Capel, B.; Zhu, R.; Matin, A.; Sarver, A.L.; Parker, K.L.; Griswold, M.D.; et al. The DM domain protein DMRT1 is a dose-sensitive regulator of fetal germ cell proliferation and pluripotency. Proc. Natl. Acad. Sci. USA 2009, 106, 22323–22328. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dai, S.; Li, M.; Yuan, J.; Wei, X.; Ma, E.; Wang, D.; Li, M. dmrt1 Is Responsible for Androgen-Induced Masculinization in Nile Tilapia. Genes 2024, 15, 1238. https://doi.org/10.3390/genes15091238
Dai S, Li M, Yuan J, Wei X, Ma E, Wang D, Li M. dmrt1 Is Responsible for Androgen-Induced Masculinization in Nile Tilapia. Genes. 2024; 15(9):1238. https://doi.org/10.3390/genes15091238
Chicago/Turabian StyleDai, Shengfei, Mei Li, Jie Yuan, Xueyan Wei, Eryan Ma, Deshou Wang, and Minghui Li. 2024. "dmrt1 Is Responsible for Androgen-Induced Masculinization in Nile Tilapia" Genes 15, no. 9: 1238. https://doi.org/10.3390/genes15091238
APA StyleDai, S., Li, M., Yuan, J., Wei, X., Ma, E., Wang, D., & Li, M. (2024). dmrt1 Is Responsible for Androgen-Induced Masculinization in Nile Tilapia. Genes, 15(9), 1238. https://doi.org/10.3390/genes15091238






