Advances in Understanding the Karyotype Evolution of Tetrapulmonata and Two Other Arachnid Taxa, Ricinulei and Solifugae
Abstract
1. Introduction
2. Material and Methods
2.1. Material Examined and Taxonomic Determination
2.2. Optimal Ontogenetic Stages and Tissues for Chromosome Preparation
2.3. Preparation of Chromosomes and Analysis of Constitutive Heterochromatin
2.4. Detection of NORs and Telomere Repeats by Fluorescence In Situ Hybridization (FISH)
2.5. Detection of Sex Chromosomes by Comparative Genomic Hybridization (CGH)
2.6. Analysis of Preparations and Images
2.7. Evolution of Chromosome Characters
3. Results
3.1. Amblypygi
3.1.1. Charontidae
3.1.2. Phrynidae
3.1.3. Phrynichidae
3.2. Thelyphonida
3.2.1. Thelyphonidae (Hypoctoninae)
3.2.2. Thelyphonidae (Mastigoproctinae)
3.2.3. Thelyphonidae (Thelyphoninae)
3.2.4. Thelyphonidae (Typopeltinae)
3.3. Schizomida
3.4. Ricinulei
3.5. Solifugae
3.5.1. Ammotrechidae and Eremobatidae
3.5.2. Daesiidae
3.5.3. Galeodidae and Rhagodidae
3.5.4. Solpugidae
4. Discussion
4.1. Evolution of Cytogenetic Characters
4.1.1. Diploid Numbers and Chromosome Morphology
Arachnid Orders
Amblypygi and Thelyphonida
Schizomida
Ricinulei
Solifugae
4.1.2. Somatic Pairing of Chromosomes
4.1.3. Constitutive Heterochromatin
4.1.4. Nucleolar Organizer Regions
4.1.5. Telomeric Repeats
4.1.6. Sex Chromosomes
4.1.7. Male Meiosis
4.2. Karyotype Evolution of Arachnids
4.2.1. Solifugae and Putative Relatives
4.2.2. Tetrapulmonata
4.2.3. Ricinulei
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ontano, A.Z.; Steiner, H.G.; Sharma, P.P. How many long branch orders occur in Chelicerata? Opposing effects of Palpigradi and Opilioacariformes on phylogenetic stability. Mol. Phylogenet. Evol. 2022, 168, 107378. [Google Scholar] [CrossRef] [PubMed]
- Harvey, M.S. The neglected cousins: What do we know about the smaller arachnid orders? J. Arachnol. 2002, 30, 357–372. [Google Scholar] [CrossRef]
- Garwood, R.J.; Dunlop, J. Three-dimensional reconstruction and the phylogeny of extinct chelicerate orders. PeerJ 2014, 2, e641. [Google Scholar] [CrossRef]
- Wheeler, W.C.; Hayashi, C.Y. The phylogeny of the extant chelicerate orders. Cladistics 1998, 14, 173–192. [Google Scholar] [CrossRef]
- Giribet, G.; Edgecombe, G.D.; Wheeler, W.C.; Babbitt, C. Phylogeny of the Arachnida and Opiliones: A combined approach using morphological and molecular sequence data. Cladistics 2002, 18, 5–70. [Google Scholar] [PubMed]
- Regier, J.C.; Shultz, J.W.; Zwick, A.; Hussey, A.; Ball, B.; Wetzer, R.; Martin, J.W.; Cunningham, C.W. Arthropod relationships revealed by phylogenomic analysis of nuclear protein coding sequences. Nature 2010, 463, 1079–1083. [Google Scholar] [CrossRef] [PubMed]
- Arabi, J.; Judein, M.L.I.; Deharveng, L.; Lourenço, W.R.; Cruaund, C.; Hussain, A. Nucleotide composition of CO1 sequences in Chelicerata (Arthropoda): Detecting new mitogenomic rearrangements. J. Mol. Evol. 2012, 74, 81–95. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.P.; Kaluziak, S.T.; Pérez-Porro, A.R.; González, V.L.; Hormiga, G.; Wheeler, W.C.; Giribet, G. Phylogenomic interrogation of Arachnida reveals systemic conflicts in phylogenetic signal. Mol. Biol. Evol. 2014, 31, 2963–2984. [Google Scholar] [CrossRef]
- Lozano-Fernandez, J.; Tanner, A.R.; Giacomelli, M.; Carton, R.; Vinther, J.; Edgecombe, G.D.; Pisani, D. Increasing species sampling in chelicerate genomic-scale datasets provides support for monophyly of Acari and Arachnida. Nat. Commun. 2019, 10, 2295. [Google Scholar] [CrossRef] [PubMed]
- Ballesteros, J.A.; Sharma, P.P. A critical appraisal of the placement of Xiphosura (Chelicerata) with account of known sources of phylogenetic error. Syst. Biol. 2019, 68, 896–917. [Google Scholar] [CrossRef]
- Howard, R.J.; Edgecombe, G.D.; Legg, D.A.; Pisani, D.; Lozano-Fernandez, J. Exploring the evolution and terrestrialization of scorpions (Arachnida: Scorpiones) with rocks and clocks. Org. Divers. Evol. 2019, 19, 71–86. [Google Scholar] [CrossRef]
- Ban, X.C.; Shao, Z.K.; Wu, L.J.; Sun, J.T.; Xue, X.F. Highly diversified mitochondrial genomes provide new evidence for interordinal relationships in the Arachnida. Cladistics 2022, 38, 452–464. [Google Scholar] [CrossRef]
- Deakin, J.E.; Pootter, S.; O’Neill, R.; Ruiz-Herrera, A.; Cioffi, M.B.; Eldridge, M.D.B.; Fukui, K.; Marshall Graves, J.A.; Griffin, D.; Grutzner, F.; et al. Chromosomics: Bridging the gap between genomes and chromosomes. Genes 2019, 10, 627. [Google Scholar] [CrossRef] [PubMed]
- Araujo, D.; Schneider, M.C.; Paula-Neto, E.; Cella, D.M. The Spider Cytogenetic Database, Version 13.0. Available online: http://www.arthropodacytogenetics.bio.br/spiderdatabase (accessed on 12 December 2024).
- Norton, R.A.; Kethley, J.B.; Johnston, D.E.; OConnor, B.M. Phylogenetic perspectives on genetic systems and reproductive modes of mites. In Evolution and Diversity of Sex Ratio in Insects and Mites; Wrensch, D.L., Ebbert, M.A., Eds.; Chapman & Hall: New York, NY, USA, 1993; pp. 8–99. [Google Scholar]
- Tsurusaki, N.; Svojanovská, H.; Schöenhofer, A.; Šťáhlavský, F. The Harvestmen Cytogenetic Database, Version 9.1. Available online: http://arthropodacytogenetics.bio.br/harvestmendatabase (accessed on 12 December 2024).
- Schneider, M.C.; Mattos, V.F.; Cella, D.M. The Scorpion Cytogenetic Database, Version 12. Available online: http://www.arthropodacytogenetics.bio.br/scorpiondatabase (accessed on 12 December 2024).
- Šťáhlavský, F. The Pseudoscorpion Cytogenetic Database, Version 10.6. Available online: http://www.arthropodacytogenetics.bio.br/pseudoscorpiondatabase (accessed on 12 December 2024).
- Harvey, M.S. Catalogue of the Smaller Arachnid Orders of the World: Amblypygi, Uropygi, Schizomida, Palpigradi, Ricinulei and Solifugae; CSIRO Publishing: Clayton, Australia, 2003. [Google Scholar]
- Wheeler, W.C.; Coddington, J.A.; Crowley, L.M.; Dimitrov, D.; Goloboff, P.A.; Griswold, C.E.; Hormiga, G.; Prendini, L.; Ramírez, M.J.; Sierwald, P.; et al. The spider tree of life: Phylogeny of Araneae based on target-gene analyses from an extensive taxon sampling. Cladistics 2017, 33, 574–616. [Google Scholar] [CrossRef] [PubMed]
- World Spider Catalog. Version 25. Natural History Museum Bern. Available online: http://wsc.nmbe.ch (accessed on 19 August 2024).
- Moreno-González, J.A.; Gutierrez-Estrada, M.; Prendini, L. Systematic revision of the whip spider family Paracharontidae (Arachnida: Amblypygi) with description of a new troglobitic genus and species from Colombia. Am. Mus. Novitates 2023, 4000, 1–36. [Google Scholar] [CrossRef]
- World Amblypygi Catalog. Natural History Museum Bern. 2022. Available online: http://wac.nmbe.ch (accessed on 19 August 2024).
- De Miranda, G.S.; Kulkarni, S.S.; Tagliatela, J.; Baker, C.M.; Giupponi, A.P.L.; Labarque, F.M.; Gavish-Regev, E.; Rix, M.G.; Carvalho, L.S.; Fusari, L.M.; et al. The rediscovery of a relict unlocks the first global phylogeny of whip spiders (Amblypygi). Syst. Biol. 2024, 73, 495–505. [Google Scholar] [CrossRef]
- World Schizomida Catalog. Natural History Museum Bern. 2022. Available online: http://wac.nmbe.ch (accessed on 19 August 2024).
- World Uropygi Catalog. Natural History Museum Bern. 2022. Available online: http://wac.nmbe.ch (accessed on 19 August 2024).
- Clouse, R.M.; Branstetter, M.G.; Buenavente, P.; Crowley, L.M.; Czekanski-Moir, J.; General, D.E.M.; Giribet, G.; Harvey, M.S.; Janies, D.A.; Mohagan, A.B.; et al. First global molecular phylogeny and biogeographical analysis of two arachnid orders (Schizomida and Uropygi) supports a tropical Pangean origin and mid-Cretaceous diversification. J. Biogeogr. 2017, 44, 2660–2672. [Google Scholar] [CrossRef]
- Pepato, A.R.; da Rocha, C.A.F.; Dunlop, J.A. Phylogenetic position of the acariform mites: Sensitivity to homology assessment under total evidence. BMC Evol. Biol. 2010, 10, 235. [Google Scholar] [CrossRef]
- Weygoldt, P.; Paulus, H.F. Untersuchungen zur Morphologie, Taxonomie und Phylogenie der Chelicerata. Z. Zool. Syst. Evol. 1979, 17, 177–200. [Google Scholar] [CrossRef]
- Petrunkevitch, A.I. Arachnida. In Treatise on Invertebrate Palaeontology, Part P. Arthropoda 2; Moore, R.C., Ed.; Geological Society of America & University of Kansas Press: Lawrence, KS, USA, 1955; pp. 42–162. [Google Scholar]
- Van der Hammen, L. An Introduction to Comparative Arachnology; SPB Academic Publishing: Hague, The Netherlands, 1989. [Google Scholar]
- Shultz, J.W. Muscular anatomy of a whipspider, Phrynus longipes (Amblypygi), and its evolutionary significance. Zool. J. Linn. Soc. 1999, 126, 81–116. [Google Scholar] [CrossRef]
- Shultz, J.W. A phylogenetic analysis of the arachnid orders based on morphological characters. Zool. J. Linn. Soc. 2007, 150, 221–265. [Google Scholar] [CrossRef]
- Shear, W.A.; Selden, P.A.; Rolfe, W.D.I.; Bonamo, P.M.; Grierson, J.D. New terrestrial arachnids from the Devonian of Gilboa, New York (Arachnida, Trigonotarbida). Am. Mus. Novit. 1987, 2901, 1–74. [Google Scholar]
- Bu, Y.; Gao, Y.; Nunes Godeiro, N. The first complete mitochondrial genome of the micro-whip-scorpion Schizomus zhensis (Arachnida: Schizomida) and phylogenetic analysis. Mitochondrial DNA B Resour. 2022, 7, 709–711. [Google Scholar] [CrossRef]
- Selden, P.A.; Shear, W.A.; Bonamo, P.M. A spider and other arachnids from the Devonian of New York, and reinterpretations of Devonian Araneae. Palaeontology 1991, 34, 241–281. [Google Scholar]
- Dunlop, J.A. Evidence for a sister group relationship between Ricinulei and Trigonotarbida. Bull. Brit. Arachnol. Soc. 1996, 10, 193–204. [Google Scholar]
- World Ricinulei Catalog. Natural History Museum Bern. 2022. Available online: http://wac.nmbe.ch (accessed on 19 August 2024).
- Murienne, J.; Benavides, L.R.; Prendini, L.; Hormiga, G.; Giribet, G. Forest refugia in Western and Central Africa as ‘museums’ of Mesozoic biodiversity. Biol. Lett. 2013, 9, 20120932. [Google Scholar] [CrossRef] [PubMed]
- Fernández, R.; Giribet, G. Unnoticed in the tropics: Phylogenomic resolution of the poorly known arachnid order Ricinulei (Arachnida). R. Soc. Open Sci. 2015, 2, 150065. [Google Scholar] [CrossRef] [PubMed]
- Benavides, L.R.; Daniels, S.R.; Giribet, G. Understanding the real magnitude of the arachnid order Ricinulei through deep Sanger sequencing across its distribution range and phylogenomics, with the formalization of the first species from the Lesser Antilles. J. Zool. Syst. Evol. Res. 2021, 59, 1850–1873. [Google Scholar] [CrossRef]
- Sato, S.; Derkarabetian, S.; Valdez-Mondragón, A.; Pérez-González, A.; Benavides, L.R.; Daniels, S.R.; Giribet, G. Under the hood: Phylogenomics of hooded tick spiders (Arachnida, Ricinulei) uncovers discordance between morphology and molecules. Mol. Phylogenet. Evol. 2024, 193, 108026. [Google Scholar] [CrossRef]
- Schwager, E.E.; Sharma, P.P.; Clarke, T.; Leite, D.J.; Wierschin, T.; Pechmann, M.; Akiyama-Oda, Y.; Esposito, L.; Bechsgaard, J.; Bilde, T.; et al. The house spider genome reveals an ancient whole-genome duplication during arachnid evolution. BMC Biol. 2017, 15, 62. [Google Scholar] [CrossRef] [PubMed]
- Nolan, E.D.; Santibáñez-López, C.E.; Sharma, P.P. Developmental gene expression as a phylogenetic data class: Support for the monophyly of Arachnopulmonata. Dev. Genes Evol. 2020, 230, 137–153. [Google Scholar] [CrossRef] [PubMed]
- Harper, A.; Baudouin Gonzalez, L.; Schönauer, A.; Janssen, R.; Seiter, M.; Holzem, M.; Arif, S.; McGregor, A.P.; Sumner-Rooney, L. Widespread retention of ohnologs in key developmental gene families following whole-genome duplication in arachnopulmonates. G3 2021, 11, jkab299. [Google Scholar] [CrossRef]
- Ontano, A.Z.; Gainett, G.; Aharon, S.; Ballesteros, J.A.; Benavides, L.R.; Corbett, K.F.; Gavish-Regev, E.; Harvey, M.S.; Monsma, S.; Santibáñez-López, C.E.; et al. Taxonomic sampling and rare genomic changes overcome long-branch attraction in the phylogenetic placement of pseudoscorpions. Mol. Biol. Evol. 2021, 38, 2446–2467. [Google Scholar] [CrossRef]
- Ballesteros, J.A.; Santibáñez López, C.E.; Kováč, L.; Gavish-Regev, E.; Sharma, P.P. Ordered phylogenomic subsampling enables diagnosis of systematic errors in the placement of the enigmatic arachnid order Palpigradi. Proc. R. Soc. B 2019, 286, 2426. [Google Scholar] [CrossRef]
- Kulkarni, S.S.; Yamasaki, T.; Thi Hong Phung, L.; Karuaera, N.; Daniels, S.R.; Gavish-Regev, E.; Sharma, P.P. Phylogenomic data reveal three new families of poorly studied Solifugae (camel spiders). Mol. Phylogenet. Evol. 2024, 191, 107989. [Google Scholar] [CrossRef]
- Dunlop, J.A.; Krüger, J.; Alberti, G. The sejugal furrow in camel spiders and acariform mites. Arachnol. Mitt. 2012, 43, 8–15. [Google Scholar] [CrossRef]
- Dabert, M.; Witalinski, W.; Kazmierski, A.; Olszanowski, Z.; Dabert, J. Molecular phylogeny of acariform mites (Acari, Arachnida): Strong conflict between phylogenetic signal and long-branch attraction artifacts. Mol. Phylogenet. Evol. 2010, 56, 222–241. [Google Scholar] [CrossRef] [PubMed]
- Ballesteros, J.A.; Santibáñez-López, C.E.; Baker, C.M.; Benavides, L.R.; Cunha, T.J.; Gainett, G.; Ontano, A.Z.; Setton, E.V.W.; Arango, C.P.; Gavish-Regev, E.; et al. Comprehensive species sampling and sophisticated algorithmic approaches refute the monophyly of Arachnida. Mol. Biol. Evol. 2022, 39, msac021. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, S.S.; Steiner, H.G.; Garcia, E.L.; Iuri, H.; Jones, R.R.; Ballesteros, J.A.; Gainett, G.; Graham, M.R.; Harms, D.; Lyle, R.; et al. Neglected no longer: Phylogenomic resolution of higher level relationships in Solifugae. iScience 2023, 26, 107684. [Google Scholar] [CrossRef] [PubMed]
- Král, J.; Forman, M.; Kořínková, T.; Reyes Lerma, A.C.; Haddad, C.R.; Musilová, J.; Řezáč, M.; Ávila Herrera, I.M.; Thakur, S.; Dippenaar-Schoeman, A.S.; et al. Insights into the karyotype and genome evolution of haplogyne spiders indicate a polyploid origin of lineage with holokinetic chromosomes. Sci. Rep. 2019, 9, 3001. [Google Scholar] [CrossRef] [PubMed]
- Kořínková, T.; Král, J. Karyotypes, sex chromosomes, and meiotic division in spiders. In Spider Ecophysiology; Nentwig, W., Ed.; Springer-Verlag: Berlin/Heidelberg, Germany, 2013; pp. 159–171. [Google Scholar]
- Král, J.; Kořínková, T.; Krkavcová, L.; Musilová, J.; Forman, M.; Ávila Herrera, I.M.; Haddad, C.R.; Vítková, M.; Henriques, S.; Palacios Vargas, J.G.; et al. Evolution of the karyotype, sex chromosomes, and meiosis in mygalomorph spiders (Araneae: Mygalomorphae). Biol. J. Linn. Soc. 2013, 109, 377–408. [Google Scholar] [CrossRef]
- Král, J. Evolution of multiple sex chromosomes in the spider genus Malthonica (Araneae: Agelenidae) indicates unique structure of the spider sex chromosome systems. Chromosome Res. 2007, 15, 863–879. [Google Scholar] [CrossRef] [PubMed]
- Král, J.; Kořínková, T.; Forman, M.; Krkavcová, L. Insights into the meiotic behavior and evolution of multiple sex chromosome systems in spiders. Cytogenet. Genome Res. 2011, 133, 43–66. [Google Scholar] [CrossRef]
- Sember, A.; Pappová, M.; Forman, M.; Nguyen, P.; Marec, F.; Dalíková, M.; Divišová, K.; Doležálková-Kaštánková, M.; Zrzavá, M.; Sadílek, D.; et al. Patterns of sex chromosome differentiation in spiders: Insights from comparative genomic hybridisation. Genes 2020, 11, 849. [Google Scholar] [CrossRef]
- Forman, M.; Nguyen, P.; Hula, V.; Král, J. Sex chromosome pairing and extensive NOR polymorphism in Wadicosa fidelis (Araneae: Lycosidae). Cytogenet. Genome Res. 2013, 141, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Seiter, M.; Reyes Lerma, A.C.; Král, J.; Sember, A.; Divišová, K.; Palacios-Vargas, J.G.; Colmenares, P.; Loria, S.F.; Prendini, L. Cryptic diversity in the whip spider genus Paraphrynus (Amblypygi: Phrynidae): Integrating morphology, karyotype and DNA. Arthropod Syst. Phylogeny 2020, 78, 265–285. [Google Scholar]
- Paula-Neto, E.; Araujo, D.; Carvalho, L.S.; Cella, D.M.; Schneider, M.C. Chromosomal characteristics of a Brazilian whip spider (Amblypygi) and evolutionary relationships with other arachnid orders. Genet. Mol. Res. 2013, 12, 3726–3734. [Google Scholar] [CrossRef] [PubMed]
- Reyes Lerma, A.C.; Šťáhlavský, F.; Seiter, M.; Carabajal Paladino, L.Z.; Divišová, K.; Forman, M.; Sember, A.; Král, J. Insights into the karyotype evolution of Charinidae, the early-diverging clade of whip spiders (Arachnida: Amblypygi). Animals 2021, 11, 3233. [Google Scholar] [CrossRef]
- Millot, J.; Tuzet, O. La spermatogenèse chez les Pédipalpes. Bull. Biol. Fr. Belg. 1934, 68, 77–83. [Google Scholar]
- Kasturi Bai, A.R.; Parthasarathy, M.D. The chromosomes of Thelyphonus indicus Stoliczka. Proc. Indian Acad. Sci. B 1957, 24, 9–22. [Google Scholar]
- Jayarama, K.S.; Gowda, L.S. Karyological and meiotic studies in whipscorpion Thelyphonus sepiaris Butler. Cytologia 1994, 59, 87–91. [Google Scholar] [CrossRef][Green Version]
- Santos-Costa, R.C.; Carvalho, L.S.; Schneider, M.C. The first cytogenetic description in Schizomida, a micro-diverse arachnid order. 21st International Chromosome Conference. Cytogenet. Genome Res. 2016, 148, 91. [Google Scholar]
- Vítková, M.; Král, J.; Traut, W.; Zrzavý, J.; Marec, F. The evolutionary origin of insect telomeric repeats, (TTAGG)n. Chromosome Res. 2005, 13, 145–156. [Google Scholar] [CrossRef]
- Warren, E. The genital system of Hypoctonus formosus (Butler). Ann. Natal Mus. 1939, 9, 307–344. [Google Scholar]
- Traut, W. Pachytene mapping in the female silkworm, Bombyx mori L. (Lepidoptera). Chromosoma 1976, 58, 275–284. [Google Scholar] [CrossRef]
- Dolejš, P.; Kořínková, T.; Musilová, J.; Opatová, V.; Kubcová, L.; Buchar, J.; Král, J. Karyotypes of central European spiders of the genera Arctiosa, Tricca, and Xerolycosa (Araneae: Lycosidae). Eur. J. Entomol. 2011, 108, 1–16. [Google Scholar] [CrossRef]
- Král, J.; Kováč, Ľ.; Šťáhlavský, F.; Lonský, P.; Ľuptáčik, P. The first karyotype study in palpigrades, a primitive order of arachnids (Arachnida: Palpigradi). Genetica 2008, 134, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Sola, L.; Rossi, A.R.; Iaselli, V.; Rasch, E.M.; Monaco, P.J. Cytogenetics of bisexual/unisexual species of Poecilia. II. Analysis of heterochromatin and nucleolar organizer regions in Poecilia mexicana mexicana by C-banding and DAPI, quinacrine, chromomycin A3, and silver staining. Cytogenet. Cell Genet. 1992, 60, 229–235. [Google Scholar] [CrossRef] [PubMed]
- Ávila Herrera, I.M.; Král, J.; Pastuchová, M.; Forman, M.; Musilová, J.; Kořínková, T.; Šťáhlavský, F.; Zrzavá, M.; Nguyen, P.; Just, P.; et al. Evolutionary pattern of karyotypes and meiosis in pholcid spiders (Araneae: Pholcidae): Implications for reconstructing chromosome evolution of araneomorph spiders. BMC Ecol. Evol. 2021, 21, 75. [Google Scholar]
- Sahara, K.; Marec, F.; Traut, W. TTAGG telomeric repeats in chromosomes of some insects and other arthropods. Chromosome Res. 1999, 7, 449–460. [Google Scholar] [CrossRef] [PubMed]
- Mediouni, J.; Fuková, I.; Frydrychová, R.; Dhouibi, M.H.; Marec, F. Karyotype, sex chromatin and sex chromosome differentiation in the carob moth, Ectomyelois ceratoniae (Lepidoptera: Pyralidae). Caryologia 2004, 57, 184–194. [Google Scholar] [CrossRef]
- Traut, W.; Eickhoff, U.; Schorch, J.C. Identification and analysis of sex chromosomes by comparative genomic hybridization (CGH). Methods Cell Sci. 2001, 23, 157–163. [Google Scholar] [CrossRef]
- Graham, D.E. The isolation of high molecular weight DNA from whole organisms of large tissue masses. Anal. Biochem. 1978, 85, 609–613. [Google Scholar] [CrossRef] [PubMed]
- Traut, W.; Sahara, K.; Otto, T.D.; Marec, F. Molecular differentiation of sex chromosomes probed by comparative genomic hybridization. Chromosoma 1999, 108, 173–180. [Google Scholar] [CrossRef] [PubMed]
- Levan, A.K.; Fredga, K.; Sandberg, A.A. Nomenclature for centromeric position on chromosomes. Hereditas 1964, 52, 201–220. [Google Scholar] [CrossRef]
- Oliver, J.H., Jr. Cytogenetics of mites and ticks. Annu. Rev. Entomol. 1977, 22, 407–429. [Google Scholar] [CrossRef] [PubMed]
- Tsurusaki, N. Chapter 6. Cytogenetics. In The Harvestmen: The Biology of Opiliones; Pinto da Rocha, R., Machado, G., Giribet, G., Eds.; Harvard University Press: Cambridge, MA, USA, 2007; pp. 266–279. [Google Scholar]
- Šťáhlavský, F.; Král, J.; Harvey, M.S.; Haddad, C.R. A karyotype study on the pseudoscorpion families Geogarypidae, Garypinidae and Olpiidae (Arachnida: Pseudoscorpiones). Eur. J. Entomol. 2006, 103, 277–289. [Google Scholar] [CrossRef]
- Schneider, M.C.; Zacaro, A.A.; Pinto-da-Rocha, R.; Candido, D.M.; Cella, D.M. A comparative cytogenetic analysis of 2 Bothriuridae species and overview of the chromosome data of Scorpiones. J. Hered. 2009, 100, 545–555. [Google Scholar] [CrossRef] [PubMed]
- Stávale, L.M.; Schneider, M.C.; Brescovit, A.D.; Cella, D.M. Chromosomal characteristics and karyotype evolution of Oxyopidae spiders (Araneae, Entelegynae). Genet. Mol. Res. 2011, 10, 752–763. [Google Scholar] [CrossRef] [PubMed]
- De Miranda, G.S.; Giupponi, A.P.L.; Scharff, N.; Prendini, L. Phylogeny and biogeography of the pantropical whip spider family Charinidae (Arachnida: Amblypygi). Zool. J. Linn. Soc. 2022, 194, 136–180. [Google Scholar] [CrossRef]
- Suzuki, S. Cytological studies in spiders III. Studies on the chromosomes of fifty-seven species of spiders belonging to seventeen families with general considerations on chromosomal evolution. J. Sci. Hiroshima Univ. B 1954, 15, 23–136. [Google Scholar]
- Král, J.; Dulíková, L.; Kořínková, T.; Musilová, J.; Vítková, M.; Hedin, M.; Henriques, S.S.; Haddad, C.R. Evolution of the karyotype and sex chromosome systems in mesothelid and mygalomorph spiders. In Book of Abstracts, 18th International Congress of Arachnology; Żabka, M., Ed.; University of Podlasie & International Society of Arachnology: Siedlce, Poland, 2010; pp. 227–229. [Google Scholar]
- Rowland, J.M.; Cooke, J.A.L. Systematics of the arachnid order Uropygida (=Thelyphonida). J. Arachnol. 1973, 1, 55–71. [Google Scholar]
- Liu, Y.; Yi, C.; Fan, C.; Liu, Q.; Liu, S.; Shen, L.; Zhang, K.; Huang, Y.; Liu, C.; Wang, Y.; et al. Pan-centromere reveals widespread centromere repositioning of soybean genomes. Proc. Natl. Acad. Sci. USA 2023, 120, e2310177120. [Google Scholar] [CrossRef]
- Noor, M.A.; Grams, K.L.; Bertucci, L.A.; Reiland, J. Chromosomal inversions and the reproductive isolation of species. Proc. Natl. Acad. Sci. USA 2001, 98, 12084–12088. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, A.A.; Riesenberg, L.H. Revisiting the impact of inversions in evolution: From population genetic markers to drivers of adaptive shifts and speciation? Annu. Rev. Ecol. Evol. Syst. 2008, 39, 21–42. [Google Scholar] [CrossRef] [PubMed]
- Rieseberg, L.H. Chromosomal rearrangements and speciation. Trends Ecol. Evol. 2001, 16, 351–358. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.; He, X. Centromere repositioning causes inversion of meiosis and generates a reproductive barrier. Proc. Natl. Acad. Sci. USA 2019, 116, 21580–21591. [Google Scholar] [CrossRef]
- John, B. Meiosis; Cambridge University Press: Cambridge, MA, USA, 1990. [Google Scholar]
- Lukaszewski, A.J.D.; Kopecky, G. Inversions of chromosome arms 4AL and 2BS in wheat invert the patterns of chiasma distribution. Chromosoma 2012, 121, 201–208. [Google Scholar] [CrossRef] [PubMed]
- King, M. Species Evolution: The Role of Chromosome Change; Cambridge University Press: Cambridge, MA, USA, 1993. [Google Scholar]
- Zhang, L.; Reifová, R.; Halenková, Z.; Gompert, Z. How important are structural variants for speciation? Genes 2021, 12, 1084. [Google Scholar] [CrossRef]
- Lysak, M.A. Celebrating Mendel, McClintock, and Darlington: On end-to-end chromosome fusions and nested chromosome fusions. Plant Cell 2022, 34, 2475–2491. [Google Scholar] [CrossRef]
- Warren, E. Spermatogenesis in spiders and the chromosome hypothesis of heredity. Nature 1926, 117, 82–83. [Google Scholar] [CrossRef]
- Hale, D.W.; Greenbaum, I.F. Synapsis of a chromosomal pair heterozygous for a pericentric inversion and the presence of a heterochromatic short arm. Cytogenet. Cell Genet. 1988, 48, 55–57. [Google Scholar] [CrossRef]
- Sharma, T.; Garg, G.S. Constitutive heterochromatin and karyotype variation in Indian pygmy mouse, Mus dunni. Genet. Res. 1975, 25, 189–191. [Google Scholar] [CrossRef]
- Prakhongcheep, O.; Chaiprasertsri, N.; Terada, S.; Hirai, Y.; Srikulnath, K.; Hirai, H.; Koga, A. Heterochromatin blocks constituting the entire short arms of acrocentric chromosomes of Azara’s owl monkey: Formation processes inferred from chromosomal locations. DNA Res. 2013, 20, 461–470. [Google Scholar] [CrossRef] [PubMed]
- Masta, S.E.; Klann, A.E.; Podsiadlowski, L. A comparison of the mitochondrial genomes from two families of Solifugae (Arthropoda: Chelicerata): Eremobatidae and Ammotrechidae. Gene 2008, 417, 35–42. [Google Scholar] [CrossRef]
- Cushing, P.E.; Graham, M.R.; Prendini, L.; Brookhart, J.O. A multilocus molecular phylogeny of the endemic North American camel spider family Eremobatidae (Arachnida: Solifugae). Mol. Phylogenet. Evol. 2015, 92, 280–293. [Google Scholar] [CrossRef] [PubMed]
- Golic, M.M.; Golic, K.G. A quantitative measure of the mitotic pairing of alleles in Drosophila melanogaster and the influence of structural heterozygosity. Genetics 1996, 143, 385–400. [Google Scholar] [CrossRef]
- Tsurusaki, N. Geographic variation of the number of B-chromosomes in Metagagrella tenuipes (Opiliones, Phalangiidae, Gagrellinae). Mem. Queensl. Mus. 1993, 33, 659–665. [Google Scholar]
- Gunn, S.J.; Hilburn, L.R. Differential staining of tick chromosomes: Techniques for C-banding and silver-staining and karyology of Rhipicephalus sanguineus (Latreille). J. Parasitol. 1989, 75, 239–245. [Google Scholar] [CrossRef] [PubMed]
- Gunn, S.J.; Hilburn, L.R. Cytosystematics of five North American Amblyomma (Acarina: Ixodidae) species. J. Parasitol. 1995, 81, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Adilardi, R.S.; Ojanguren-Affilastro, A.A.; Rodriguez-Gil, S.G.; Scioscia, C.L.; Mola, L.M. Meiotic studies in Brachistosternus alienus (Scorpiones; Bothriuridae). J. Arachnol. 2013, 41, 222–226. [Google Scholar] [CrossRef]
- Vitturi, R.; Colomba, M.S.; Caputo, V.; Sparacio, I.; Barbieri, R. High heterochromatin content in somatic chromosomes of two unrelated species of Diplopoda (Myriapoda). Chromosome Res. 1997, 5, 407–412. [Google Scholar] [CrossRef] [PubMed]
- Lopes, D.M.; Fernandes, A.; Diniz, D.; Scudeler, P.E.S.; Foresti, F.; Campos, L.A.D.O. Similarity of heterochromatic regions in the stingless bees (Hymenoptera: Meliponini) revealed by chromosome painting. Caryologia 2014, 67, 222–226. [Google Scholar] [CrossRef]
- Hughes, S.E.; Hawley, R.S. Heterochromatin: A rapidly evolving species barrier. PLoS Biol. 2009, 7, e1000233. [Google Scholar] [CrossRef]
- Ferree, P.M.; Barbash, D.A. Species-specific heterochromatin prevents mitotic chromosome segregation to cause hybrid lethality in Drosophila. PLoS Biol. 2009, 7, e1000234. [Google Scholar] [CrossRef] [PubMed]
- Wolf, K.W. How meiotic cells deal with non-exchange chromosomes. BioEssays 1994, 16, 107–114. [Google Scholar] [CrossRef]
- Gorlov, I.P.; Tsurusaki, N. Morphology and meiotic/mitotic behavior of B chromosomes in a Japanese harvestman, Metagagrella tenuipes (Arachnida: Opiliones): No evidence for B accumulation mechanisms. Zool. Sci. 2000, 17, 349–355. [Google Scholar]
- Oliveira, R.M.; Zacaro, A.A.; Gnaspin, I.P.; Cella, D.M. Cytogenetics of three Brazilian Goniosoma species: A new record for diploid number in Laniatores (Opiliones, Gonyleptidae, Goniosomatinae). J. Arachnol. 2006, 34, 435–443. [Google Scholar] [CrossRef]
- Schneider, M.C.; Zacaro, A.A.; Oliveira, R.M.; Gnaspini, P.; Cella, D.M. Conventional and ultrastructural analyses of the chromosomes of Discocyrtus pectinifemur (Opiliones, Laniatores, Gonyleptidae). J. Zool. Syst. Evol. Res. 2008, 47, 203–207. [Google Scholar] [CrossRef]
- Svojanovská, H.; Nguyen, P.; Hiřman, M.; Tuf, I.H.; Wahab, R.A.; Haddad, C.R.; Šťáhlavský, F. Karyotype evolution in harvestmen of the suborder Cyphophthalmi (Opiliones). Cytogenet. Genome Res. 2016, 148, 227–236. [Google Scholar] [CrossRef]
- Šťáhlavský, F.; Opatova, V.; Just, P.; Lotz, L.N.; Haddad, C.R. Molecular technique reveals high variability of 18S rDNA distribution in harvestmen (Opiliones, Phalangiidae) from South Africa. Comp. Cytogen. 2018, 12, 41–59. [Google Scholar] [CrossRef] [PubMed]
- Jindrová, H.; Hiřman, M.; Sadílek, D.; Bezděčka, P.; Št’áhlavský, F. Distribution of 18S rDNA clusters in Central European harvestmen of the suborder Eupnoi (Arachnida: Opiliones). Eur. J. Entomol. 2020, 117, 282–288. [Google Scholar] [CrossRef]
- Gunn, S.J.; Hilburn, L.R. Cytosystematics of five North American Dermacentor (Acari: Ixodidae) species. J. Med. Entomol. 1990, 27, 620–627. [Google Scholar] [CrossRef]
- Meyer, J.M.; Kurtti, T.J.; Van Zee, J.P.; Hill, C.A. Genome organization of major tandem repeats in the hard tick, Ixodes scapularis. Chromosome Res. 2010, 18, 357–370. [Google Scholar] [CrossRef] [PubMed]
- Christophoryová, J.; Krajčovičová, K.; Šťáhlavský, F.; Španiel, S.; Opatova, V. Integrative taxonomy approach reveals cryptic diversity within the phoretic pseudoscorpion genus Lamprochernes (Pseudoscorpiones: Chernetidae). Insects 2023, 14, 122. [Google Scholar] [CrossRef] [PubMed]
- Schneider, M.C.; Cella, D.M. Karyotype conservation in 2 populations of the parthenogenetic scorpion Tityus serrulatus (Buthidae): rDNA and its associated heterochromatin are concentrated on only one chromosome. J. Hered. 2010, 101, 491–496. [Google Scholar] [CrossRef] [PubMed]
- Adilardi, R.S.; Ojanguren-Affilastro, A.A.; Mola, L.M. Sex-linked chromosome heterozygosity in males of Tityus confluens (Buthidae): A clue about the presence of sex chromosomes in scorpions. PLoS ONE 2016, 11, e0164427. [Google Scholar] [CrossRef] [PubMed]
- Almeida, B.R.R.; Milhomem-Paixão, S.S.R.; Noronha, R.C.R.; Nagamachi, C.Y.; Costa, M.J.R.D.; Pardal, P.P.O.; Coelho, J.S.; Pieczarka, J.C. Karyotype diversity and chromosomal organization of repetitive DNA in Tityus obscurus (Scorpiones, Buthidae). BMC Genet. 2017, 18, 35. [Google Scholar] [CrossRef]
- Šťáhlavský, F.; Nguyen, P.; Sadílek, D.; Štundlová, J.; Just, P.; Haddad, C.R.; Koç, H.; Ranawana, K.B.; Stockmann, M.; Yağmur, E.A.; et al. Evolutionary dynamics of rDNA clusters on chromosomes of buthid scorpions (Chelicerata: Arachnida). Biol. J. Linn. Soc. 2020, 131, 547–565. [Google Scholar] [CrossRef]
- Just, P.; Šťáhlavský, F.; Kovařík, F.; Štundlová, J. Tracking the trends of karyotype differentiation in the phylogenetic context of Gint, a scorpion genus endemic to the Horn of Africa (Scorpiones: Buthidae). Zool. J. Linn. Soc. 2022, 196, 885–901. [Google Scholar] [CrossRef]
- Araujo, D.; Cella, D.M.; Brescovit, A.D. Cytogenetic analysis of the neotropical spider Nephilengys cruentata (Araneomorphae, Tetragnathidae): Standard staining, NORs, C-bands and base-specific fluorochromes. Braz. J. Biol. 2005, 65, 193–202. [Google Scholar] [CrossRef] [PubMed]
- Král, J.; Musilová, J.; Šťáhlavský, F.; Řezáč, M.; Akan, Z.; Edwards, R.L.; Coyle, F.A.; Almerje, C.R. Evolution of the karyotype and sex chromosome systems in basal clades of araneomorph spiders. Chromosome Res. 2006, 14, 859–880. [Google Scholar] [CrossRef] [PubMed]
- Araujo, D.; Oliveira, E.G.; Giroti, A.M.; Mattos, V.F.; Paula-Neto, E.; Brescovit, A.D.; Schneider, M.C.; Cella, D.M. Comparative cytogenetics of seven Ctenidae species (Araneae). Zool. Sci. 2014, 31, 83–88. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, R.M.; de Jesus, A.C.; Brescovit, A.D.; Cella, D.M. Chromosomes of Crossopriza lyoni (Blackwall 1867), intraindividual numerical chromosome variation in Physocyclus globosus (Taczanowski 1874), and the distribution pattern of NORs (Araneomorphae, Haplogynae, Pholcidae). J. Arachnol. 2007, 35, 293–306. [Google Scholar] [CrossRef]
- Šťáhlavský, F.; Forman, M.; Just, P.; Denič, F.; Haddad, C.R.; Opatova, V. Cytogenetics of entelegyne spiders (Arachnida, Araneae) from southern Africa. Comp. Cytogenet. 2020, 14, 107–138. [Google Scholar] [CrossRef] [PubMed]
- Miller, D.A.; Dew, V.; Tantravahi, R.; Miller, O. Supression of human nucleolar organizer activity in mouse human somatic hybrid cells. Exp. Cell Res. 1976, 101, 235–243. [Google Scholar] [CrossRef] [PubMed]
- Dobigny, G.; Ozouf-Costaz, C.; Bonillo, C.; Volobouev, V. Evolution of rRNA gene clusters and telomeric repeats during explosive genome repatterning in Taterillus X (Rodentia, Gerbillinae). Cytogenet. Genome Res. 2003, 103, 94–103. [Google Scholar] [CrossRef]
- Grzywacz, B.; Maryańska-Nadachowska, A.; Chobanov, D.P.; Karamysheva, T.; Warchałowska-Śliwa, E. Comparative analysis of the location of rDNA in the Palaearctic bushcricket genus Isophya (Orthoptera: Tettigoniidae: Phaneropterinae). Eur. J. Entomol. 2011, 108, 509–517. [Google Scholar] [CrossRef]
- Frydrychová, R.; Mason, J. Telomeres: Their structure and maintenance. In The Mechanisms of DNA Replication; Stuart, D., Ed.; InTech: Rijeka, Croatia, 2013; pp. 423–443. [Google Scholar]
- Araujo, D.; Schneider, M.C.; Paula-Neto, E.; Cella, D.M. Sex chromosomes and meiosis in spiders: A review. In Meiosis—Molecular Mechanisms and Cytogenetic Diversity; Swan, A., Ed.; InTech: Rijeka, Croatia, 2012; pp. 87–108. [Google Scholar]
- Oliver, J.H., Jr. Cytogenetics of ticks (Acari: Ixodoidea) II. Multiple sex chromosomes. Chromosoma 1965, 17, 323–327. [Google Scholar] [CrossRef]
- Štundlová, J.; Hospodářská, M.; Lukšíková, K.; Voleníková, A.; Pavlica, T.; Altmanová, M.; Richter, A.; Reichard, M.; Dalíková, M.; Pelikánová, Š.; et al. Sex chromosome differentiation via changes in the Y chromosome repeat landscape in African annual killifishes Nothobranchius furzeri and N. kadleci. Chromosome Res. 2022, 30, 309–333. [Google Scholar] [CrossRef] [PubMed]
- Chung Voleníková, A.; Sahara, K.; Štundlová, J.; Dalíková, M.; Koutecký, P.; Grof-Tisza, P.; Simonsen, T.J.; Žurovec, M.; Provazníková, I.; Walters, J.R.; et al. Ghost W chromosomes and unique genome architecture in ghost moths of the family Hepialidae. bioRxiv 2023, bioRxiv:09.03.556148. [Google Scholar]
- Dunlop, J.A.; Penney, D. Fossil Arachnids; Monograph Series Volume 2; Siri Scientific Press: Manchester, UK, 2012. [Google Scholar]
- Charlesworth, D.; Charlesworth, B.; Marais, G. Steps in the evolution of heteromorphic sex chromosomes. Heredity 2005, 95, 118–128. [Google Scholar] [CrossRef] [PubMed]
- Phillips, R.B.; Ihssen, P.E. Identification of sex chromosomes in lake trout (Salvelinus namaycush). Cytogenet. Cell Genet. 1985, 39, 14–18. [Google Scholar] [CrossRef]
- Kratochvíl, L.; Stöck, M.; Rovatsos, M.; Bullejos, M.; Herpin, A.; Jeffries, D.L.; Peichel, C.L.; Perrin, N.; Valenzuela, N.; Johnson Pokorná, M. Expanding the classical paradigm: What we have learnt from vertebrates about sex chromosome evolution. Phil. Trans. R. Soc. B 2021, 376, 20200097. [Google Scholar] [CrossRef] [PubMed]
- Pigozzi, M.I. Diverse stages of sex-chromosome differentiation in tinamid birds: Evidence from crossover analysis in Eudromia elegans and Crypturellus tataupa. Genetica 2011, 139, 771–777. [Google Scholar] [CrossRef]
- Matsunaga, S. Junk DNA promotes sex chromosome evolution. Heredity 2009, 102, 525–526. [Google Scholar] [CrossRef] [PubMed]
- Cuevas, C.C.; Formas, J.R. Heteromorphic sex chromosomes in Eupsophus insularis (Amphibia: Anura: Leptodactylidae). Chromosome Res. 1996, 4, 467–470. [Google Scholar] [CrossRef] [PubMed]
- Reed, K.M.; Phillips, R.B. Polymorphism of the nucleolus organizer region (NOR) on the putative sex chromosomes of Arctic char (Salvelinus alpinus) is not sex related. Chromosome Res. 1997, 5, 221–227. [Google Scholar] [CrossRef] [PubMed]
- Schmid, M.; Sims, S.H.; Haaf, T.; Macgregor, H.C. Chromosome banding in Amphibia. X. 18S and 28S ribosomal RNA genes, nucleolus organizers and nucleoli in Gastrotheca riobambae. Chromosoma 1986, 94, 139–145. [Google Scholar] [CrossRef]
- Schmid, M.; Ohta, S.; Steinlein, C.; Guttenbach, M. Chromosome banding in Amphibia. XIX. Primitive ZW/ZZ sex chromosomes in Buergeria buergeri (Anura, Rhacophoridae). Cytogenet. Cell Genet. 1993, 62, 238–246. [Google Scholar] [CrossRef]
- Cioffi, M.B.; Bertollo, L.A.C. Initial steps in XY chromosome differentiation in Hoplias malabaricus and the origin of an X1X2Y sex chromosome system in this fish group. Heredity 2010, 105, 554–561. [Google Scholar] [CrossRef]
- McKee, B.D.; Handel, M.A. Sex chromosomes, recombination, and chromatin conformation. Chromosoma 1993, 102, 71–80. [Google Scholar] [CrossRef] [PubMed]
- Šťáhlavský, F.; Boyer, S.L.; Harvey, M.S.; Giribet, G. First cytogenetic study of a member of the harvestman family Pettalidae (Opiliones: Cyphophthalmi). Austr. J. Entomol. 2012, 51, 299–302. [Google Scholar] [CrossRef]
- Qumsiyeh, M.B. Evolution of number and morphology of mammalian chromosomes. J. Hered. 1994, 85, 455–465. [Google Scholar] [CrossRef]
- Shultz, J.W. Evolutionary morphology and phylogeny of Arachnida. Cladistics 1990, 6, 1–38. [Google Scholar] [CrossRef] [PubMed]
- Alberti, G.; Peretti, A.V. Fine structure of male genital system and sperm in Solifugae does not support a sister-group relationship with Pseudoscorpiones (Arachnida). J. Arachnol. 2002, 30, 268–274. [Google Scholar] [CrossRef]
- Sokolov, I.I. The chromosome complex of mites and its importance for systematics and phylogeny. Trud. Leningr. Obshch. Estestvois, Otd. Zool. 1954, 72, 124–159. (In Russian) [Google Scholar]
- Pepato, A.R.; Dos, S.; Costa, S.G.; Harvey, M.S.; Klimov, P.B. One-way ticket to the blue: A large-scale, dated phylogeny revealed asymmetric land-to-water transitions in acariform mites (Acari: Acariformes). Mol. Phylogenet. Evol. 2022, 177, 107626. [Google Scholar] [CrossRef] [PubMed]
- Blackmon, H.; Hardy, N.B.; Ross, L. The evolutionary dynamics of haplodiploidy: Genome architecture and haploid viability. Evolution 2015, 69, 2971–2978. [Google Scholar] [CrossRef] [PubMed]
- Helle, W.; Pijnacker, L.P. Parthenogenesis, chromosomes and sex. In Spider Mites: Their Biology, Natural Enemies and Control; Helle, W., Sabelis, M.S., Eds.; Elsevier: Amsterdam, The Netherlands, 1985; Volume 1a, pp. 129–139. [Google Scholar]
- Gregory, T.R. Animal Genome Size Database, Release 2.0. 2023. Available online: http://www.genomesize.com (accessed on 28 August 2023).
- Král, J. Holokinetic (holocentric) chromosomes. Biologické listy 1994, 59, 191–217, (In Czech, with English summary). [Google Scholar]
- Bureš, P.; Zedek, F.; Marková, M. Holocentric chromosomes. In Plant Genome Diversity. Physical Structure of Plant Genomes; Wendel, J., Greilhuber, J., Doležel, J., Leitch, I.J., Eds.; Springer Verlag: Berlin/Heidelberg, Germany, 2013; Volume 2, pp. 187–208. [Google Scholar]
- Diaz, M.O.; Maynard, R.; Brum-Zorrilla, N. Diffuse centromere and chromosome polymorphism in haplogyne spiders of the families Dysderidae and Segestriidae. Cytogenet. Genome Res. 2010, 128, 131–138. [Google Scholar] [CrossRef]
- Heinemann, R.L.; Hughes, R.D. Reproduction, reproductive organs, and meiosis in the bisexual non-parthenogenetic mite Caloglyphus mycophagus, with reference to oocyte degeneration in virgins (Sarcoptiformes: Acaridae). J. Morphol. 1970, 130, 93–102. [Google Scholar] [CrossRef]
- Gainett, G.; Sharma, P.P. Genomic resources and toolkits for developmental study of whip spiders (Amblypygi) provide insights into arachnid genome evolution and antenniform leg patterning. EvoDevo 2020, 11, 18. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, S.S.; Klementz, B.C.; Sharma, P.P. A chromosome-level genome of the giant vinegaroon Mastigoproctus giganteus exhibits the signature of pre-Silurian whole genome duplication. bioRxiv 2024, bioRxiv:11.08.622695. [Google Scholar] [CrossRef]
- Telford, M.J.; Thomas, R.H. Expression of homeobox genes shows chelicerate arthropods retain their deutocerebral segment. Proc. Natl. Acad. Sci. USA 1998, 95, 10671–10675. [Google Scholar] [CrossRef] [PubMed]
- Gainett, G.; Klementz, B.C.; Setton, E.V.W.; Simian, C.; Iuri, H.A.; Edgecombe, G.D.; Peretti, A.V.; Sharma, P.P. A plurality of morphological characters need not equate with phylogenetic accuracy: A rare genomic change refutes the placement of Solifugae and Pseudoscorpiones in Haplocnemata. Evol. Dev. 2024, 26, e12467. [Google Scholar] [CrossRef]
- Gunn, S.J.; Hilburn, L.R. Parthenogenesis and karyotypic evolution in the Cayenne tick, model for the production of karyotypic changes through a parthenogenetic pathway. J. Med. Entomol. 1991, 28, 340–349. [Google Scholar] [CrossRef]
- Oliver, J.H., Jr.; Tanaka, K.; Sawada, M. Cytogenetics of ticks (Acari: Ixodoidea). 12. Chromosome and hybridization studies of bisexual and parthenogenetic Haemaphysalis longicornis races from Japan and Korea. Chromosoma 1973, 42, 269–288. [Google Scholar] [CrossRef] [PubMed]
- Goroschenko, Y.L. Karyotypes of argasid ticks of USSR fauna in connection with their taxonomy. Tsitologiya 1962, 4, 1–15. (In Russian) [Google Scholar]
- Ribeiro, J.M.; Alarcon-Chaidez, F.; Francischetti, I.M.; Mans, B.J.; Mather, T.N.; Valenzuela, J.G.; Wikel, S.K. An annotated catalog of salivary gland transcripts from Ixodes scapularis ticks. Insect Biochem. Mol. Biol. 2006, 36, 111–129. [Google Scholar] [CrossRef] [PubMed]
- Geraci, N.S.; Johnston, J.S.; Robinson, J.P.; Wikel, S.K.; Hill, C.A. Variation in genome size of argasid and ixodid ticks. Insect Biochem. Mol. Biol. 2007, 37, 399–408. [Google Scholar] [CrossRef] [PubMed]
- Šťáhlavský, F.; Král, J.; Harvey, M.S.; Haddad, C.R. The first cytogenetic characterization of atemnids: Pseudoscorpions with the highest chromosome numbers (Arachnida: Pseudoscorpiones). Cytogenet. Genome Res. 2012, 137, 22–30. [Google Scholar] [CrossRef]
- Beukeboom, L.; Perrin, N. The Evolution of Sex Determination; Oxford University Press: Oxford, UK, 2014. [Google Scholar]
- Comai, L. The advantages and disadvantages of being polyploid. Nat. Rev. Genet. 2005, 6, 836–846. [Google Scholar] [CrossRef]
- McGregor, H.C. Introduction to Animal Cytogenetics; Chapman & Hall: London, UK, 1993. [Google Scholar]
- Yant, L.; Hollister, J.D.; Wright, K.M.; Arnold, B.J.; Higgins, J.D.; Franklin, F.C.H.; Bomblies, K. Meiotic adaptation to genome duplication in Arabidopsis arenosa. Curr. Biol. 2013, 23, 2151–2156. [Google Scholar] [CrossRef]
- Cifuentes, M.; Grandont, L.; Moore, G.; Chevre, A.M.; Jenczewski, E. Genetic regulation of meiosis in polyploid species: New insights into an old question. New Phytol. 2010, 186, 29–36. [Google Scholar] [CrossRef]
- Kinn, D.N. The life cycle and behaviour of Cercoleipus coelonotus (Acarina: Mesostigmata). Univ. Calif. Publ. Entomol. 1971, 65, 1–62. [Google Scholar]
- Schneider, M.C.; Zacaro, A.A.; Pinto-da-Rocha, R.; Candido, D.M.; Cella, D.M. Complex meiotic configuration of the holocentric chromosomes: The intriguing case of the scorpion Tityus bahiensis. Chromosome Res. 2009, 17, 883–898. [Google Scholar] [CrossRef] [PubMed]
- Soleglad, M.E.; Fet, V. High-level systematics and phylogeny of the extant scorpions (Scorpiones: Orthosterni). Euscorpius 2003, 11, 1–175. [Google Scholar] [CrossRef]
- Santibáñez-López, C.E.; Aharon, S.; Ballesteros, J.A.; Gainett, G.; Baker, C.M.; González-Santillán, E.; Harvey, M.S.; Hassan, M.K.; Abu Almaaty, A.H.; Aldeyarbi, S.M.; et al. Phylogenomics of scorpions reveal contemporaneous diversification of scorpion mammalian predators and mammal-active sodium channel toxins. Syst. Biol. 2022, 71, 1281–1289. [Google Scholar] [CrossRef]
- Šťáhlavský, F.; Král, J. Karyotype analysis and achiasmatic meiosis in pseudoscorpions of the family Chthoniidae (Arachnida: Pseudoscorpiones). Hereditas 2004, 140, 49–60. [Google Scholar] [CrossRef]
- Araujo, D.; Paula-Neto, E.; Brescovit, A.D.; Cella, D.M.; Schneider, M.C. Chromosomal similarities between Nephilidae and Tetragnathidae indicate unique evolutionary traits among Araneoidea. Ital. J. Zool. 2015, 82, 513–520. [Google Scholar] [CrossRef]
- Piza, S.T. Notas sobre cromossômios de alguns escorpiões brasileiros. An. Esc. Super. Agric. Luiz Queiroz 1947, 62, 169–176. [Google Scholar] [CrossRef]
- Rodríguez-Gil, S.G.; Merani, M.S.; Scioscia, C.L.; Mola, L.M. Cytogenetics in three species of Polybetes Simon 1897 from Argentina (Araneae, Sparassidae) I. Karyotype and chromosome banding pattern. J. Arachnol. 2007, 35, 227–237. [Google Scholar] [CrossRef]
- Troiano, G. Karyotype and male meiosis of four species of Roncus L. Koch, 1873 (Pseudoscorpionida, Neobisiidae). Boll. Zool. 1990, 57, 1–9. [Google Scholar] [CrossRef]
- Paula-Neto, E.; Cella, D.M.; Araujo, D.; Brescovit, A.D.; Schneider, M.C. Comparative cytogenetic analysis among filistatid spiders (Araneomorphae: Haplogynae). J. Arachnol. 2017, 45, 123–128. [Google Scholar] [CrossRef]
- Mattos, V.F.; Cella, D.M.; Carvalho, L.S.; Candido, D.M.; Schneider, M.C. High chromosome variability and the presence of multivalent associations in buthid scorpions. Chromosome Res. 2013, 21, 121–136. [Google Scholar] [CrossRef]

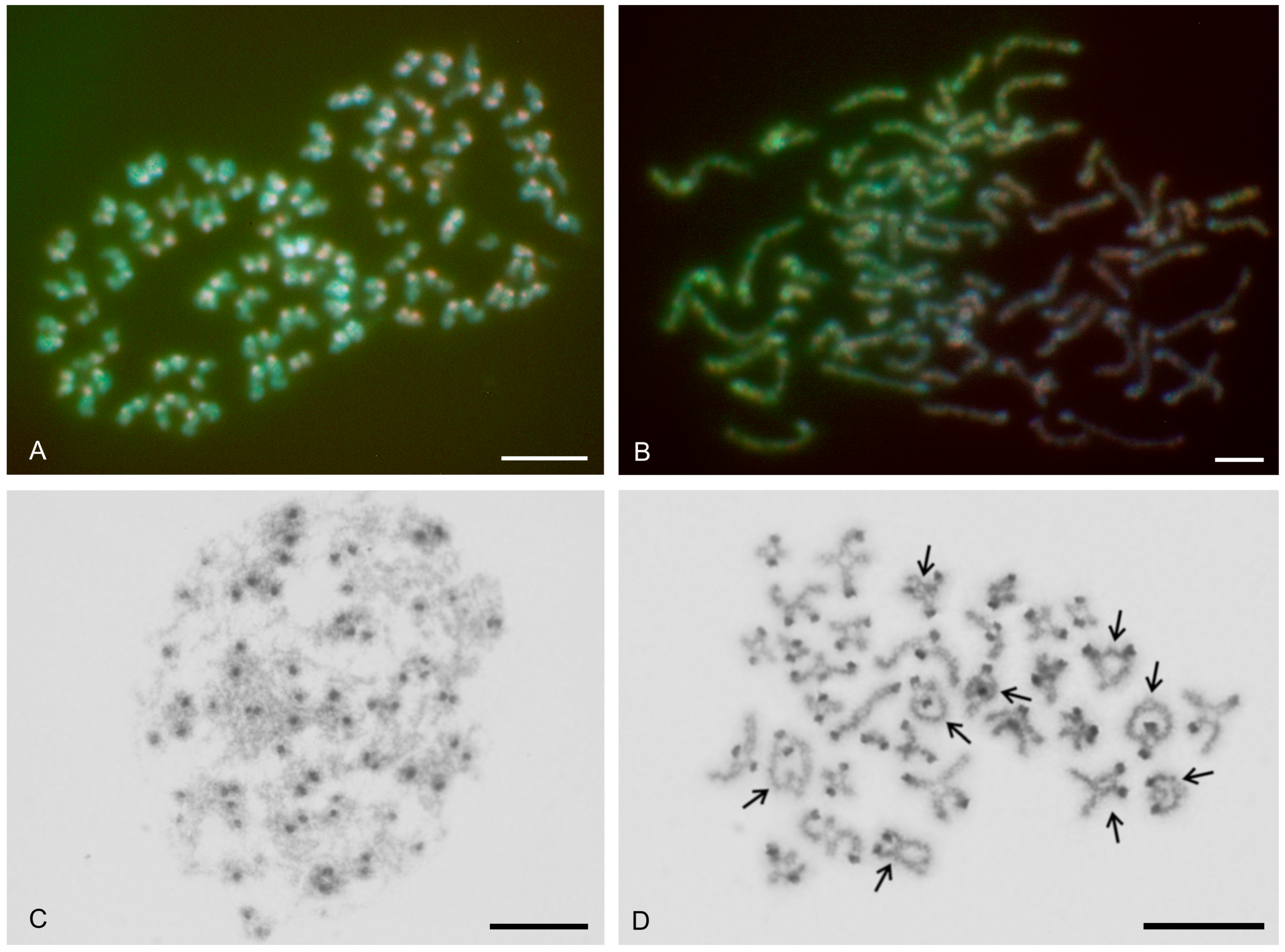
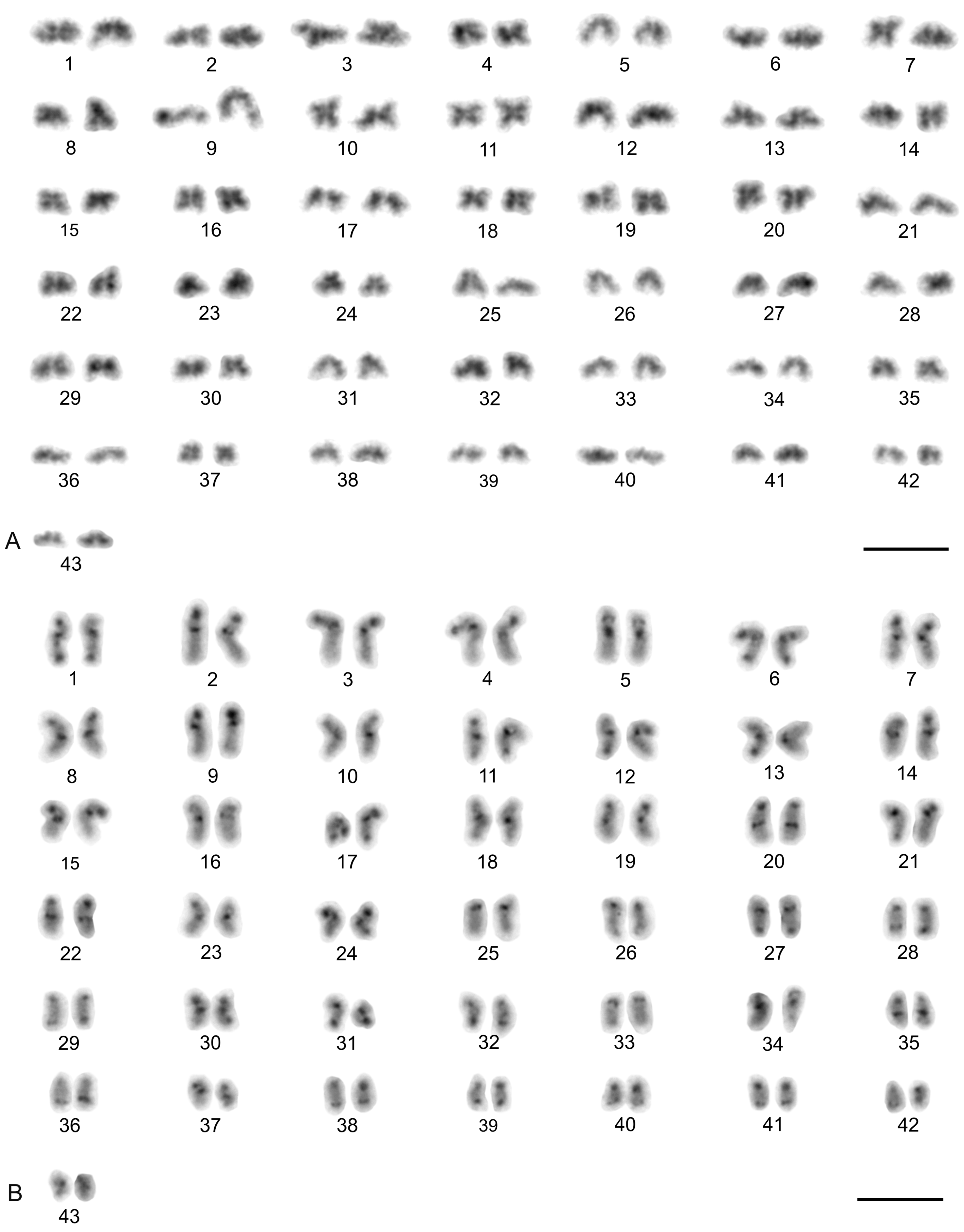
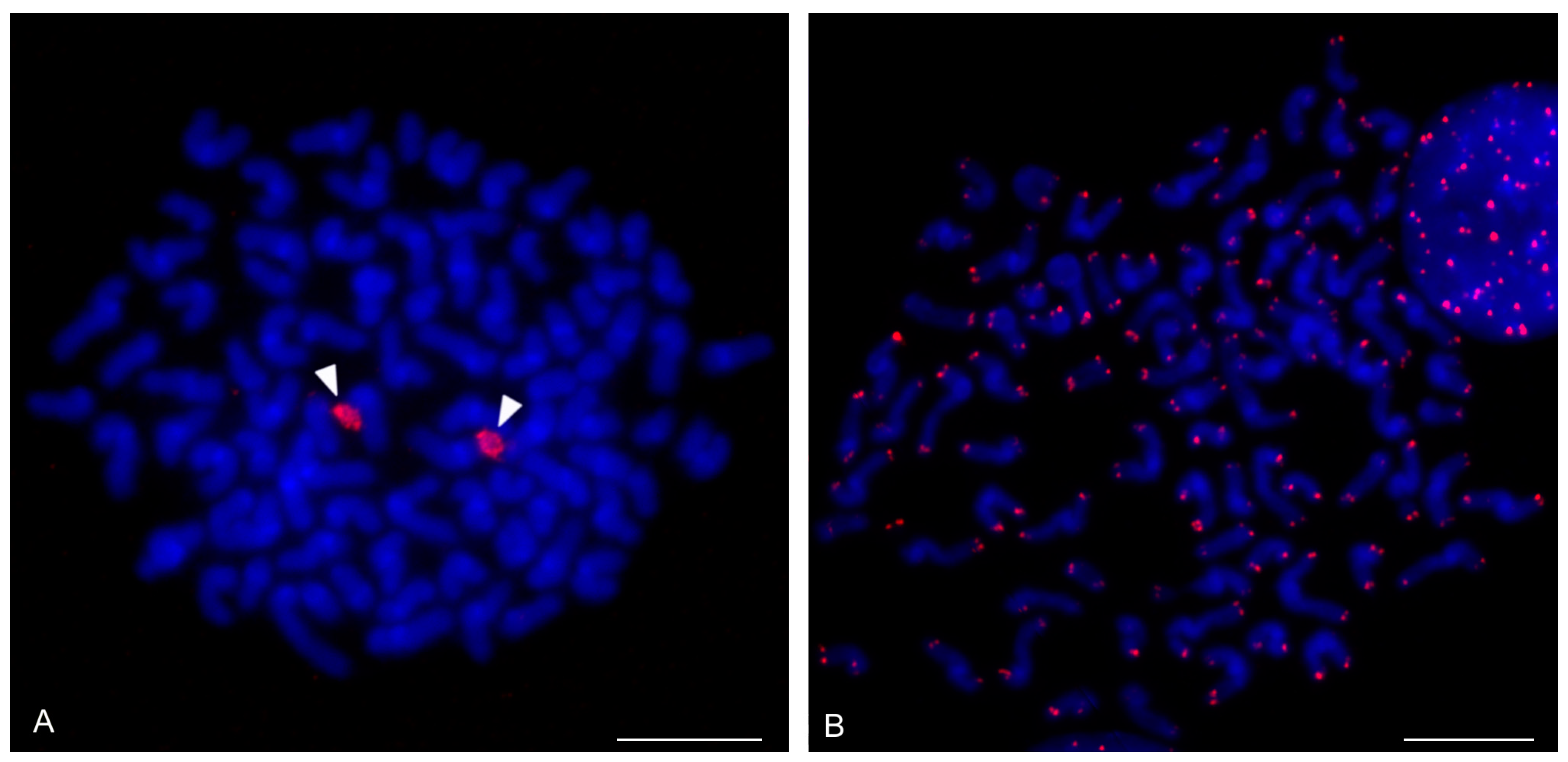

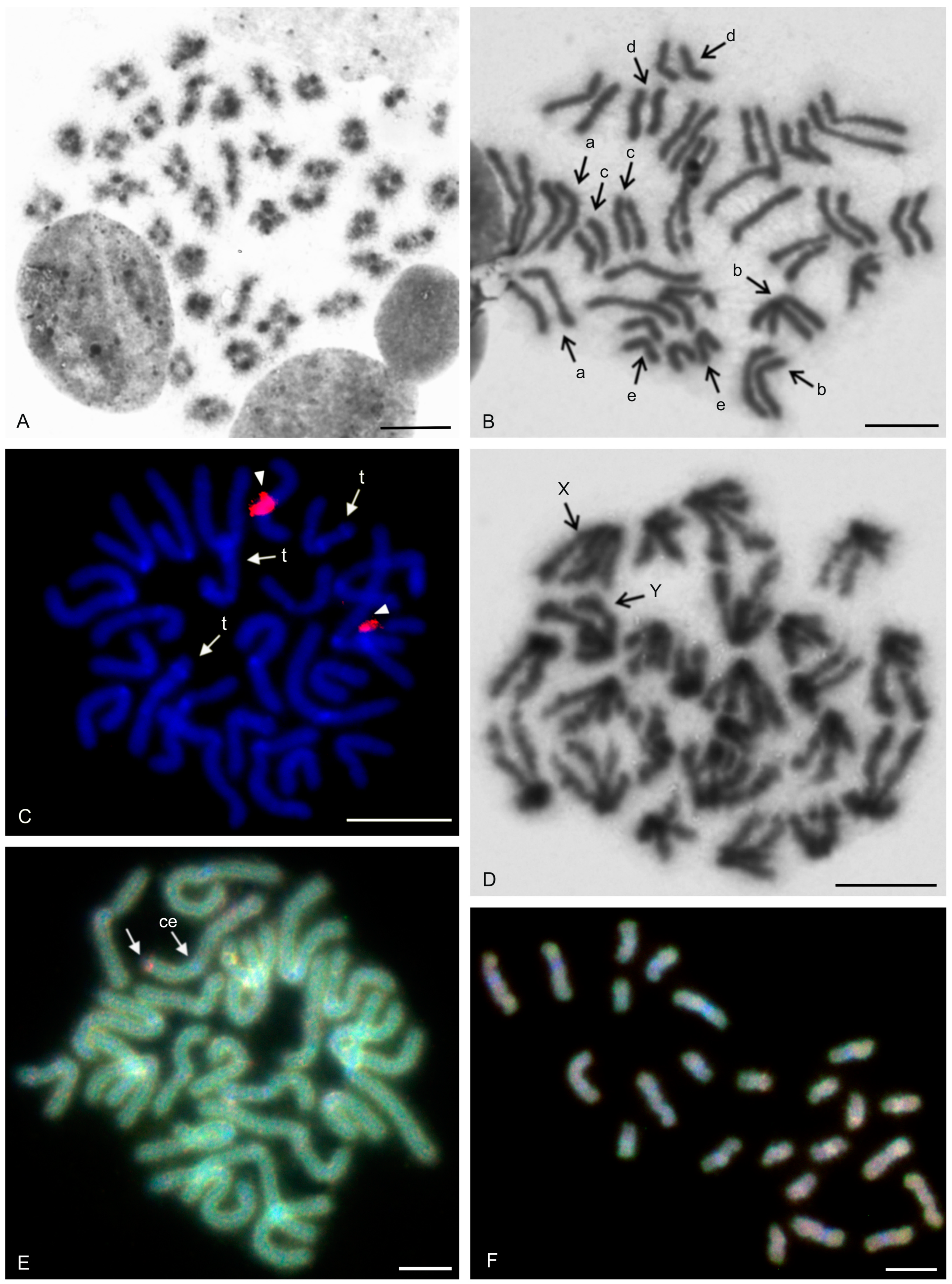
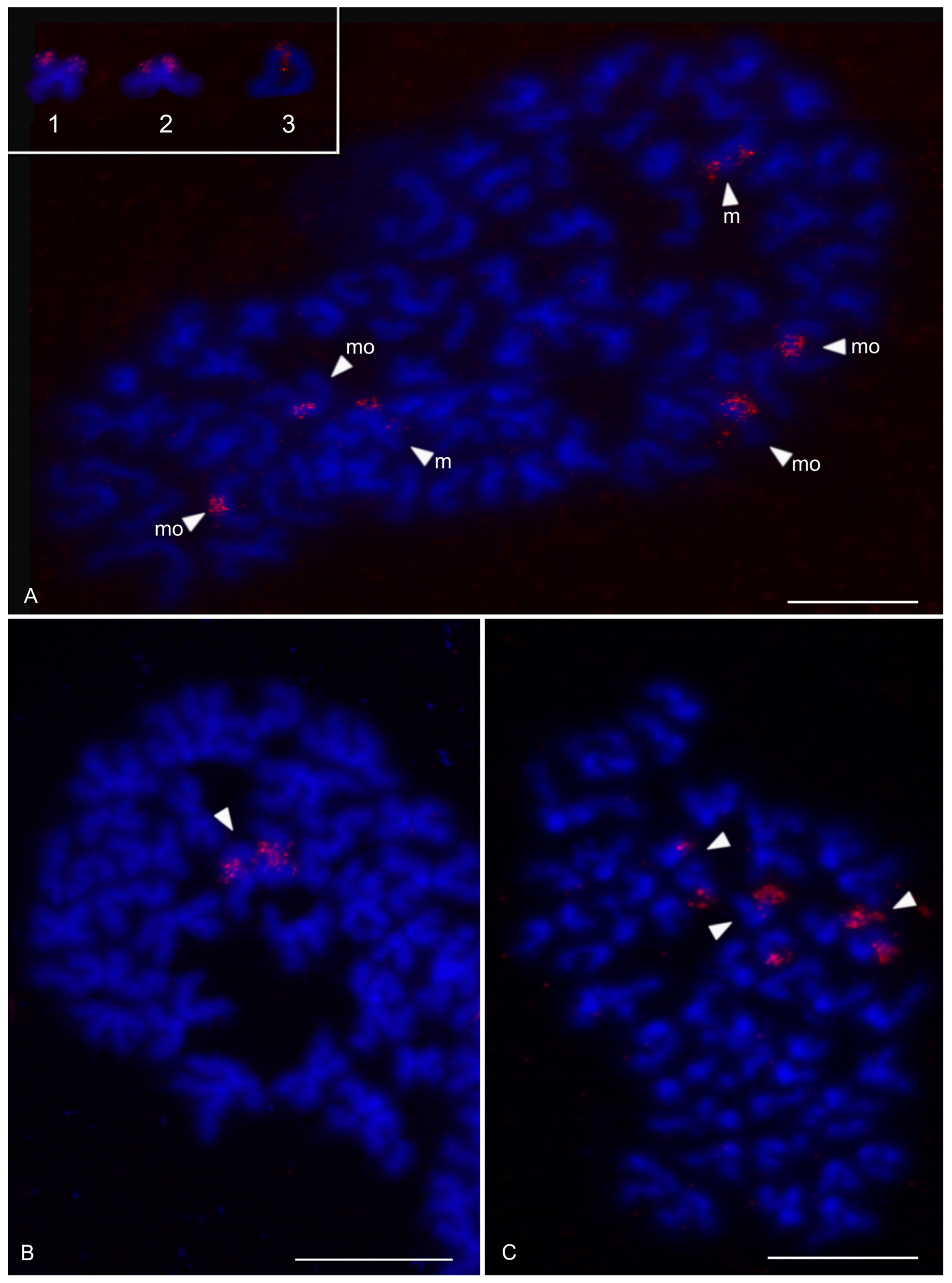
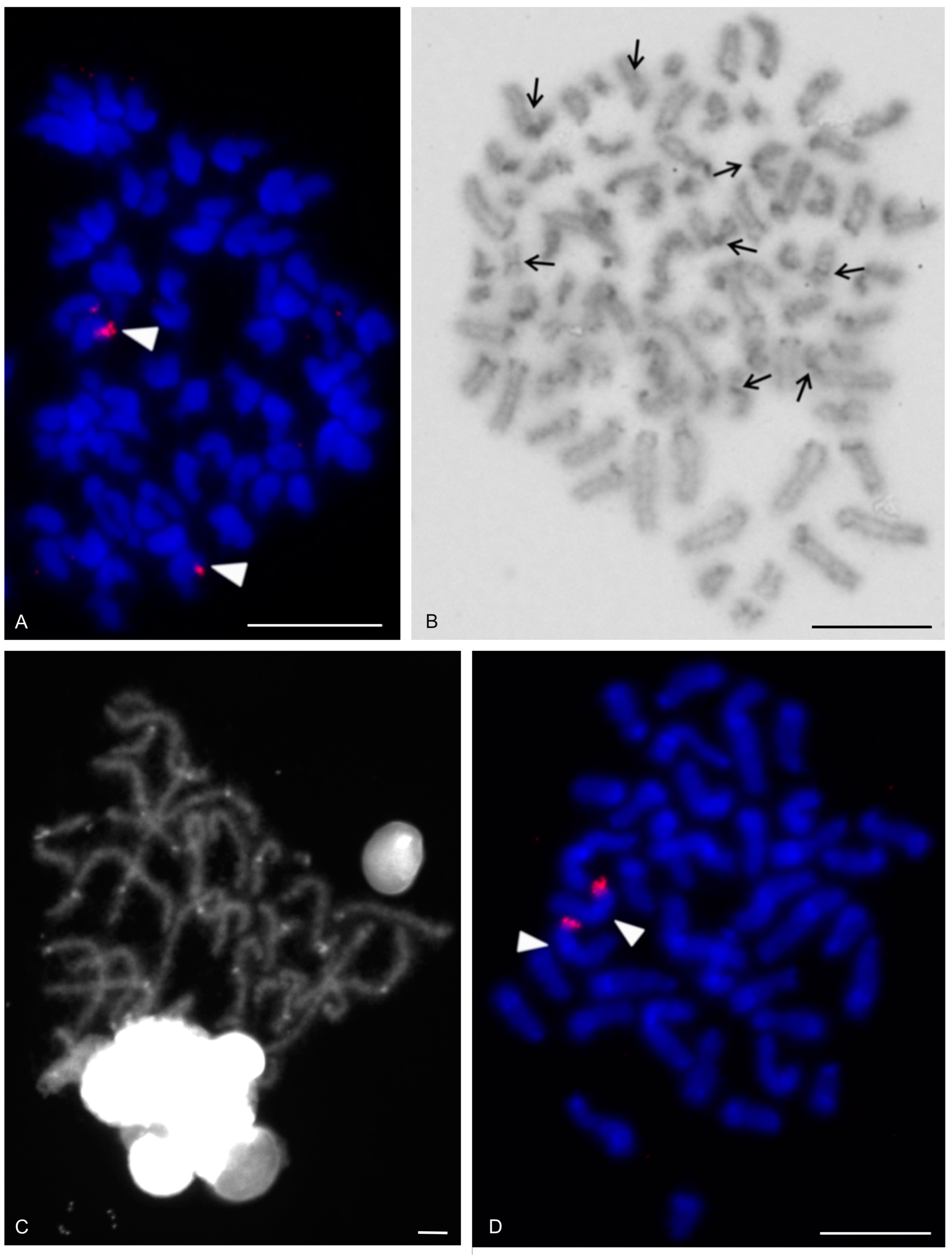


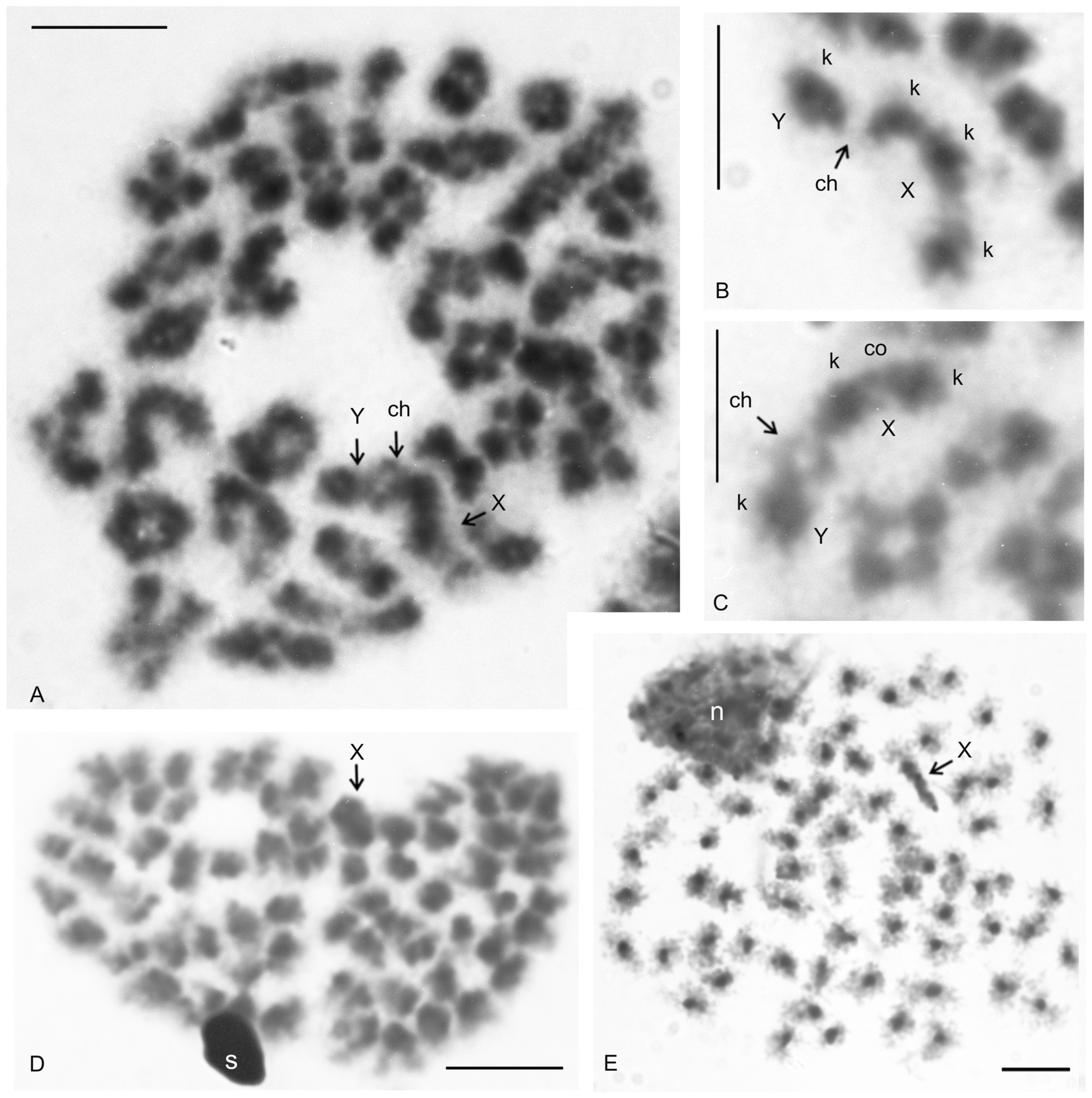
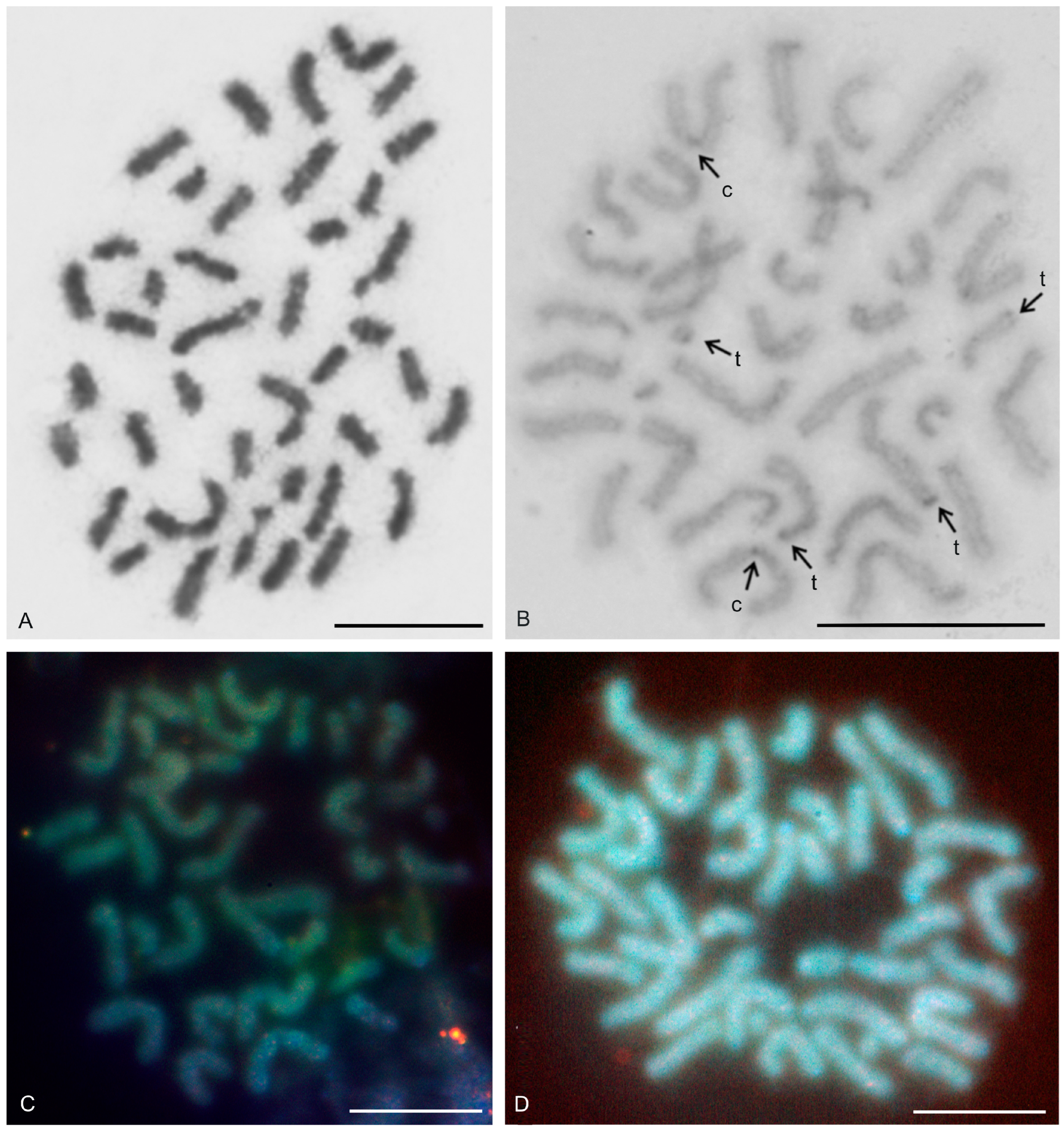


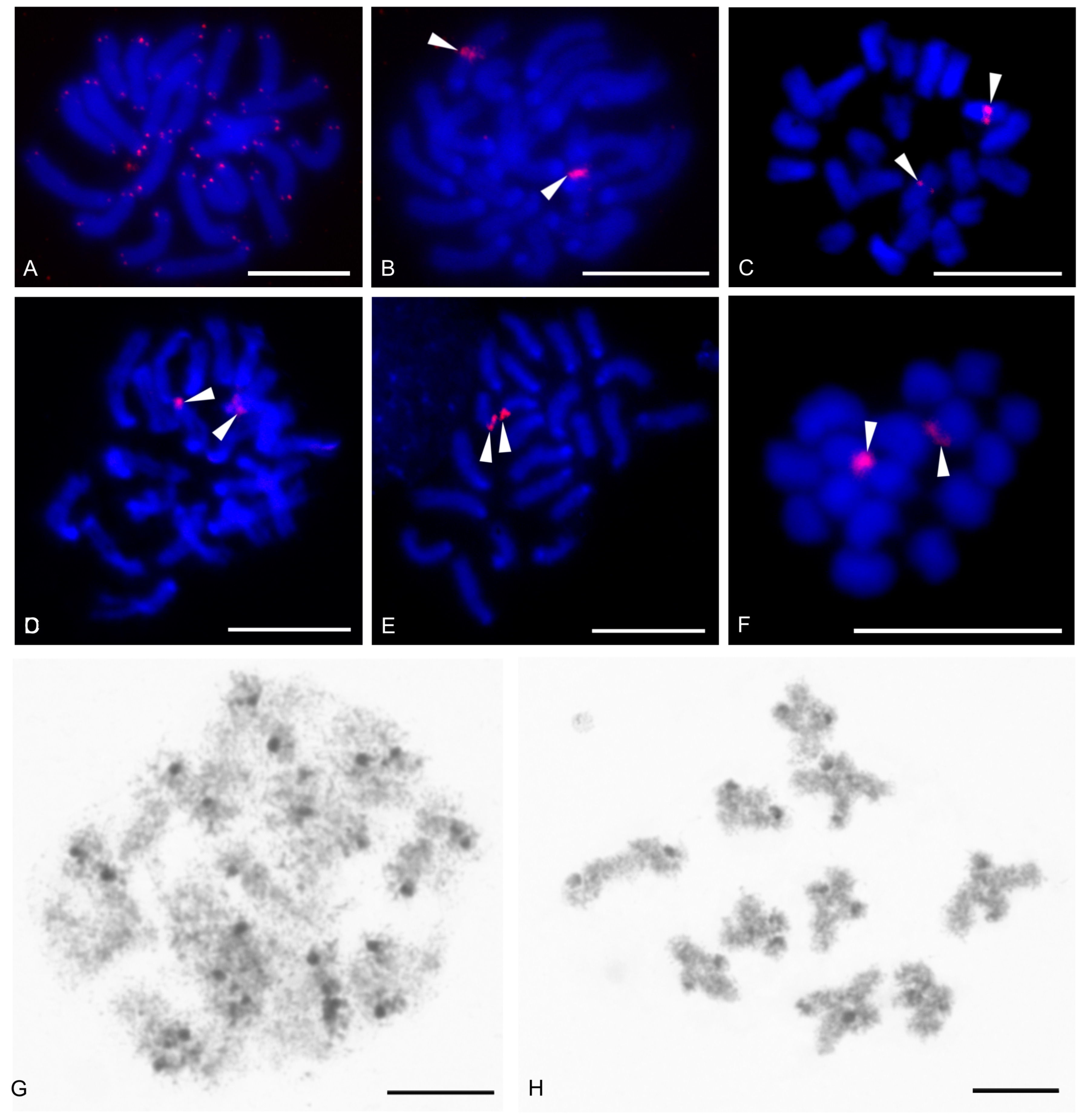
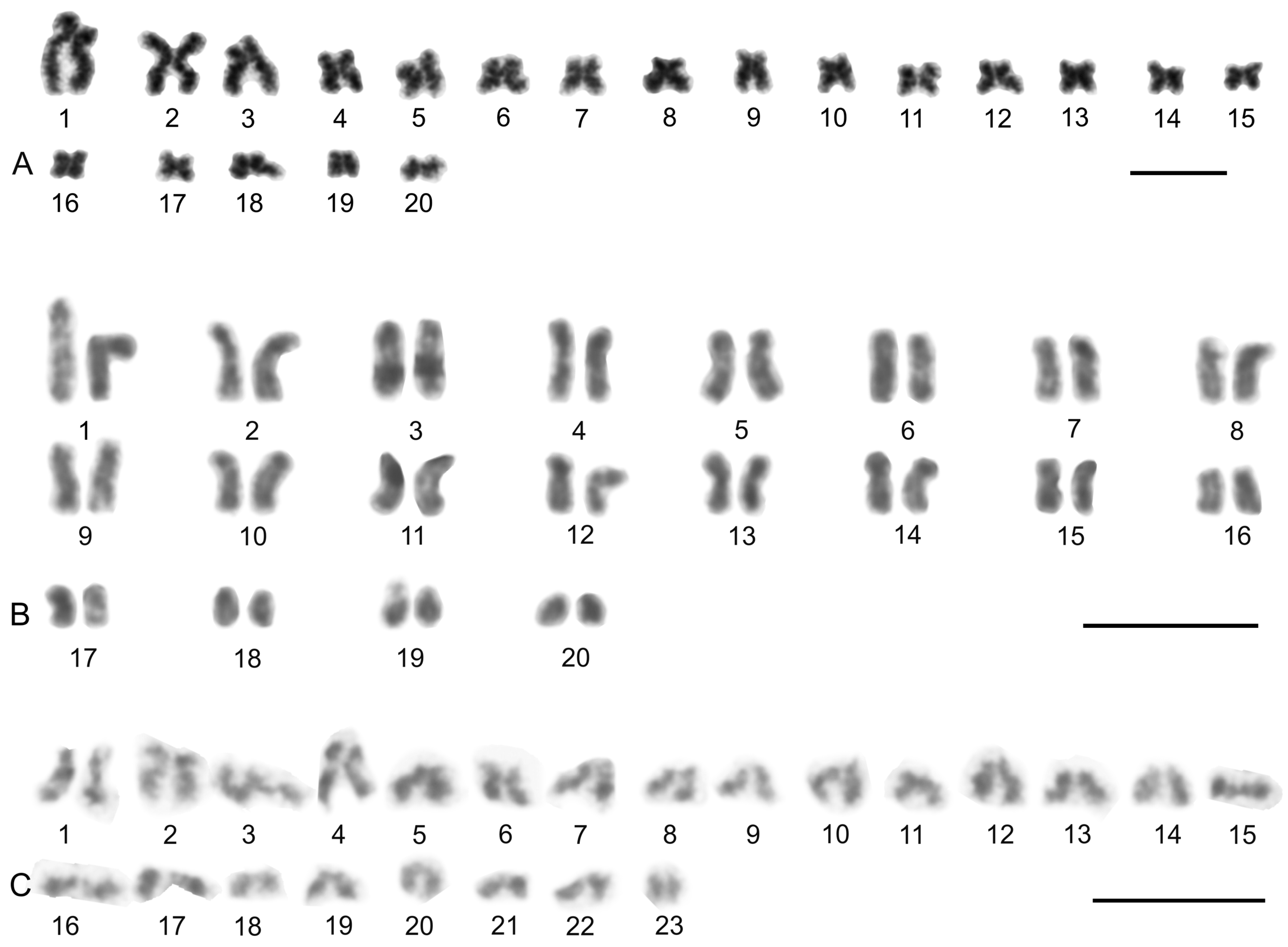
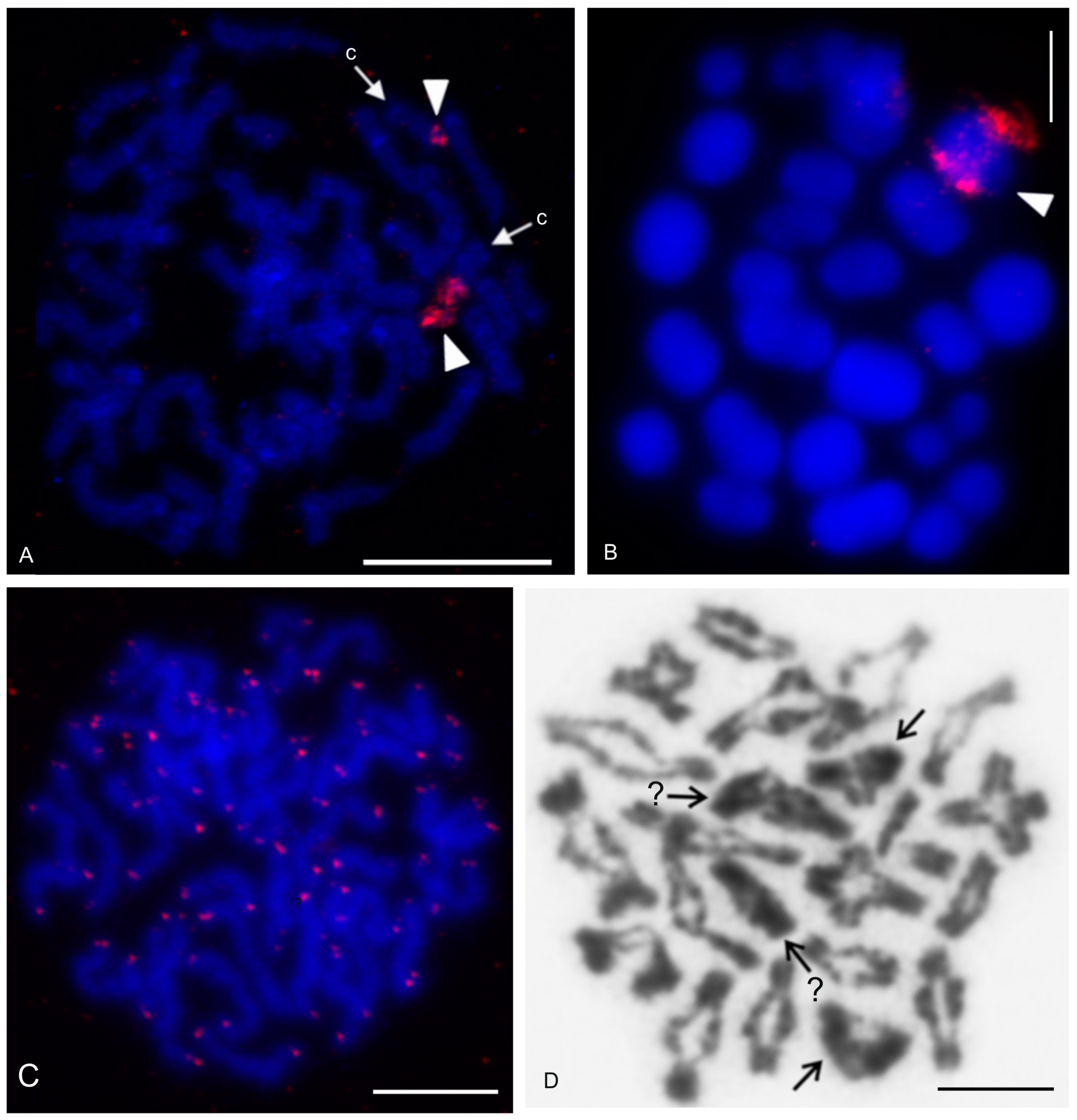






| Species | Source of Data | 2n | Morphology of Autosome Pairs | SC | NOR Number | Number and Morphology of NOR-Bearing Chromosomes/Pairs (NOR Location) | TS (TTAGG) |
|---|---|---|---|---|---|---|---|
| Amblypygi | |||||||
| Charontidae | |||||||
| Charinus cavernicolus Weygoldt, 2006 | [62] | 76♂, j | 5 mp, 9 smp, 18 stp, 6 ap | ||||
| Charinus dominicanus Armas and González, 2002 | [62] | 42♂ | 16 mp, 5 smp | ||||
| Charinus neocaledonicus Simon, 1895 | [62] | 74♂ | 9 mp, 2 smp, 2 stp, 24 ap | 1 | 1 p unknown morphology (NORt) | ||
| Charinus pescotti Dunn, 1949 | [62] | 74♂ | 9 mp, 5 smp, 23 ap | 1 | 1 p unknown morphology (NORt) | ||
| Charon cf. grayi (Gervais, 1842) | this study | 70♂♀ | 4 mp, 3 smp, 27 monop | XY (Xa, Ya) | 2 | 1 ap (NORpt), 1 Xa (NORpt) | |
| Sarax aff. batuensis Roewer, 1962 | [62] | 56♂ | 5 mp, 8 smp, 6 stp, 9 ap | ||||
| Sarax huberi Seiter et al., 2015 | [62] | 50♂ | 15 mp, 2 stp, 8 ap | ||||
| Sarax ioanniticus Kritscher, 1959 | [62] | 72♀ | monop predominant | ||||
| Sarax seychellarum Kraepelin, 1898 | [62] | 22♂♀ | 9 mp, 2 smp | 1 | 1 mp (NORi) | + | |
| Sarax sp. (brachydactylus group) | [62] | 74♂ | 9 mp, 3 smp, 2 stp, 23 ap | ||||
| Phrynichidae | |||||||
| Damon medius (Herbst, 1797) | this study | ♂ | bip and monop | 2 | 2 smp (each pair with NORpt) | ||
| [67] | 70♂ | + | |||||
| Euphrynichus amanica (Werner, 1916) | this study | 78♂ | 15 mp, 4 smp, 20 monop | 3 | 1 mp (NORt), 1 stp (NORpt), 1 ap (NORpt) | ||
| Euphrynichus bacillifer (Gerstaecker,1873) | this study | 56♂ | 22 mp, 2 smp, 1 stp, 3 ap | 1 | 1 mp (NORperic) | ||
| Phrynichus ceylonicus (C.L. Koch, 1843) | this study | 52♂♀ | 17 mp, 3 mp/smp, 5 smp, 1 stp | 3 | 3 mp (each pair with NORt) | ||
| Phrynidae | |||||||
| Acanthophrynus coronatus (Butler, 1873) | this study | 86♂ | 27 bip, 6 stp, 10 ap | 1 | 1 stp (NORpt) | + | |
| Heterophrynus cf. elaphus Pocock, 1903 | this study | 76♂ | 17 bip, 21 monop | 1 | 1 ap (NORqt) | ||
| Heterophrynus longicornis Butler, 1873 | [61] | 66♂♀ | 24 mp, 3 smp, 6 stp | 2 | 2 mp (each pair with NORt) | ||
| Paraphrynus aztecus Pocock, 1894 | [60] | 36♂ | 14 mp, 4 smp | ||||
| Paraphrynus carolynae Armas, 2012 | [60] | 30♂ | 12 mp, 3 smp | ||||
| Paraphrynus cubensis Quintero, 1983 | [60] | 34♂ | 16 mp, 1 sm | ||||
| Paraphrynus mexicanus (Bilimek, 1867) | this study | 24♂♀ | 11 bip | XY (Xm, Ysm) | 1 | 1 mp (NORperic) | |
| [60] | 24♂ | 9 mp, 3 smp | |||||
| Paraphrynus pseudomexicanus Seiter et al., 2020 | [60] | 32♂ | 8 mp, 1 smp, 7 ap | ||||
| Paraphrynus robustus Franganillo, 1931 | [60] | 64♂ | 16 mp, 6 smp, 2 st, 8 ap | ||||
| Phrynus marginemaculatus C.L. Koch, 1840 | [60] | 68♂ | 14 mp, 5 smp, 5 stp, 10 ap | ||||
| Thelyphonida | |||||||
| Thelyphonidae (Hypoctoninae) | |||||||
| Hypoctonus cf. gastrostictus Kraepelin, 1897 | this study | 66♂ | 22 bip, 2 stp, 9 ap | 2 | 2 mp (each pair with NORt) | ||
| Labochirus proboscideus (Butler, 1872) | this study | 78♂♀ | 6 mp, 2 mp/smp, 3 smp, 28 monop *1 | ||||
| Yekuana venezolensis (Haupt, 2009) | this study | 38♂ | 4 mp, 1 mp/smp, 4 smp, 1 stp, 9 ap | 1 | 1 smp (NORpt) | ||
| Thelyphonidae (Mastigoproctinae) | |||||||
| Mastigoproctus giganteus (Lucas, 1835) | this study | 28♂♀ | 7 mp, 1 smp, 1 smp/stp, 1 stp, 4 ap | 2 | 2 mp (each pair with NORt) | ||
| Uroproctus assamensis (Stoliczka, 1869) | this study | 72♂♀ | 4 mp, 3 smp,1 smp/stp, 28 monop | 3 | 3 p unknown morphology (NORt) | ||
| Thelyphonidae (Thelyphoninae) | |||||||
| Ginosigma sp. | this study | 40♂ | 4 mp, 4 smp, 1 smp/stp, 4 stp, 7 ap | 3 | 1 mp (NORpt), 1 smp/stp (NORpt), 1 stp (NORpt) | + | |
| Thelyphonus cf. linganus C.L. Koch, 1843 | this study | 66♂♀ | 19 bip, 3 stp, 10 ap | XY (Xsm, Ym) | |||
| Thelyphonus sepiaris Butler, 1873 | [64] | 44♂ | 21 ap | XY (Xa, Ya) | |||
| [65] | 42♂♀ | 1 mp, 8 stp, 12 ap | |||||
| Thelyphonidae (Typopeltinae) | |||||||
| Typopeltis crucifer Pocock, 1894 | this study | 40♀ | |||||
| Typopeltis guangxiensis Haupt and Song, 1996 | this study | 36♂♀ | 6 mp, 4 smp, 1 stp, 7 ap | 5 | 1 mp (NORt), 1 mp (NORpt + NORqt)*2, 2 ap (each pair with NORqt) | ||
| Schizomida | |||||||
| Hubbardiidae | |||||||
| Clavizomus sp. | this study | 22♂ | 1 mp, 3 stp, 7 ap | ||||
| Notozomus sp. | this study | 22♂ | 2 stp, 9 ap | ||||
| Orientzomus sp. (Luzon) | this study | 16♂♀ | 5 bip, 3 ap | ||||
| Orientzomus sp. (Mindanao) | this study | 16♀ | 1 | 1p unknown morphology (NORt) | |||
| Stenochrus sp. | this study | 22♂♀ | 8 stp, 3 ap | + | |||
| Olmecazomus brujo Monjaraz-Ruedas and Francke, 2017 | this study | 22♂♀ | 11 ap | 1 | 1 ap (NORqi) | ||
| Rowlandius ubajara Santos et al., 2013 | [66] | 22♀ | 1 mp, 10 monop | ||||
| Rowlandius sp. | [66] | 20♀ | 1 mp, 9 monop | ||||
| Hubbardiidae sp. (Seychelles) | this study | 22♀,j | 11 ap | 1 | 1 ap (NORt) | ||
| Hubbardiidae sp. (Cameroon) | this study | 22♀, j | 11 ap | 1 | 1 ap (NORqi) | ||
| Protoschizomidae | |||||||
| Agastoschizomus lucifer Rowland, 1971 | this study | 22♀ | 11 ap | 1 | 1 ap (NORt) | ||
| Ricinulei | |||||||
| Cryptocellus narino Platnick and Paz, 1979 | this study | 46♂♀ | 1 mp, 1 mp/smp, 2 smp, 1 stp, 18 ap | 1 | 1 p unknown morphology (NORsubt) | ||
| Pseudocellus gertschi (Márquez and Conconi, 1974) | this study | 40♂♀ | 2 mp, 4 smp, 1 smp/stp, 13 monop | 1 | 1 ap (NORqt)*3 | + | |
| Ricinoides olounoua Legg, 1978 | this study | 40♂ | 16 bip, 4 stp | 1 | 1 p unknown morphology (NORsubt) | ||
| Solifugae | |||||||
| Ammotrechidae | |||||||
| Ammotrechula mulaiki Muma, 1951 | this study | 24♂♀ | 2 stp, 10 ap | ||||
| Daesiidae | |||||||
| Eberlanzia flava Roewer, 1941 | this study | 22♂ | 2 mp, 2 stp, 7 ap | 1 | 1 stp (NORpt) | ||
| Gluvia dorsalis (Latreille, 1817) | this study | 10♂♀ | 5 ap | ||||
| Gnosippus sp. | this study | 16♂ | karyotype predominated by ap | ||||
| Eremobatidae | |||||||
| Eremobates pallipes (Say, 1823) | this study | 22♂ | 1 mp, 2 stp, 8 ap | 1 | 1 mp (NORarm) | ||
| Eremobates similis Muma, 1951 | this study | 22♂ | 1 mp, 10 ap | 1 | 1 p unknown morphology (NORt) | ||
| Galeodidae | |||||||
| Paragaleodes pallidus (Birula, 1890) | this study | 12♂♀ | 6 mp | 8 | 2 mp (each pair with NORsubt), 3 mp (each pair with NORpsubt + | ||
| NORqsubt) | |||||||
| Rhagodidae | |||||||
| Rhagodes sp. | this study | 18j | 7 mp, 2 stp | ||||
| [67] | 18j | + | |||||
| Solpugidae | |||||||
| Solpugista sp. | this study | 20♀ | 10 ap | 1 | 1 ap (NORt) *3 |
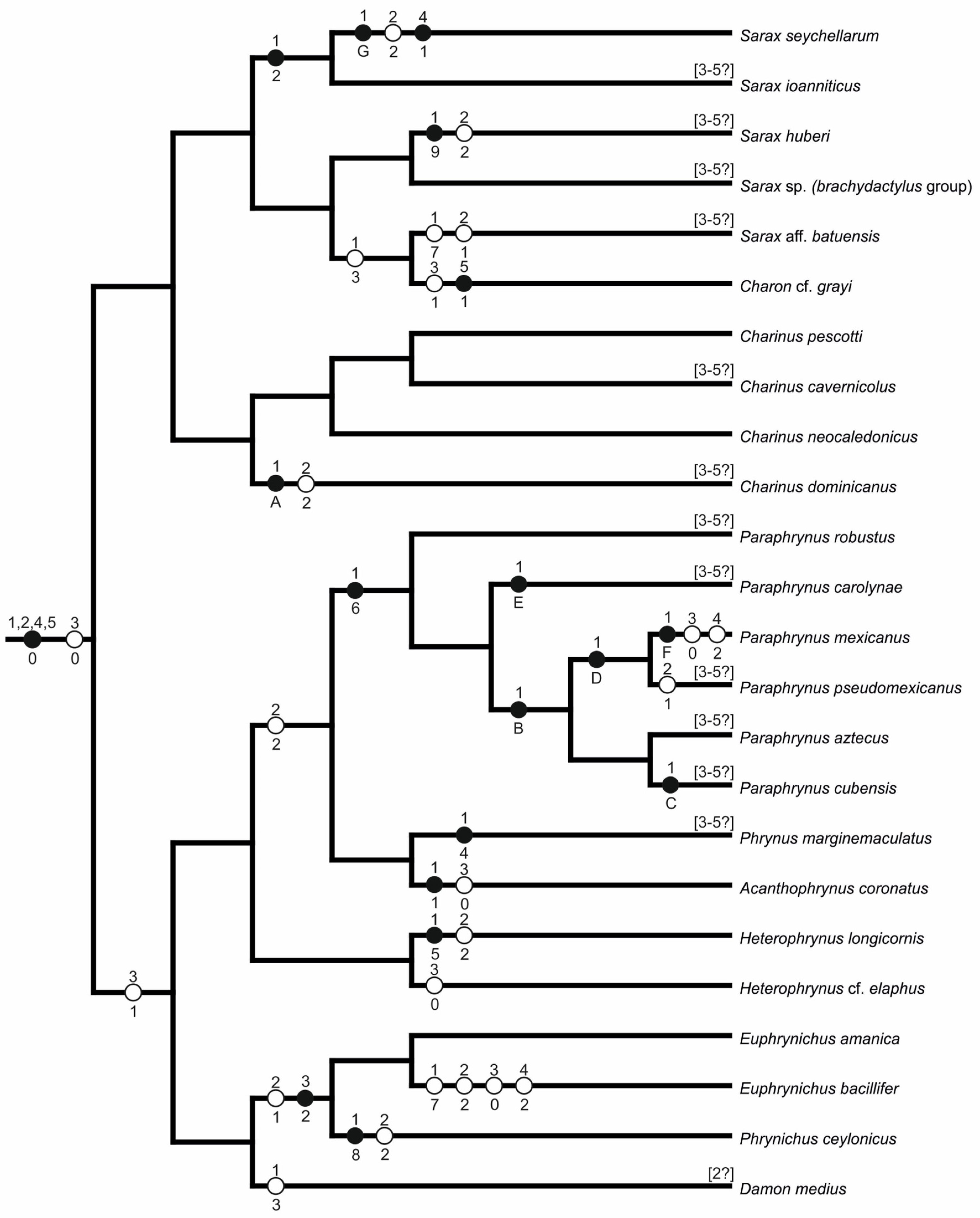
| Outgroup | 00000 |
| Acanthophrynus coronatus | 12000 |
| Charon cf. grayi | 30101 |
| Charinus cavernicolus | 00??? |
| Charinus dominicanus | A2??? |
| Charinus neocaledonicus | 00000 |
| Charinus pescotti | 00000 |
| Damon medius | 3?100 |
| Euphrynichus amanica | 01200 |
| Euphrynichus bacillifer | 72020 |
| Heterophrynus cf. elaphus | 00000 |
| Heterophrynus longicornis | 52100 |
| Paraphrynus aztecus | B2??? |
| Paraphrynus carolynae | E2??? |
| Paraphrynus cubensis | C2??? |
| Paraphrynus mexicanus | F2020 |
| Paraphrynus pseudomexicanus | D1??? |
| Paraphrynus robustus | 62??? |
| Phrynichus ceylonicus | 82200 |
| Phrynus marginemaculatus | 42??? |
| Sarax aff. batuensis | 71??? |
| Sarax huberi | 92??? |
| Sarax ioanniticus | 20??? |
| Sarax seychellarum | G2010 |
| Sarax sp. (brachydactylus group) | 00??? |
| Outgroup | 000000 |
| Ginosigma sp. | 300000 |
| Hypoctonus cf. gastrostictus | 120000 |
| Labochirus proboscideus | 000??0 |
| Mastigoproctus giganteus | 620000 |
| Thelyphonus cf. linganus | 121??0 |
| Thelyphonus sepiaris | 201??0 |
| Typopeltis crucifer | 3????? |
| Typopeltis guangxiensis | 510200 |
| Uroproctus assamensis | 000000 |
| Yekuana venezolensis | 410100 |
| Outgroup | 0000 |
| Agastoschizomus lucifer | 0000 |
| Clavizomus sp. | 00?? |
| Hubbardiidae (Cameroon) | 0001 |
| Hubbardiidae (Seychelles) | 0000 |
| Notozomus sp. | 00?? |
| Olmecazomus brujo | 0001 |
| Orientzomus sp. (Luzon) | 21?? |
| Orientzomus sp. (Mindanao) | 2?00 |
| Rowlandius ubajara | 00?? |
| Rowlandius sp. | 10?? |
| Stenochrus sp. | 00?? |
| Outgroup | 0000 |
| Cryptocellus narino | 1100 |
| Pseudocellus gertschi | 0110 |
| Ricinoides olounoua | 0010 |
| Outgroup | 00000 |
| Ammotrechula mulaiki | 00??? |
| Eberlanzia flava | 10000 |
| Eremobates pallipes | 10000 |
| Eremobates similis | 10000 |
| Gluvia dorsalis | 60??0 |
| Gnosippus sp. | 40??? |
| Paragaleodes pallidus | 51111 |
| Rhagodes sp. | 31??1 |
| Solpugista sp. | 20000 |
| Taxon | Ancestral Pattern | Hypothesis Source |
|---|---|---|
| Male diploid number or genome event | ||
| Arachnida | low to moderate 2n (30–40) {low 2n (22–24)} | this study |
| Arachnopulmonata | duplication of genome | [46] |
| Cephalosomata + Arachnopulmonata | 22–24 | this study |
| Higher Tetrapulmonata (Amblypygi + Thelyphonida + Araneae) | duplication of genome | this study |
| Panscorpiones (Pseudoscoriones + Scorpiones) | duplication of genome | this study |
| Acariformes | 18 | this study |
| Amblypygi | 74–78 | this study |
| Opiliones | 30 | [154] |
| Pseudoscorpiones | a high diploid number | [186] |
| Ricinulei | 40 | this study |
| Schizomida | 22 | this study |
| Scorpiones | a high diploid number | this study |
| Solifugae | 24 | this study |
| Amblypygi (Charontidae) | 74–76 | this study |
| Amblypygi (Phrynoidea) | 76–78 | this study |
| Araneae (Araneomorphae, Araneoidea) | 24 | [187] |
| Araneae (Araneomorphae, Caponiidae) | duplication of genome | [53] |
| Araneae (Araneomorphae, Entelegynae) | 42 | [130] |
| Araneae (Mygalomorphae, Avicularioidea) | 70–90, duplication of genome | [55] |
| Araneae (Opisthothelae) | 40–50 | [130] |
| Opiliones (Laniatores) | duplication of genome {increase in diploid number by chromosome fissions} | [117] |
| Parasitiformes (Ixodida) | 26–28 | this study |
| Pseudoscorpiones (Atemnidae) | duplication of genome {increase in diploid number by chromosome fissions} | [176] |
| Scorpiones (Buthidae) | 6–32 | [127] |
| Thelyphonida | 72–78 | this study |
| Chromosome morphology | ||
| Arachnida | monocentric chromosomes | [71] |
| Clade Cephalosomata + Arachnopulmonata | monoarmed chromosomes, predominantly acrocentric elements | this study |
| Higher Tetrapulmonata (Amblypygi + Thelyphonida + Araneae) | predominantly monoarmed chromosomes (1/3 of chromosomes biarmed) | this study |
| Acariformes | holocentric chromosomes | [15] |
| Opiliones | predominantly biarmed chromosomes | [154] |
| Ricinulei | predominantly biarmed chromosomes | this study |
| Schizomida | acrocentric chromosomes | this study |
| Solifugae | almost all or all chromosomes acrocentric | this study |
| Amblypygi (Phrynoidea) | karyotype includes 17–19 biarmed pairs | this study |
| Araneae (Araneomorphae, Dysderoidea) | holocentric chromosomes | [130] |
| Araneae (Araneomorphae, Entelegynae) | acrocentric chromosomes | [86] |
| Araneae (Opisthothelae) | predominantly biarmed chromosomes | [55] |
| Scorpiones (Buthidae) | holocentric chromosomes | [188] |
| Solifugae (Galeodidae + Rhagodidae) | predominantly biarmed chromosomes | this study |
| Chromosome behaviour throughout cell cycle | ||
| Solifugae | pairing of homologous chromosomes throughout cell cycle | this study |
| Constitutive heterochromatin | ||
| Arachnida | low to moderate content of constitutive heterochromatin | this study |
| Arachnida | constitutive heterochromatin located in centromeric and telomeric regions | this study |
| Arachnida | centromeric heterochromatin predominantly AT-rich | this study |
| Araneae | constitutive heterochromatin located at centromeric regions only | [189] |
| Solifugae (Galeodidae + Rhagodidae) | considerable expansion of constitutive heterochromatin | this study |
| NORs | ||
| Arachnida | 1 NOR | [59] |
| Arachnida | terminal position of NORs | [61] |
| Amblypygi | 1 NOR | this study |
| Thelyphonida | 2 or 3 NORs | this study |
| Amblypygi (Phrynichus + Euphrynichus) | 3 NORs | this study |
| Amblypygi (Phrynidae) | two subtelocentric NOR-bearing chromosome pairs | this study |
| Amblypygi (Phrynoidea) | 2 NORs | this study |
| Araneae (Araneomorphae, Entelegynae) | 2 autosome NORs | [70] |
| Araneae (Opisthothelae) | 1 or 2 autosome NORs | [55] |
| Schizomida (Mexican + West African clade of Hubbardiidae) | interstitial NOR | this study |
| Solifugae (Galeodidae) | a high number of NORs | this study |
| Telomeric repeats | ||
| Arachnida | insect motif | [67] |
| Araneae | absence of insect motif | [67] |
| Sex chromosomes (heterogametic sex) | ||
| Arachnida | homomorphic sex chromosomes or absence of sex chromosomes | [71] |
| Higher Tetrapulmonata (Amblypygi + Thelyphonida + Araneae) | homomorphic sex chromosomes X and Y | this study |
| Araneae | homomorphic sex chromosomes X and Y + X1 and X2 | [56] |
| Pseudoscorpiones | X0 system | [190] |
| Pseudoscorpiones | metacentric morphology of X chromosome | [190] |
| Scorpiones | homomorphic sex chromosomes X and Y | [125] |
| Araneae (Araneomorphae) | homomorphic sex chromosomes X and Y + chromosomes X1, X2, and Y | [73,191] |
| Araneae (Mygalomorphae, Avicularioidea) | two homomorphic XY pairs + X1X2X3X4 (result of genome duplication) | [55] |
| Parasitiformes (Ixodida) | sex chromosomes X and Y | this study |
| Male meiosis (heterogametic sex) | ||
| Arachnida | low frequency of chiasmata | this study |
| Tetrapulmonata | diffuse stage {diffuse stage arose several times during evolution of Tetrapulmonata} | this study |
| Araneae | specific end-to-end pairing of X chromosome univalents | [57] |
| Palpigradi | diffuse stage | [71] |
| Ricinulei | diffuse stage | this study |
| Scorpiones | achiasmatic meiosis | [192] |
| Araneae (Araneomorphae, Haplogynae) | diffuse stage | [130] |
| Pseudoscorpiones (Chthoniidae) | achiasmatic meiosis | [186] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Král, J.; Sember, A.; Divišová, K.; Kořínková, T.; Reyes Lerma, A.C.; Ávila Herrera, I.M.; Forman, M.; Šťáhlavský, F.; Musilová, J.; Torres Kalme, S.; et al. Advances in Understanding the Karyotype Evolution of Tetrapulmonata and Two Other Arachnid Taxa, Ricinulei and Solifugae. Genes 2025, 16, 207. https://doi.org/10.3390/genes16020207
Král J, Sember A, Divišová K, Kořínková T, Reyes Lerma AC, Ávila Herrera IM, Forman M, Šťáhlavský F, Musilová J, Torres Kalme S, et al. Advances in Understanding the Karyotype Evolution of Tetrapulmonata and Two Other Arachnid Taxa, Ricinulei and Solifugae. Genes. 2025; 16(2):207. https://doi.org/10.3390/genes16020207
Chicago/Turabian StyleKrál, Jiří, Alexandr Sember, Klára Divišová, Tereza Kořínková, Azucena C. Reyes Lerma, Ivalú M. Ávila Herrera, Martin Forman, František Šťáhlavský, Jana Musilová, Sabrina Torres Kalme, and et al. 2025. "Advances in Understanding the Karyotype Evolution of Tetrapulmonata and Two Other Arachnid Taxa, Ricinulei and Solifugae" Genes 16, no. 2: 207. https://doi.org/10.3390/genes16020207
APA StyleKrál, J., Sember, A., Divišová, K., Kořínková, T., Reyes Lerma, A. C., Ávila Herrera, I. M., Forman, M., Šťáhlavský, F., Musilová, J., Torres Kalme, S., Palacios Vargas, J. G., Zrzavá, M., Vrbová, I., Moreno-González, J. A., Cushing, P. E., Gromov, A. V., Šebestiánová, Š., Šlechtová, V. B., Prendini, L., & Bird, T. L. (2025). Advances in Understanding the Karyotype Evolution of Tetrapulmonata and Two Other Arachnid Taxa, Ricinulei and Solifugae. Genes, 16(2), 207. https://doi.org/10.3390/genes16020207








