Dimensionality Reduction by Similarity Distance-Based Hypergraph Embedding
Abstract
:1. Introduction
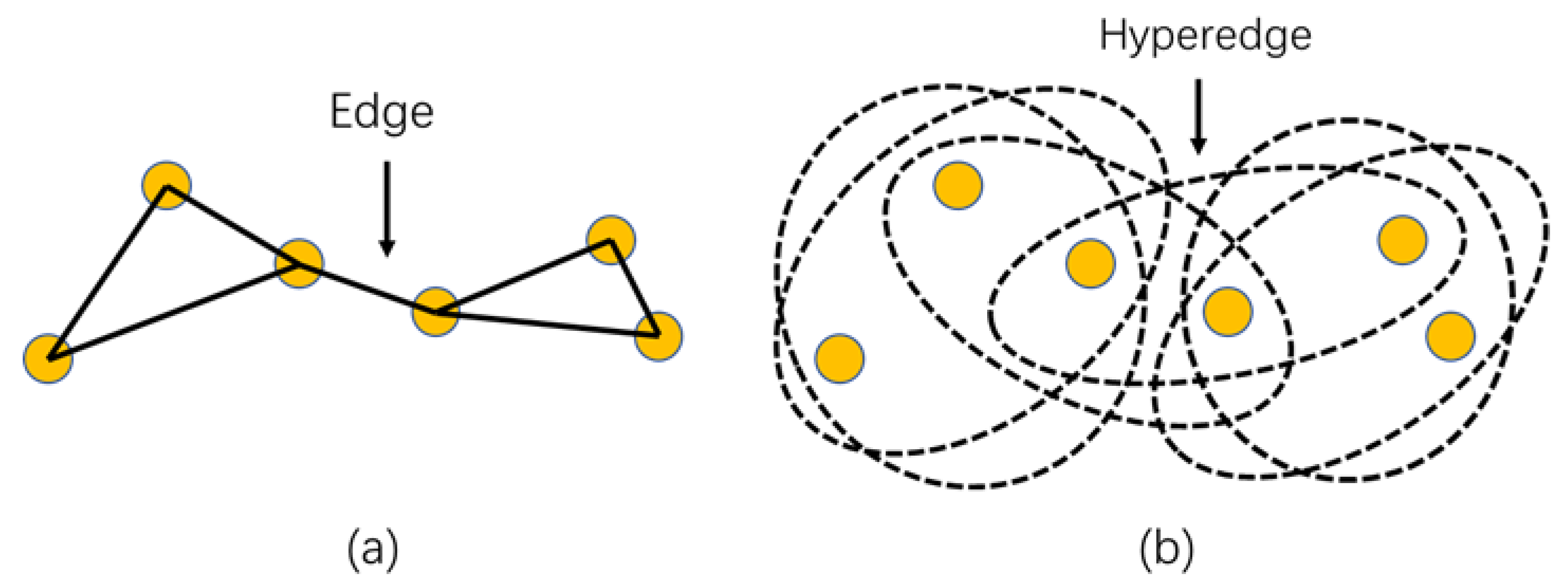
- The conventional graph embedding-based DR methods, for example, LPP, aims to preserve the local adjacent relationship of samples by constructing a weight matrix which only takes the affinity between pairwise samples into account. However, the weight matrix fails to reflect the complex relationship of samples in high order [21], leading to the loss of information.
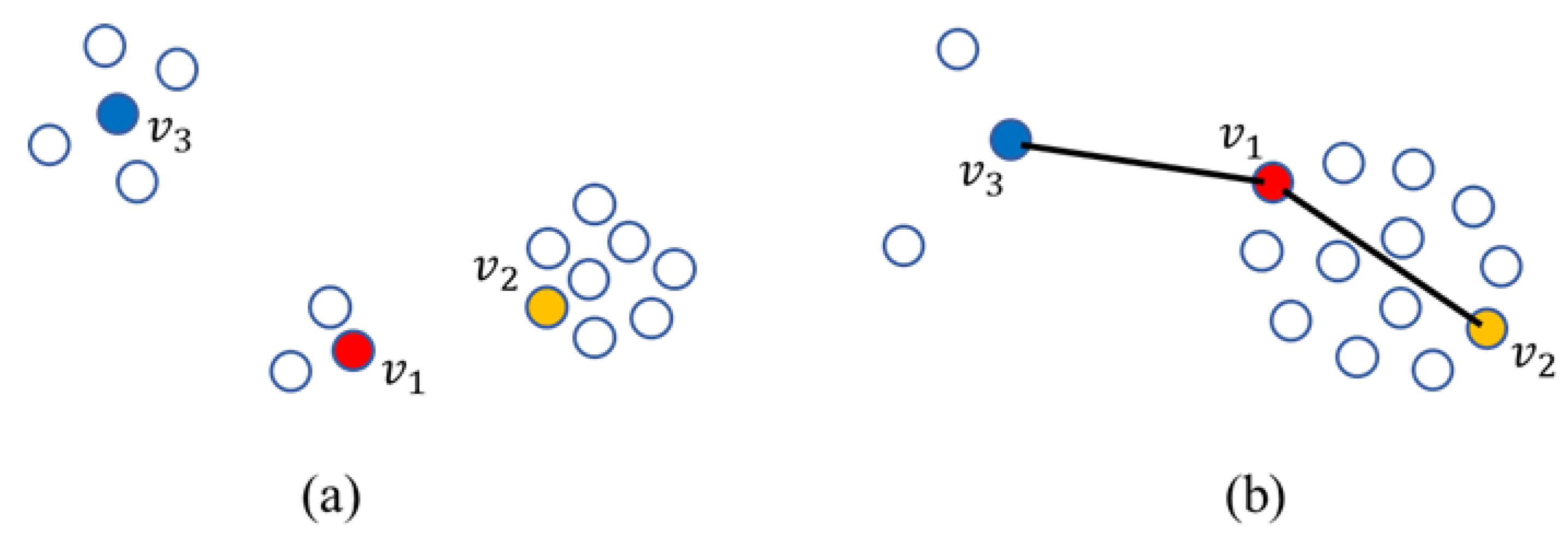
- When employed to calculate the similarity between two samples, the usual Euclidean distance is merely related to the two samples themselves but hardly considers the influence caused by their ambient samples [22,23] and ignores the distribution information of samples, which usually plays an important role for further data processing.
2. Related Work
2.1. Notations of Unsupervised Dimensionality Reduction Problem
2.2. Locality Preserving Projection (LPP)
2.3. Hypergraph Embedding
3. Proposed Method
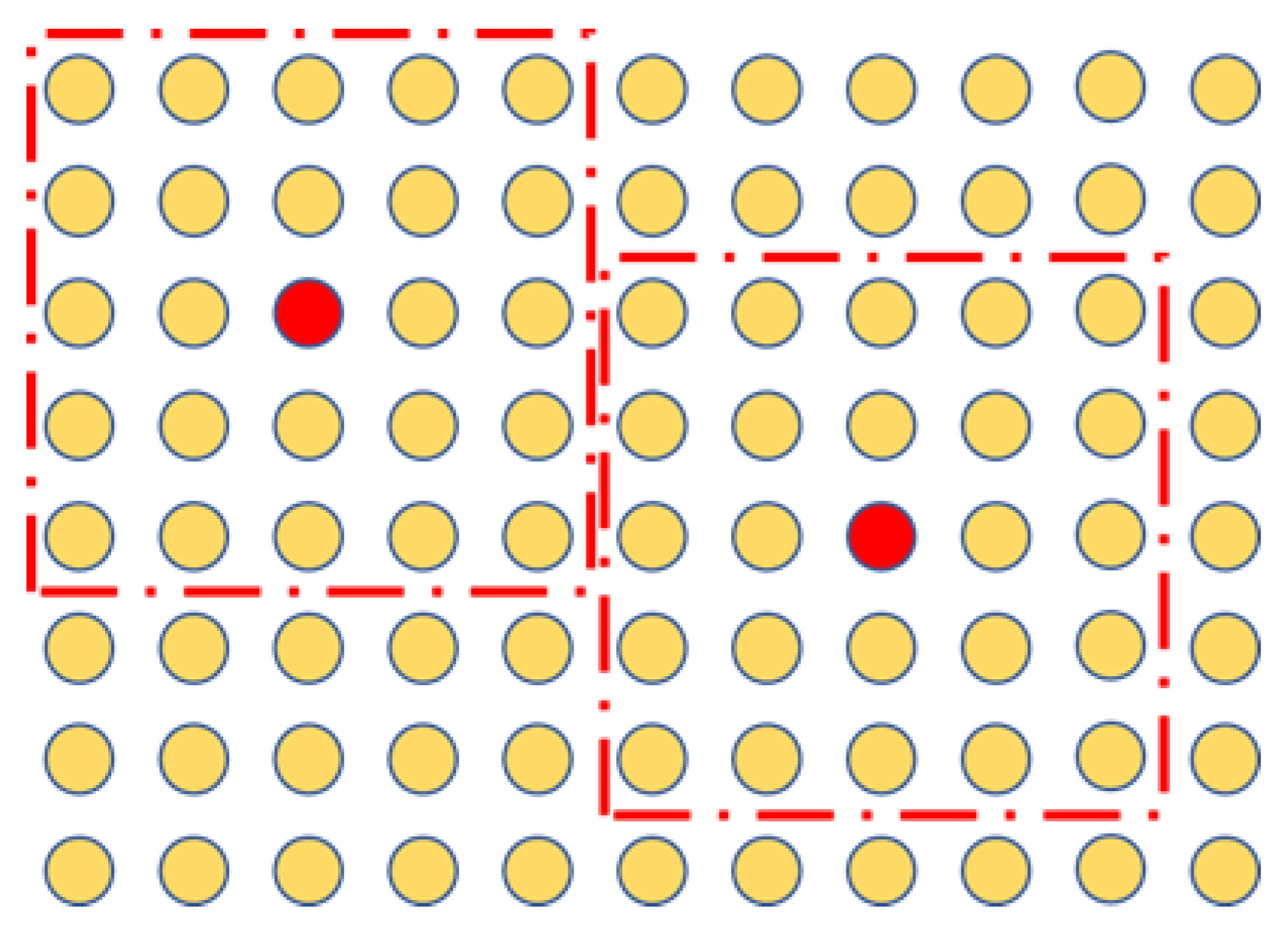
3.1. Hypergraph Embedding-Based Similarity
3.2. Similarity Distance Construction
3.3. Similarity Distance-Based Hypergraph Embedding Model
| Algorithm 1: SDHE |
| Require: Training samples , dimensionality of transformed space , the number of nearest neighbors K, the Gaussian kernel parameters h and t |
| Ensure: The optimal projection matrix . |
| Step 1: Embed hypergraph by using K nearest neighbors algorithm and get affiliation relationship according to Equation (5); Step 2: Calculate the weight of each hyperedge according to Equation (6); Step 3: Calculate the similarity by Step 4: Translate the similarity into relative similarity : Step 5: Construct the similarity distance by ; Step 6: Construct penalty factor by ; Step 7: Calculate and ; Step 8: Solve generalized eigenvalues problem Step 9: is the eigenvectors corresponded with maximum eigenvalues. |
4. Result and Discussion
4.1. Hyperspectral Images Data Set
4.1.1. Pavia University
4.1.2. Salinas
4.1.3. Kennedy Space Center
4.2. Experimental Setup
4.2.1. Training Set and Testing Set
4.2.2. Data Pre-Processing
4.2.3. Comparison and Evaluation
4.2.4. Parameter Selection
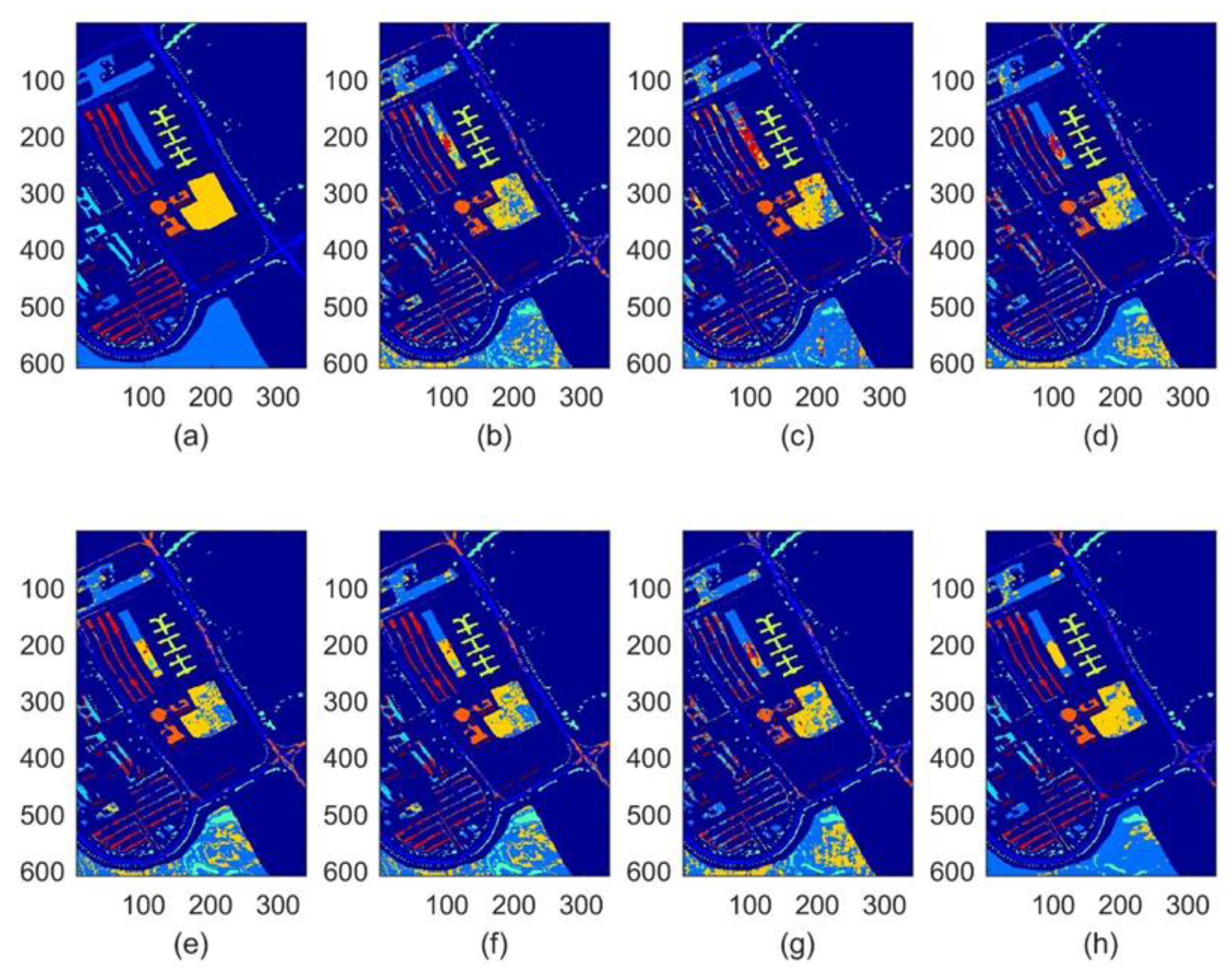
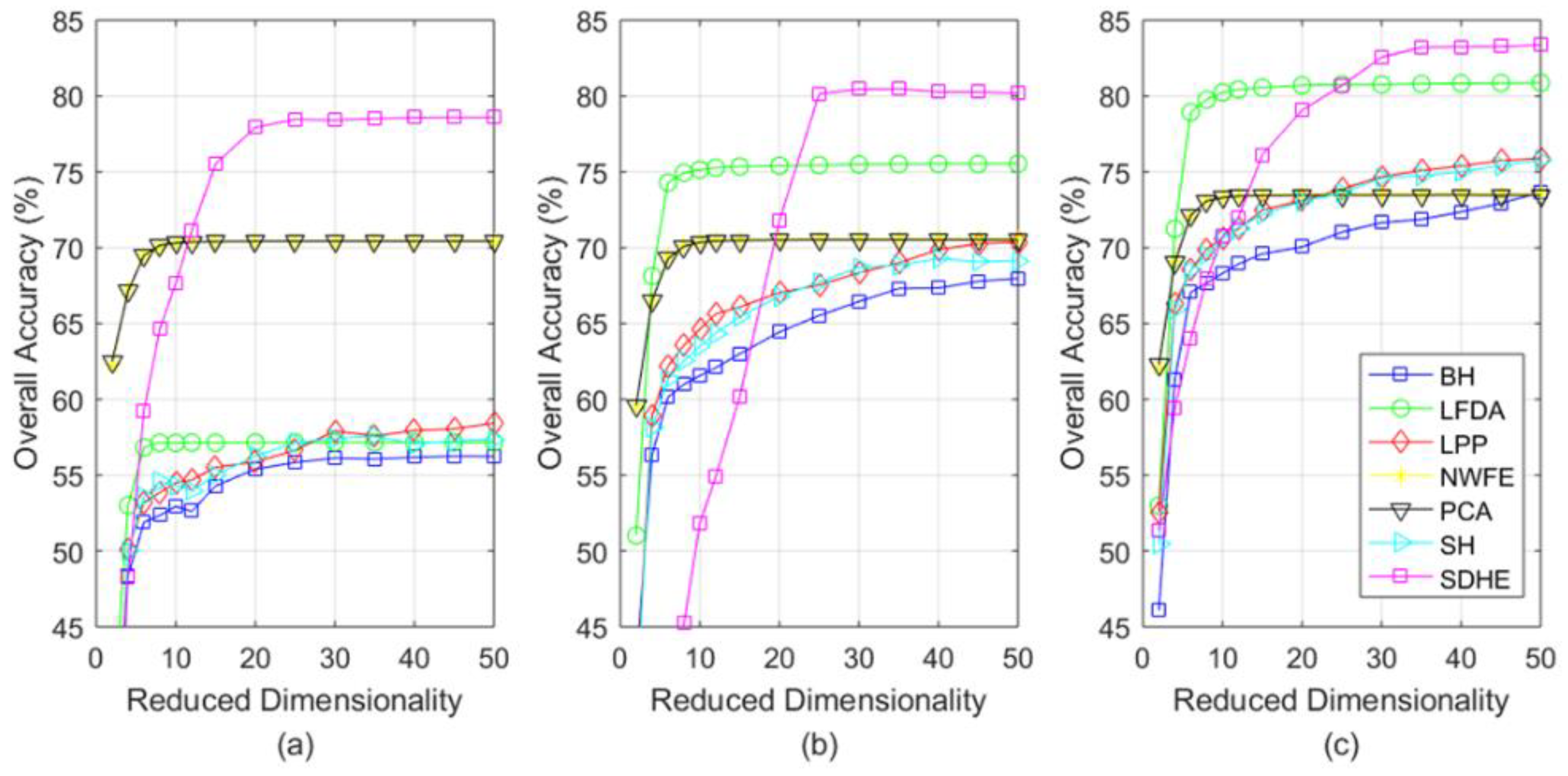
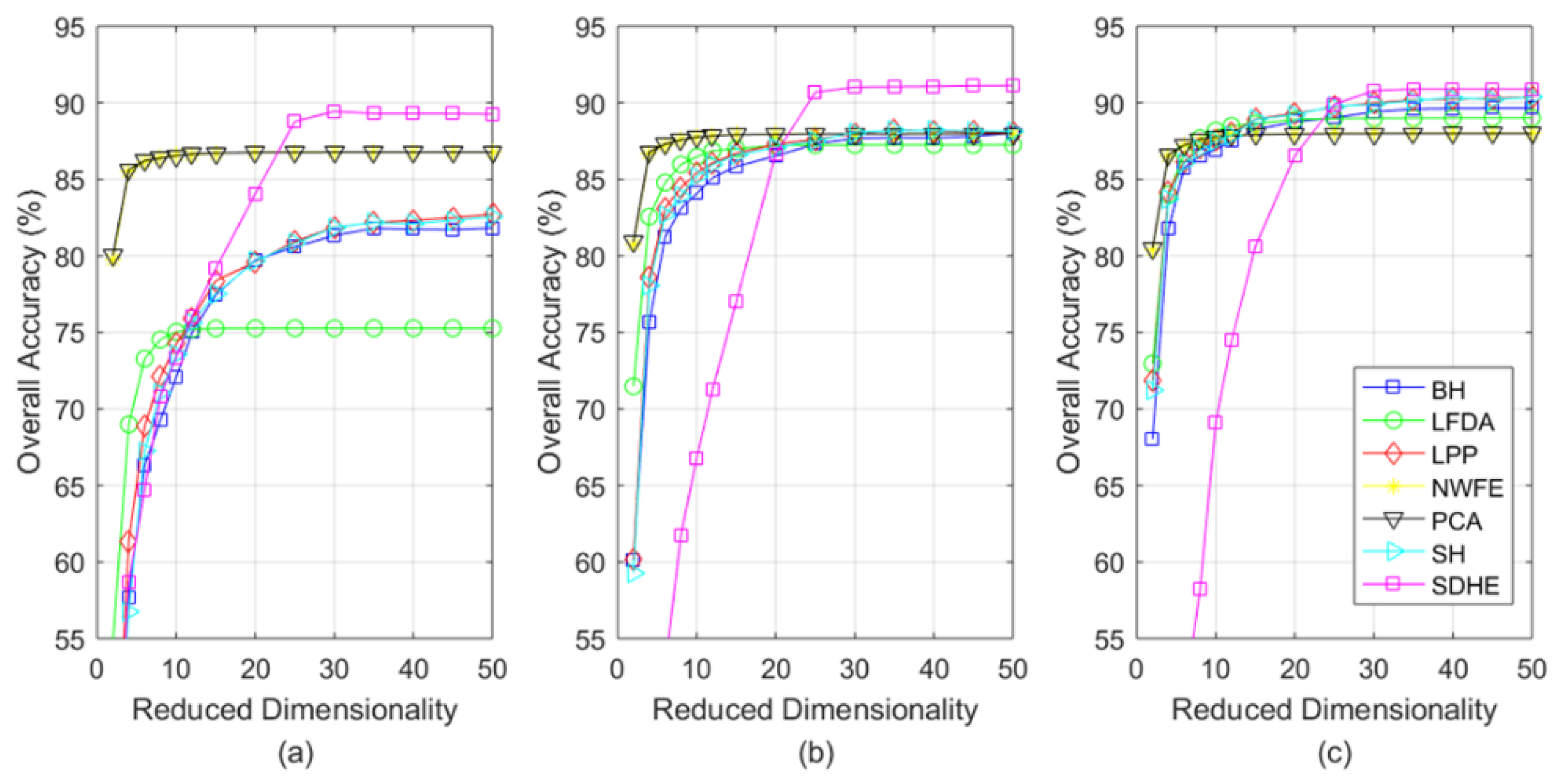
4.3. Experimental Results
5. Conclusions
- A novel similarity distance is proposed via hypergraph construction. Compared with Euclidean distance; it can make better use of the sample structure and distribution information; for the reason that it considers not only the adjacent relationship between samples but also the mutual affinity of samples in high order.
- The proposed similarity distance is employed to optimize DR problem, i.e., our proposed SDHE aims to maintain the similarity distance in a low-dimensional space. In this way, the similarity in capturing the structure and distribution information between samples is inherited in the transformed space.
- When applied for the classification task of three different hyperspectral images, our SDHE is proved to perform more effectively, especially the size of the training set is comparatively small. As shown in Table 7, Table 8 and Table 9, our method improves OA, AA, and KC by at least 2% on average on different data sets.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tan, K.; Wang, X.; Zhu, J.; Hu, J.; Li, J. A novel active learning approach for the classification of hyperspectral imagery using quasi-Newton multinomial logistic regression. Int. J. Remote Sens. 2018, 39, 3029–3054. [Google Scholar] [CrossRef]
- Zhong, Z.; Li, J.; Luo, Z.; Chapman, M. Spectral–spatial residual network for hyperspectral image classification: A 3-D deep learning framework. IEEE Trans. Geosci. Remote Sens. 2017, 56, 847–858. [Google Scholar] [CrossRef]
- Melgani, F.; Bruzzone, L. Classification of hyperspectral remote sensing images with support vector machines. IEEE Trans. Geosci. Remote Sens. 2004, 42, 1778–1790. [Google Scholar] [CrossRef]
- Chang, C.-I. Hyperspectral Data Exploitation: Theory and Applications; John Wiley & Sons: Hoboken, NJ, USA, 2007. [Google Scholar]
- Yu, C.; Lee, L.-C.; Chang, C.-I.; Xue, B.; Song, M.; Chen, J. Band-specified virtual dimensionality for band selection: An orthogonal subspace projection approach. IEEE Trans. Geosci. Remote Sens. 2018, 56, 2822–2832. [Google Scholar] [CrossRef]
- Wang, Q.; Meng, Z.; Li, X. Locality adaptive discriminant analysis for spectral–spatial classification of hyperspectral images. IEEE Geosci. Remote Sens. Lett. 2017, 14, 2077–2081. [Google Scholar] [CrossRef]
- Fan, Z.; Xu, Y.; Zuo, W.; Yang, J.; Tang, J.; Lai, Z.; Zhang, D. Modified principal component analysis: An integration of multiple similarity subspace models. IEEE Trans. Neural Netw. Learn. Syst. 2014, 25, 1538–1552. [Google Scholar] [CrossRef]
- Kuo, B.-C.; Li, C.-H.; Yang, J.-M. Kernel nonparametric weighted feature extraction for hyperspectral image classification. IEEE Trans. Geosci. Remote Sens. 2009, 47, 1139–1155. [Google Scholar]
- Sugiyama, M. Dimensionality reduction of multimodal labeled data by local fisher discriminant analysis. J. Mach. Learn. Res. 2007, 8, 1027–1061. [Google Scholar]
- Zhong, Z.; Fan, B.; Duan, J.; Wang, L.; Ding, K.; Xiang, S.; Pan, C. Discriminant tensor spectral–spatial feature extraction for hyperspectral image classification. IEEE Geosci. Remote Sens. Lett. 2014, 12, 1028–1032. [Google Scholar] [CrossRef]
- Wang, R.; Nie, F.; Hong, R.; Chang, X.; Yang, X.; Yu, W. Fast and orthogonal locality preserving projections for dimensionality reduction. IEEE Trans. Image Process. 2017, 26, 5019–5030. [Google Scholar] [CrossRef]
- Jolliffe, I.T.; Cadima, J. Principal component analysis: A review and recent developments. Philos. Trans. R. Soc. A Math. Phys. Eng. Sci. 2016, 374, 20150202. [Google Scholar] [CrossRef]
- Wang, Q.; Lin, J.; Yuan, Y. Salient band selection for hyperspectral image classification via manifold ranking. IEEE Trans. Neural Netw. Learn. Syst. 2016, 27, 1279–1289. [Google Scholar] [CrossRef]
- Belkin, M.; Niyogi, P. Laplacian eigenmaps for dimensionality reduction and data representation. Neural Comput. 2003, 15, 1373–1396. [Google Scholar] [CrossRef]
- Roweis, S.T.; Saul, L.K. Nonlinear dimensionality reduction by locally linear embedding. Science 2000, 290, 2323–2326. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.; Xu, D.; Zhang, B.; Zhang, H.-J. Graph embedding: A general framework for dimensionality reduction. In Proceedings of the 2005 IEEE Computer Society Conference on Computer Vision and Pattern Recognition (CVPR’05), San Diego, CA, USA, 20–25 June 2005; pp. 830–837. [Google Scholar]
- He, X.; Cai, D.; Yan, S.; Zhang, H.-J. Neighborhood preserving embedding. In Proceedings of the Tenth IEEE International Conference on Computer Vision (ICCV’05) Volume 1, Beijing, China, 17–21 October 2005; pp. 1208–1213. [Google Scholar]
- Zhong, F.; Zhang, J.; Li, D. Discriminant locality preserving projections based on L1-norm maximization. IEEE Trans. Neural Netw. Learn. Syst. 2014, 25, 2065–2074. [Google Scholar] [CrossRef] [PubMed]
- Soldera, J.; Behaine, C.A.R.; Scharcanski, J. Customized orthogonal locality preserving projections with soft-margin maximization for face recognition. IEEE Trans. Instrum. Meas. 2015, 64, 2417–2426. [Google Scholar] [CrossRef]
- Goyal, P.; Ferrara, E. Graph embedding techniques, applications, and performance: A survey. Knowl. -Based Syst. 2018, 151, 78–94. [Google Scholar] [CrossRef]
- Yu, J.; Tao, D.; Wang, M. Adaptive hypergraph learning and its application in image classification. IEEE Trans. Image Process. 2012, 21, 3262–3272. [Google Scholar]
- Sun, Y.; Wang, S.; Liu, Q.; Hang, R.; Liu, G. Hypergraph embedding for spatial-spectral joint feature extraction in hyperspectral images. Remote Sens. 2017, 9, 506. [Google Scholar] [CrossRef]
- Du, W.; Qiang, W.; Lv, M.; Hou, Q.; Zhen, L.; Jing, L. Semi-supervised dimension reduction based on hypergraph embedding for hyperspectral images. Int. J. Remote Sens. 2018, 39, 1696–1712. [Google Scholar] [CrossRef]
- Xiao, G.; Wang, H.; Lai, T.; Suter, D. Hypergraph modelling for geometric model fitting. Pattern Recognit. 2016, 60, 748–760. [Google Scholar] [CrossRef]
- Armanfard, N.; Reilly, J.P.; Komeili, M. Local feature selection for data classification. IEEE Trans. Pattern Anal. Mach. Intell. 2015, 38, 1217–1227. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Bai, L.; Liang, Y.; Hancock, E. Joint hypergraph learning and sparse regression for feature selection. Pattern Recognit. 2017, 63, 291–309. [Google Scholar] [CrossRef] [Green Version]
- Tenenbaum, J.B.; Silva, V.D.; Langford, J.C. A global geometric framework for nonlinear dimensionality reduction. Science 2000, 290, 2319–2323. [Google Scholar] [CrossRef]
- Zhang, L.; Gao, Y.; Hong, C.; Feng, Y.; Zhu, J.; Cai, D. Feature correlation hypergraph: Exploiting high-order potentials for multimodal recognition. IEEE Trans. Cybern. 2013, 44, 1408–1419. [Google Scholar] [CrossRef] [PubMed]
- Du, D.; Qi, H.; Wen, L.; Tian, Q.; Huang, Q.; Lyu, S. Geometric hypergraph learning for visual tracking. IEEE Trans. Cybern. 2016, 47, 4182–4195. [Google Scholar] [CrossRef] [PubMed]
- Feng, F.; Li, W.; Du, Q.; Zhang, B. Dimensionality reduction of hyperspectral image with graph-based discriminant analysis considering spectral similarity. Remote Sens. 2017, 9, 323. [Google Scholar] [CrossRef]
- Camps-Valls, G.; Gomez-Chova, L.; Muñoz-Marí, J.; Vila-Francés, J.; Calpe-Maravilla, J. Composite kernels for hyperspectral image classification. IEEE Geosci. Remote Sens. Lett. 2006, 3, 93–97. [Google Scholar] [CrossRef]
- Yuan, H.; Tang, Y.Y. Learning with hypergraph for hyperspectral image feature extraction. IEEE Geosci. Remote Sens. Lett. 2015, 12, 1695–1699. [Google Scholar] [CrossRef]
- Stanković, L.; Mandic, D.; Daković, M.; Brajović, M.; Scalzo, B.; Li, S.; Constantinides, A.G. Data analytics on graphs Part I: Graphs and spectra on graphs. Found. Trends® Mach. Learn. 2020, 13, 1–157. [Google Scholar] [CrossRef]
- Cichocki, A.; Mandic, D.; De Lathauwer, L.; Zhou, G.; Zhao, Q.; Caiafa, C.; Phan, H.A. Tensor decompositions for signal processing applications: From two-way to multiway component analysis. IEEE Signal Process. Mag. 2015, 32, 145–163. [Google Scholar] [CrossRef]
- Stanković, L.; Mandic, D.; Daković, M.; Brajović, M.; Scalzo, B.; Li, S.; Constantinides, A.G. Data analytics on graphs part III: Machine learning on graphs, from graph topology to applications. Found. Trends® Mach. Learn. 2020, 13, 332–530. [Google Scholar] [CrossRef]
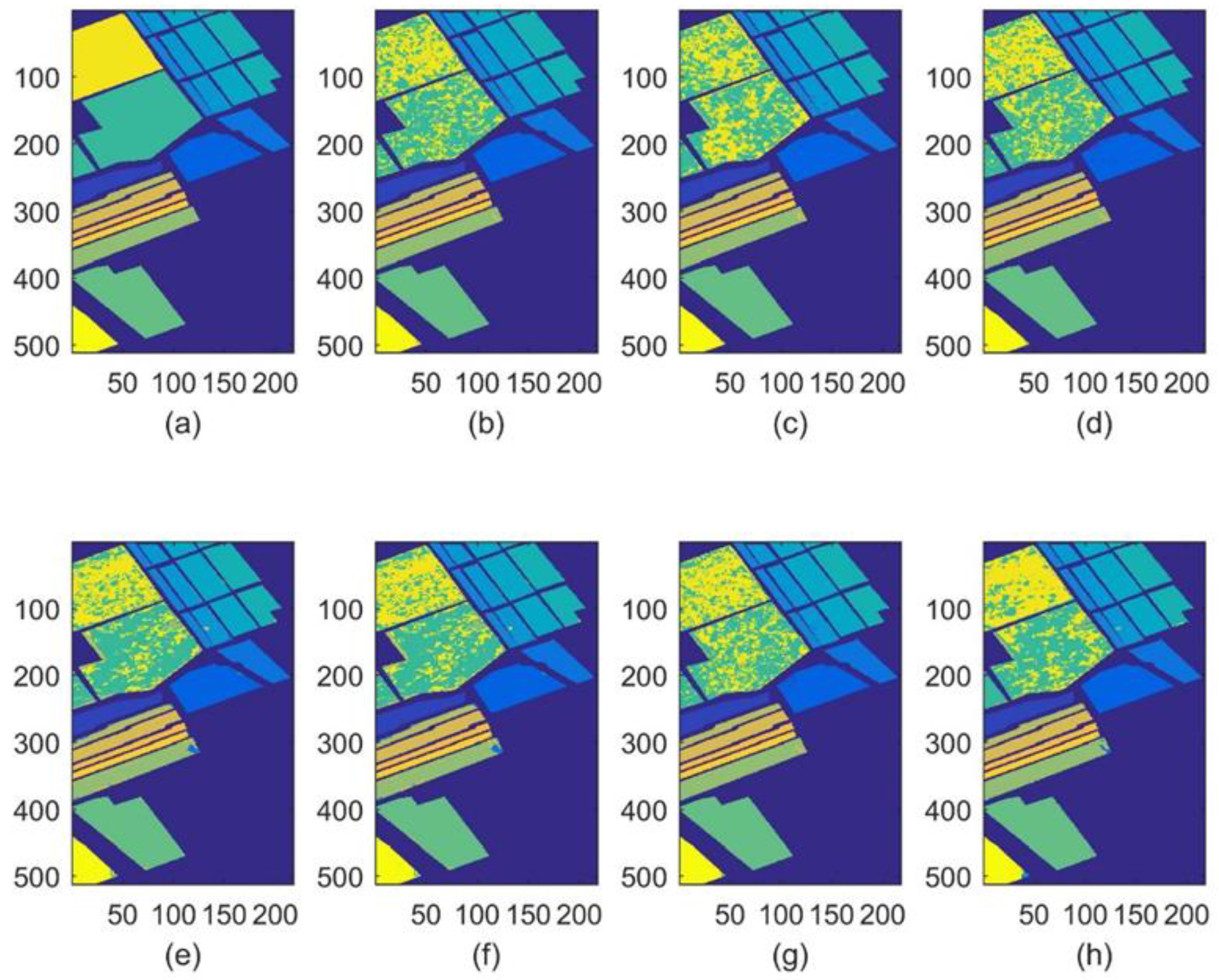


| Number | Class | Samples |
|---|---|---|
| 1 | Asphalt | 6631 |
| 2 | Meadows | 18,649 |
| 3 | Gravel | 2099 |
| 4 | Trees | 3064 |
| 5 | Painted metal sheets | 1345 |
| 6 | Bare Soil | 5029 |
| 7 | Bitumen | 1330 |
| 8 | Self-Blocking Bricks | 3682 |
| 9 | Shadows | 947 |
| Total | 42,776 |
| Number | Class | Samples |
|---|---|---|
| 1 | Brocoil-green-weeds-1 | 2009 |
| 2 | Brocoil-green-weeds-2 | 3726 |
| 3 | Fallow | 1976 |
| 4 | Fallow-rough-plow | 1394 |
| 5 | Fallow-smooth | 2678 |
| 6 | Stubble | 3959 |
| 7 | Celery | 3579 |
| 8 | Grapes-untrained | 11,271 |
| 9 | Soil-vinyard-develop | 6203 |
| 10 | Corn-senesced-green-weeds | 3278 |
| 11 | Lettuce-romaine-4wk | 1068 |
| 12 | Lettuce-romaine-5wk | 1927 |
| 13 | Lettuce-romaine-6wk | 916 |
| 14 | Lettuce-romaine-7wk | 1070 |
| 15 | Vinyard-untrained | 7268 |
| 16 | Vinyard-vertical-trellis | 1807 |
| Total | 54,129 |
| Number | Class | Samples |
|---|---|---|
| 1 | Scrub | 761 |
| 2 | Willow swamp | 243 |
| 3 | CP hammock | 256 |
| 4 | CP/Oak hammock | 252 |
| 5 | Slash pine | 161 |
| 6 | Oak/Broadleaf hammock | 229 |
| 7 | Hardwood swamp | 105 |
| 8 | Graminoid marsh | 431 |
| 9 | Spartina marsh | 520 |
| 10 | Cattail marsh | 404 |
| 11 | Salt marsh | 419 |
| 12 | Mud flats | 503 |
| 13 | Water | 927 |
| Total | 5211 |
| Class | RAW | BH | LFDA | LPP | NWFE | PCA | SH | SDHE |
|---|---|---|---|---|---|---|---|---|
| 1 | 67.82 ± 5.17 | 61.51 ± 5.88 | 70.34 ± 6.08 | 56.46 ± 6.69 | 67.99 ± 5.15 | 67.84 ± 5.16 | 57.88 ± 5.01 | 73.68 ± 8.17 |
| 2 | 63.54 ± 5.55 | 60.97 ± 5.78 | 75.98 ± 4.01 | 69.00 ± 8.34 | 63.56 ± 5.52 | 63.54 ± 5.55 | 69.40 ± 8.23 | 79.54 ± 5.75 |
| 3 | 62.37 ± 5.10 | 53.46 ± 6.34 | 65.97 ± 6.19 | 50.26 ± 5.25 | 62.46 ± 5.17 | 62.34 ± 5.13 | 48.84 ± 5.79 | 69.74 ± 6.78 |
| 4 | 86.70 ± 3.13 | 84.76 ± 4.87 | 89.01 ± 4.86 | 89.14 ± 3.28 | 86.76 ± 3.11 | 86.70 ± 3.13 | 89.08 ± 3.94 | 87.53 ± 6.02 |
| 5 | 99.52 ± 0.36 | 100.00 ± 0 | 99.92 ± 0.11 | 100.00 ± 0 | 99.52 ± 0.36 | 99.52 ± 0.36 | 100.00 ± 0 | 99.77 ± 0.40 |
| 6 | 75.52 ± 6.47 | 72.68 ± 4.49 | 74.53 ± 8.99 | 74.13 ± 4.98 | 75.58 ± 6.47 | 75.52 ± 6.47 | 73.33 ± 5.36 | 86.70 ± 3.70 |
| 7 | 80.05 ± 3.58 | 81.23 ± 5.15 | 84.25 ± 7.46 | 66.92 ± 9.20 | 79.98 ± 3.66 | 80.02 ± 3.57 | 67.10 ± 4.81 | 88.93 ± 5.30 |
| 8 | 74.10 ± 5.79 | 61.16 ± 5.08 | 59.92 ± 6.00 | 52.92 ± 5.70 | 74.22 ± 5.84 | 74.09 ± 5.79 | 54.36 ± 5.37 | 74.18 ± 8.81 |
| 9 | 99.08 ± 0.46 | 99.15 ± 0.44 | 98.79 ± 0.71 | 98.91 ± 0.70 | 99.09 ± 0.47 | 99.08 ± 0.46 | 98.88 ± 0.80 | 99.26 ± 0.35 |
| OA | 70.52 ± 2.77 | 66.45 ± 2.48 | 75.49 ± 2.21 | 68.35 ± 3.53 | 70.58 ± 2.77 | 70.52 ± 2.77 | 68.71 ± 3.07 | 80.45 ± 3.68 |
| AA | 78.74 ± 1.40 | 74.99 ± 1.22 | 79.86 ± 1.28 | 73.08 ± 1.85 | 78.80 ± 1.42 | 78.74 ± 1.40 | 73.21 ± 1.61 | 84.37 ± 2.96 |
| KC | 60.88 ± 3.68 | 55.48 ± 3.29 | 67.48 ± 2.94 | 58.00 ± 4.69 | 60.96 ± 3.68 | 60.87 ± 3.68 | 58.47 ± 4.08 | 74.06 ± 4.88 |
| Class | RAW | BH | LFDA | LPP | NWFE | PCA | SH | SDHE |
|---|---|---|---|---|---|---|---|---|
| 1 | 98.49 ± 0.59 | 99.79 ± 0.43 | 98.93 ± 1.51 | 99.43 ± 0.60 | 98.49 ± 0.59 | 98.49 ± 0.59 | 99.61 ± 0.38 | 99.50 ± 0.76 |
| 2 | 99.61 ± 0.46 | 99.88 ± 0.24 | 99.78 ± 0.50 | 99.09 ± 1.61 | 99.62 ± 0.46 | 99.61 ± 0.46 | 99.59 ± 0.63 | 99.90 ± 0.16 |
| 3 | 97.07 ± 1.70 | 98.56 ± 1.41 | 98.30 ± 1.28 | 99.36 ± 0.99 | 97.11 ± 1.67 | 97.06 ± 1.70 | 99.18 ± 0.81 | 99.16 ± 1.55 |
| 4 | 97.90 ± 1.60 | 98.53 ± 1.43 | 98.15 ± 0.64 | 99.02 ± 0.58 | 97.93 ± 1.57 | 97.89 ± 1.62 | 98.96 ± 0.61 | 98.52 ± 0.91 |
| 5 | 93.92 ± 1.26 | 96.58 ± 0.87 | 93.67 ± 2.02 | 96.86 ± 0.92 | 93.91 ± 1.28 | 93.92 ± 1.27 | 96.90 ± 0.85 | 95.64 ± 1.79 |
| 6 | 99.55 ± 0.55 | 99.84 ± 0.43 | 99.74 ± 0.54 | 99.97 ± 0.07 | 99.55 ± 0.55 | 99.55 ± 0.55 | 99.96 ± 0.07 | 99.77 ± 0.43 |
| 7 | 98.76 ± 0.54 | 99.47 ± 0.55 | 99.64 ± 0.37 | 99.70 ± 0.20 | 98.76 ± 0.55 | 98.76 ± 0.54 | 99.68 ± 0.21 | 99.54 ± 0.30 |
| 8 | 68.68 ± 3.58 | 62.60 ± 5.21 | 67.41 ± 5.72 | 65.74 ± 5.01 | 68.64 ± 3.54 | 68.64 ± 3.59 | 65.45 ± 5.33 | 75.46 ± 4.40 |
| 9 | 98.70 ± 0.55 | 99.87 ± 0.20 | 98.80 ± 2.19 | 99.54 ± 1.30 | 98.71 ± 0.54 | 98.70 ± 0.55 | 99.78 ± 0.56 | 99.80 ± 0.22 |
| 10 | 86.22 ± 4.13 | 94.67 ± 1.89 | 92.28 ± 2.52 | 95.28 ± 1.88 | 86.27 ± 4.14 | 86.22 ± 4.13 | 95.43 ± 1.69 | 92.38 ± 1.77 |
| 11 | 95.29 ± 2.19 | 98.46 ± 1.17 | 98.44 ± 1.26 | 98.94 ± 0.83 | 95.33 ± 2.20 | 95.29 ± 2.19 | 98.89 ± 0.72 | 98.38 ± 1.41 |
| 12 | 99.94 ± 0.08 | 99.51 ± 0.51 | 98.22 ± 1.83 | 99.63 ± 0.44 | 99.95 ± 0.08 | 99.94 ± 0.08 | 99.27 ± 1.48 | 99.44 ± 1.51 |
| 13 | 98.67 ± 1.78 | 99.01 ± 1.33 | 98.95 ± 0.87 | 99.23 ± 0.91 | 98.65 ± 1.77 | 98.67 ± 1.78 | 99.11 ± 1.13 | 99.74 ± 0.44 |
| 14 | 95.70 ± 2.87 | 97.00 ± 1.77 | 97.15 ± 2.28 | 96.86 ± 2.38 | 95.71 ± 2.85 | 95.70 ± 2.87 | 96.84 ± 2.44 | 97.88 ± 1.88 |
| 15 | 73.89 ± 4.75 | 72.64 ± 5.66 | 65.79 ± 6.05 | 69.82 ± 5.47 | 73.80 ± 4.83 | 73.87 ± 4.75 | 70.47 ± 6.15 | 78.80 ± 3.70 |
| 16 | 96.56 ± 1.82 | 98.82 ± 0.58 | 98.63 ± 0.44 | 99.13 ± 0.37 | 96.56 ± 1.82 | 96.55 ± 1.82 | 99.17 ± 0.33 | 98.04 ± 0.73 |
| OA | 87.98 ± 0.76 | 87.67 ± 0.74 | 87.24 ± 1.53 | 87.99 ± 0.79 | 87.97 ± 0.75 | 87.97 ± 0.76 | 88.07 ± 0.78 | 91.01 ± 1.37 |
| AA | 93.68 ± 0.35 | 94.70 ± 0.22 | 93.99 ± 0.85 | 94.85 ± 0.32 | 93.69 ± 0.35 | 93.68 ± 0.35 | 94.89 ± 0.33 | 95.75 ± 0.59 |
| KC | 86.61 ± 0.85 | 86.27 ± 0.83 | 85.79 ± 1.71 | 86.62 ± 0.88 | 86.59 ± 0.84 | 86.59 ± 0.85 | 86.71 ± 0.87 | 89.98 ± 1.53 |
| Class | RAW | BH | LFDA | LPP | NWFE | PCA | SH | SDHE |
|---|---|---|---|---|---|---|---|---|
| 1 | 94.55 ± 3.90 | 90.20 ± 6.86 | 86.13 ± 7.04 | 92.47 ± 3.33 | 94.55 ± 3.90 | 94.55 ± 3.90 | 92.46 ± 4.31 | 95.03 ± 2.75 |
| 2 | 90.45 ± 4.25 | 89.78 ± 4.99 | 89.06 ± 5.86 | 91.75 ± 4.59 | 90.49 ± 4.25 | 90.45 ± 4.25 | 91.84 ± 5.23 | 94.39 ± 3.84 |
| 3 | 92.63 ± 1.60 | 88.18 ± 7.74 | 86.86 ± 6.80 | 86.44 ± 5.88 | 92.63 ± 1.62 | 92.58 ± 1.61 | 86.10 ± 9.55 | 95.89 ± 3.22 |
| 4 | 61.51 ± 5.50 | 54.05 ± 6.12 | 72.76 ± 8.26 | 43.97 ± 7.92 | 61.72 ± 5.64 | 61.42 ± 5.53 | 51.98 ± 6.95 | 81.64 ± 4.18 |
| 5 | 72.84 ± 4.93 | 74.47 ± 7.36 | 90.00 ± 6.43 | 68.01 ± 7.00 | 72.70 ± 4.98 | 72.70 ± 5.04 | 71.28 ± 11.8 | 94.04 ± 3.89 |
| 6 | 80.86 ± 2.85 | 84.74 ± 6.36 | 90.38 ± 7.89 | 83.11 ± 6.25 | 80.86 ± 2.85 | 80.81 ± 2.87 | 82.49 ± 5.68 | 94.59 ± 3.30 |
| 7 | 99.18 ± 1.83 | 97.29 ± 4.50 | 96.82 ± 5.98 | 97.65 ± 3.11 | 99.18 ± 1.83 | 99.18 ± 1.83 | 97.65 ± 2.68 | 99.53 ± 0.94 |
| 8 | 88.44 ± 3.88 | 85.23 ± 8.67 | 90.24 ± 3.39 | 91.05 ± 6.76 | 88.44 ± 3.88 | 88.44 ± 3.88 | 89.81 ± 6.01 | 96.45 ± 3.02 |
| 9 | 96.20 ± 2.13 | 95.14 ± 3.29 | 93.60 ± 4.68 | 96.82 ± 2.86 | 96.20 ± 2.13 | 96.18 ± 2.16 | 96.66 ± 3.34 | 99.92 ± 0.13 |
| 10 | 93.54 ± 4.47 | 94.48 ± 2.39 | 92.60 ± 2.17 | 95.10 ± 2.66 | 93.72 ± 4.51 | 93.52 ± 4.44 | 95.78 ± 2.07 | 99.14 ± 1.22 |
| 11 | 98.97 ± 1.29 | 99.22 ± 0.77 | 97.72 ± 2.85 | 99.25 ± 0.68 | 98.97 ± 1.29 | 98.97 ± 1.29 | 99.10 ± 1.08 | 99.17 ± 1.43 |
| 12 | 92.88 ± 4.37 | 80.70 ± 6.26 | 79.36 ± 5.98 | 84.16 ± 7.35 | 93.21 ± 4.36 | 92.88 ± 4.37 | 83.35 ± 6.64 | 94.95 ± 2.92 |
| 13 | 100.00 ± 0 | 98.64 ± 0.82 | 98.69 ± 0.85 | 97.76 ± 2.48 | 100.00 ± 0 | 100.00 ± 0 | 98.24 ± 0.77 | 99.99 ± 0.03 |
| OA | 92.38 ± 1.18 | 89.60 ± 2.43 | 90.32 ± 2.07 | 90.11 ± 1.76 | 92.44 ± 1.14 | 92.37 ± 1.17 | 90.46 ± 1.83 | 96.61 ± 0.78 |
| AA | 89.39 ± 1.08 | 87.09 ± 2.34 | 89.56 ± 1.79 | 86.73 ± 1.66 | 89.44 ± 1.05 | 89.36 ± 1.07 | 87.44 ± 1.92 | 95.75 ± 0.74 |
| KC | 91.49 ± 1.32 | 88.39 ± 2.72 | 89.19 ± 2.31 | 88.95 ± 1.96 | 91.55 ± 1.28 | 91.47 ± 1.31 | 89.35 ± 2.05 | 96.22 ± 0.87 |
| Method | The Size of Training Set | ||
|---|---|---|---|
| 15 | 20 | 25 | |
| RAW | 70.43 ± 1.75 | 70.52 ± 2.77 | 73.47 ± 1.18 |
| BH | 56.14 ± 1.72 | 66.45 ± 2.48 | 71.65 ± 3.09 |
| LFDA | 57.16 ± 8.41 | 75.49 ± 2.21 | 80.76 ± 1.25 |
| LPP | 57.92 ± 2.57 | 68.35 ± 3.54 | 74.68 ± 2.51 |
| NWFE | 70.46 ± 1.75 | 70.58 ± 2.77 | 73.56 ± 1.18 |
| PCA | 70.42 ± 1.75 | 70.52 ± 2.77 | 73.47 ± 1.18 |
| SH | 57.43 ± 1.94 | 68.71 ± 3.08 | 74.60 ± 2.94 |
| SDHE | 78.41 ± 5.13 | 80.45 ± 3.67 | 82.56 ± 2.87 |
| Method | The Size of Training Set | ||
|---|---|---|---|
| 15 | 20 | 25 | |
| RAW | 86.77 ± 1.83 | 87.98 ± 0.76 | 88.00 ± 0.92 |
| BH | 81.32 ± 1.29 | 87.67 ± 0.74 | 89.40 ± 0.76 |
| LFDA | 75.27 ± 3.32 | 87.24 ± 1.53 | 88.99 ± 0.98 |
| LPP | 81.87 ± 1.05 | 87.99 ± 0.79 | 89.97 ± 1.11 |
| NWFE | 86.78 ± 1.85 | 87.97 ± 0.75 | 88.00 ± 0.92 |
| PCA | 86.76 ± 1.83 | 87.97 ± 0.76 | 87.98 ± 0.92 |
| SH | 81.86 ± 1.57 | 88.07 ± 0.78 | 89.90 ± 1.24 |
| SDHE | 89.43 ± 1.07 | 91.01 ± 1.37 | 90.78 ± 0.87 |
| Method | The Size of Training Set | ||
|---|---|---|---|
| 15 | 20 | 25 | |
| RAW | 91.16 ± 0.58 | 92.38 ± 1.18 | 93.39 ± 0.57 |
| BH | 73.23 ± 2.35 | 89.60 ± 2.43 | 93.99 ± 0.61 |
| LFDA | 60.05 ± 11.54 | 90.32 ± 2.07 | 94.56 ± 1.03 |
| LPP | 74.05 ± 2.88 | 90.11 ± 1.76 | 93.91 ± 0.92 |
| NWFE | 91.11 ± 0.58 | 92.44 ± 1.14 | 93.38 ± 0.56 |
| PCA | 91.15 ± 0.58 | 92.37 ± 1.17 | 93.38 ± 0.58 |
| SH | 73.73 ± 1.80 | 90.46 ± 1.83 | 94.53 ± 0.70 |
| SDHE | 95.88 ± 1.04 | 96.61 ± 0.92 | 97.49 ± 0.44 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shen, X.; Fang, S.; Qiang, W. Dimensionality Reduction by Similarity Distance-Based Hypergraph Embedding. Atmosphere 2022, 13, 1449. https://doi.org/10.3390/atmos13091449
Shen X, Fang S, Qiang W. Dimensionality Reduction by Similarity Distance-Based Hypergraph Embedding. Atmosphere. 2022; 13(9):1449. https://doi.org/10.3390/atmos13091449
Chicago/Turabian StyleShen, Xingchen, Shixu Fang, and Wenwen Qiang. 2022. "Dimensionality Reduction by Similarity Distance-Based Hypergraph Embedding" Atmosphere 13, no. 9: 1449. https://doi.org/10.3390/atmos13091449
APA StyleShen, X., Fang, S., & Qiang, W. (2022). Dimensionality Reduction by Similarity Distance-Based Hypergraph Embedding. Atmosphere, 13(9), 1449. https://doi.org/10.3390/atmos13091449






