Nitrifying and Denitrifying Microbial Communities in Centralized and Decentralized Biological Nitrogen Removing Wastewater Treatment Systems
Abstract
1. Introduction
2. Materials and Methods
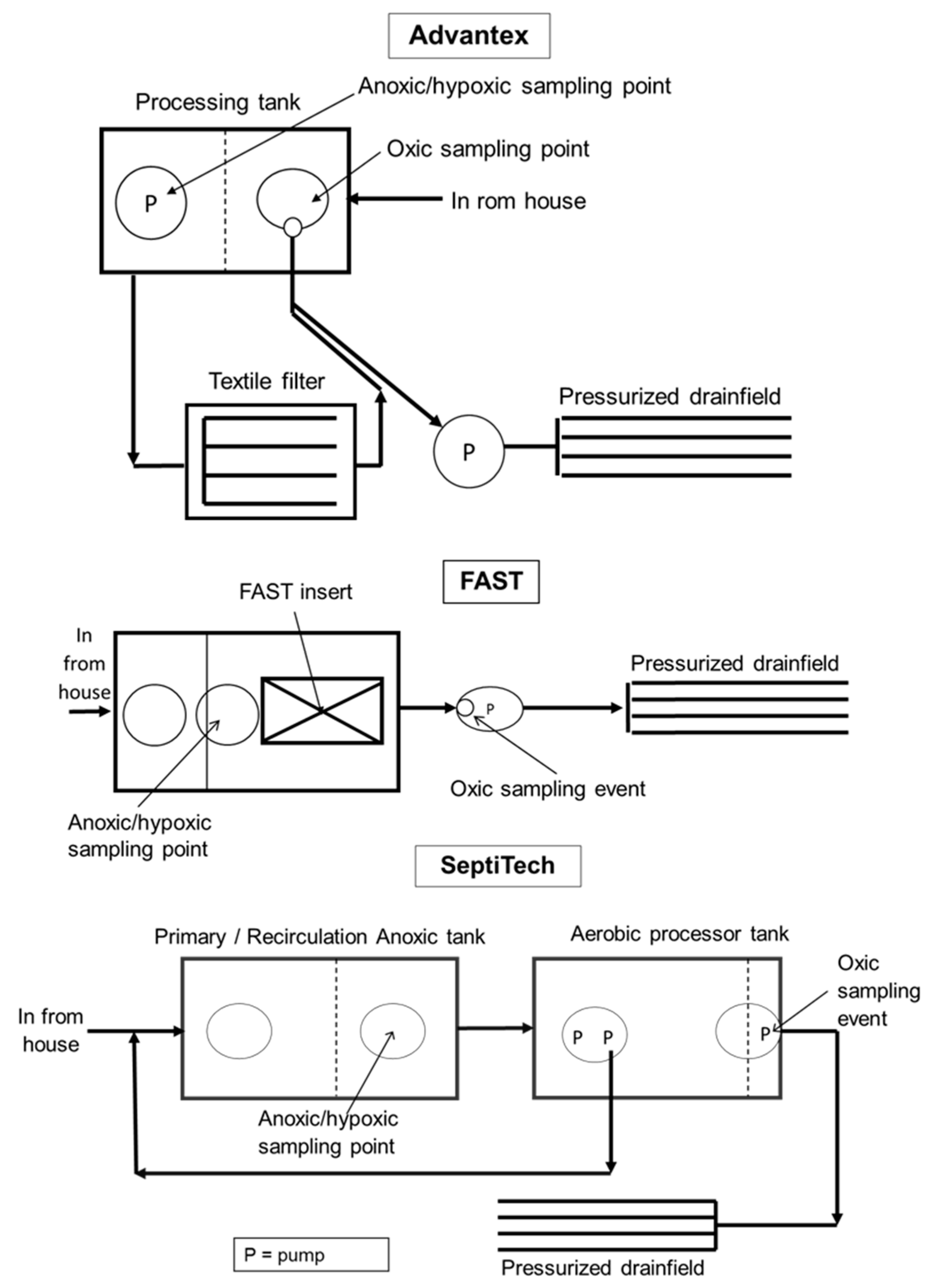
2.1. Study Systems
2.2. Sample Collection and DNA Extraction
2.3. Miseq Illumina Sequencing
2.4. Data Analyses
3. Results and Discussion
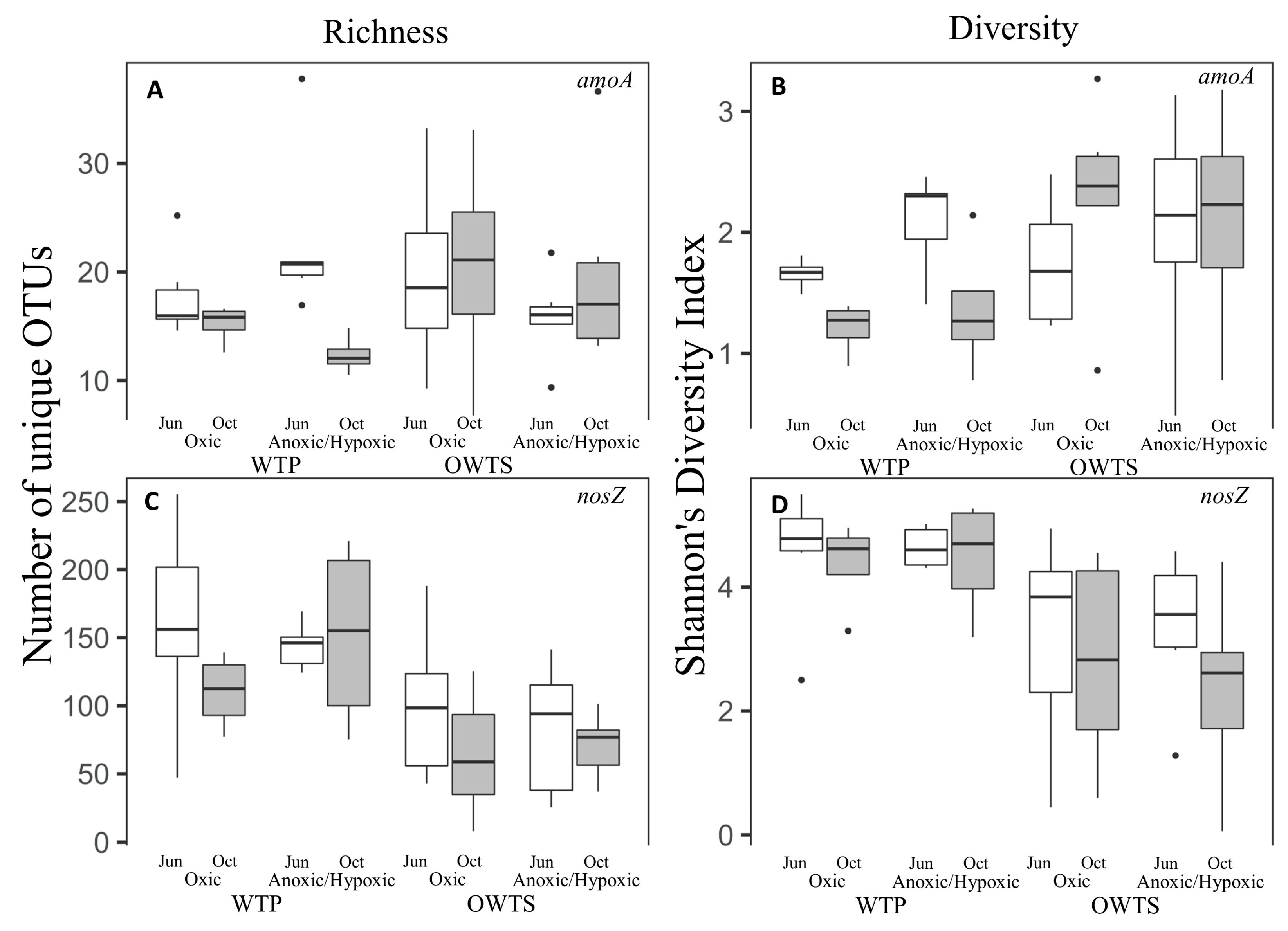
3.1. Species Richness and Diversity
3.1.1. amoA
3.1.2. nosZ
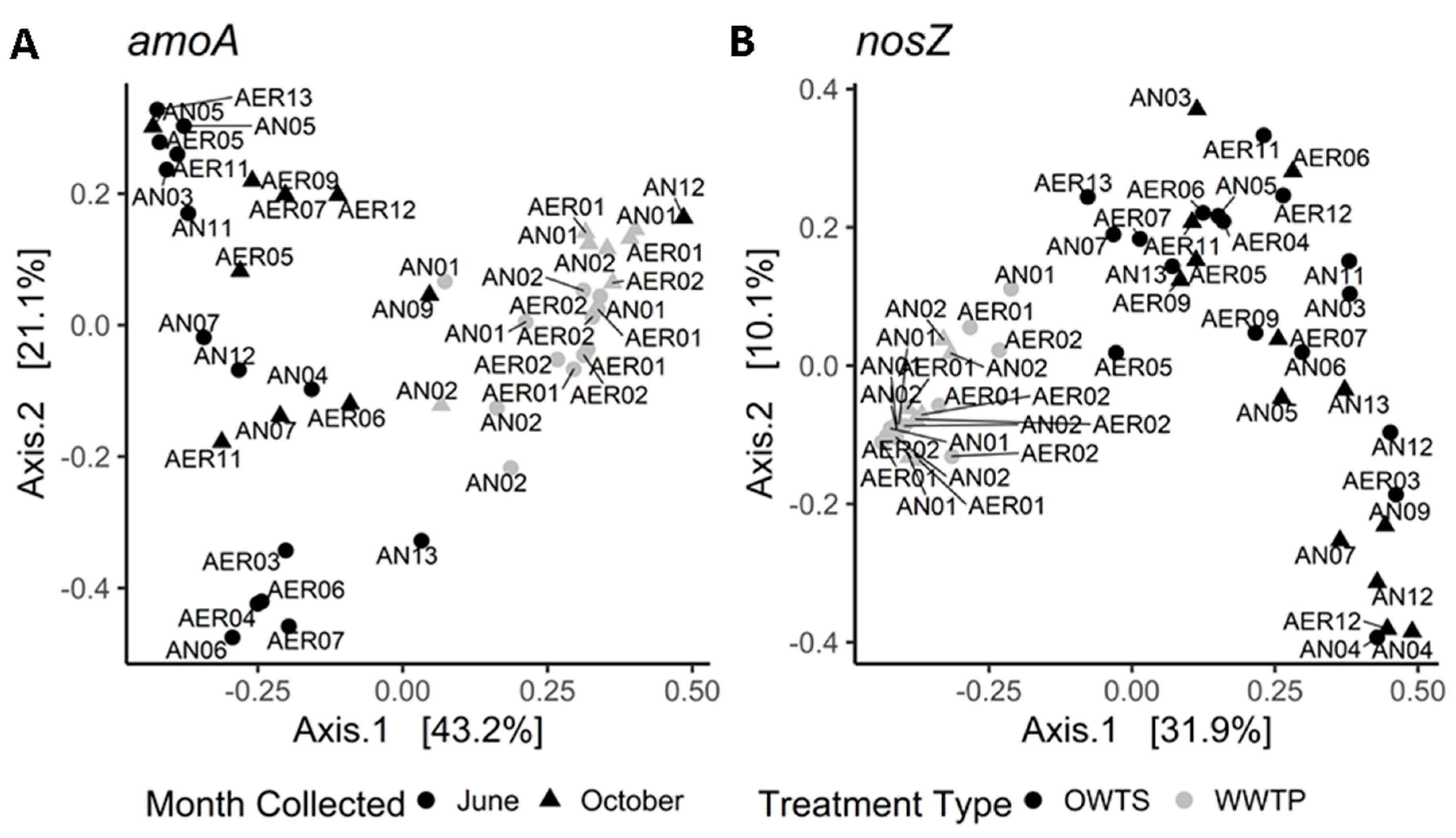
3.2. Community Structure
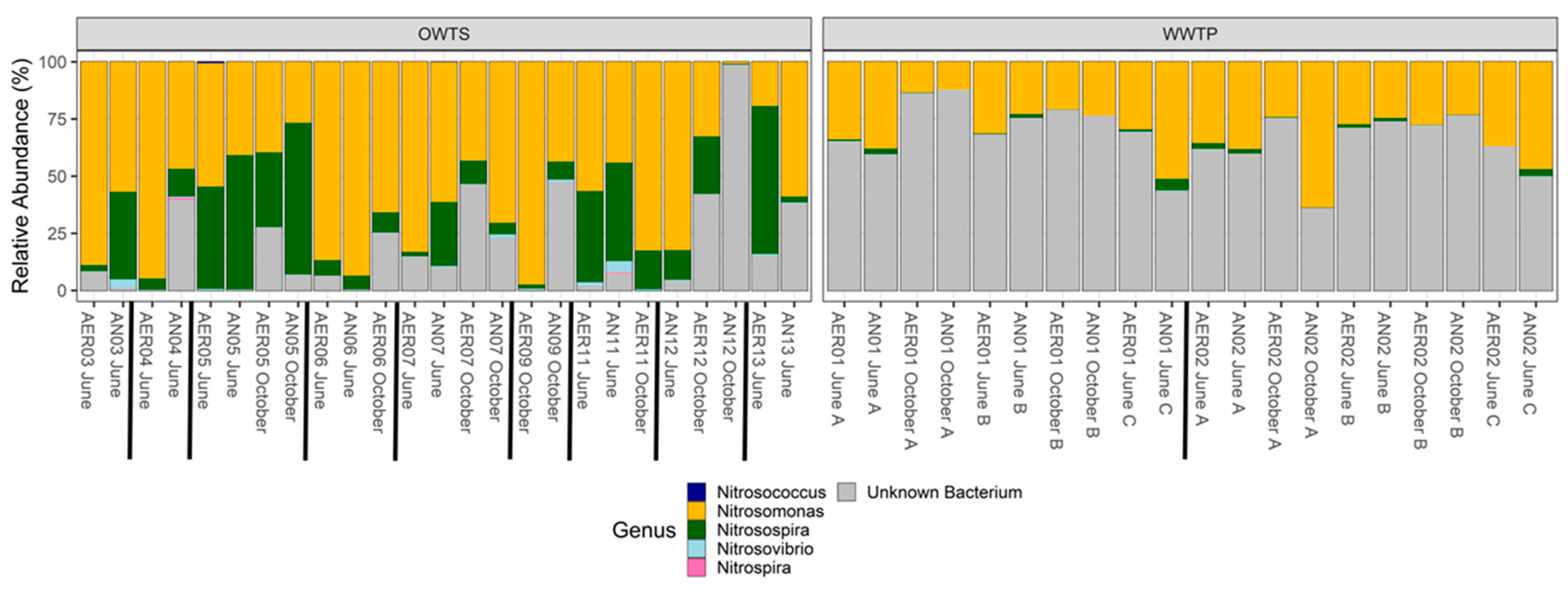
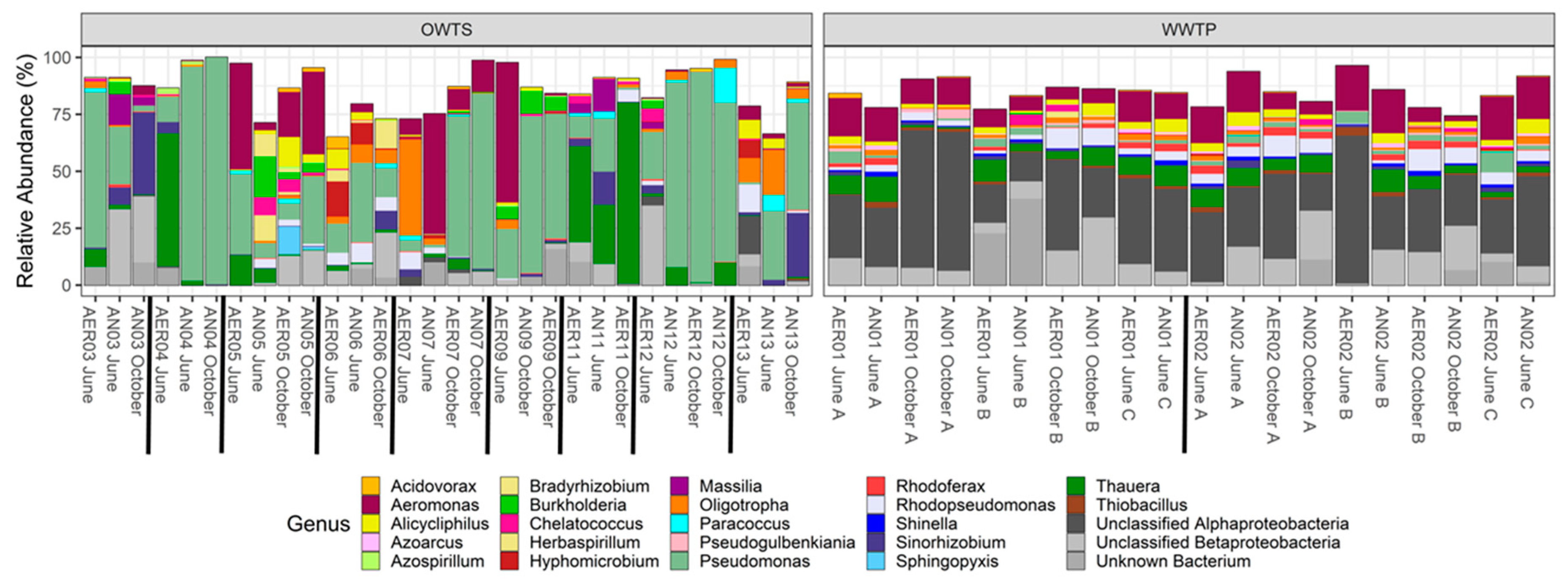
3.3. Taxonomy
3.3.1. amoA
3.3.2. nosZ
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
| System and Zone/Compartment | Water Flow Rate (MGD) | Water Temp. (°C) | DO (mg/L) | pH | Total Inorganic N (mg N/L) | Ammonium (mg N/L) | Nitrate (mg N/L) | BOD5 (mg/L) |
|---|---|---|---|---|---|---|---|---|
| WWTP | 31.9 | 211 ± 5.0 | ||||||
| Pre-Anoxic | -- a | 0.3 ± 0.0 b | 6.7 ± 0.0 b | 7.2 ± 0.6 | 4.8 ± 0.6 | 0.3 ± 0.1 | -- a | |
| Aerated IFAS | 20.3 ± 0.8 | 2.8 ± 2.4 b | 6.7 ± 0.0 b | 3.3 ± 0.3 | 1.1 ± 0.6 | 2.0 ± 0.6 | -- a | |
| Post Anoxic | -- a | 1.6 ± 1.4 b | 6.5 ± 0.1 b | 2.1 ± 1.6 | 3.0 ± 1.8 | 0.2 ± 0.1 | -- a | |
| Re-Aeration | -- a | 0.5 ± 0.1 b | 6.6 ± 0.0 b | 0.5 ± 0.1 | 0.0 ± 0.0 | 0.5 ± 0.1 | -- a | |
| Advantex | 2.1 × 10 −4 | |||||||
| Anoxic/hypoxic | 19.9 ± 0.2 | 0.2 ± 0.2 | 6.4 ± 0.1 | 15.9 ± 3.1 | 14.6 ± 3.0 | 1.3 ± 0.3 | 94.4 ± 76.9 | |
| Oxic | 18.6 ± 0.4 | 1.8 ± 1.0 | 6.4 ± 0.1 | 15.7 ± 4.7 | 9.1 ± 4.4 | 6.6 ± 2.8 | 16.9 ± 12.2 | |
| FAST | 9.4 × 10 −5 | |||||||
| Anoxic/hypoxic | 20.3 ± 0.6 | 5.1 ± 1.3 | 7.2 ± 0.2 | 19.7 ± 9.2 | 8.4 ± 4.9 | 15.0 ± 8.4 | 0.0 ± 0.0 | |
| Oxic | 18.5 ± 0.4 | 2.3 ± 0.9 | 7.0 ± 0.2 | 11.4 ± 2.1 | 1.7 ± 0.6 | 8.4 ± 1.8 | 6.0 ± 4.2 | |
| SeptiTech | 1.2 × 10 −4 | |||||||
| Anoxic/hypoxic | 21.4 ± 1.0 | 0.1 ± 0.1 | 7.2 ± 0.2 | 15.0 ± 5.0 | 11.5 ± 4.8 | 3.5 ± 0.7 | 10.3 ± 9.9 | |
| Oxic | 22.1 ± 1.1 | 4.7 ± 1.4 | 7.1 ± 0.1 | 9.2 ± 1.9 | 3.1 ± 1.7 | 6.0 ± 1.9 | 3.7 ± 1.8 |

References
- Amador, J.A.; Loomis, G.W. Soil-Based Wastewater Treatment; American Society of Agronomy, Incorporated; Soil Science Society of America, Incorporated; Crop Science Society of America, Incorporated: Madison, WI, USA, 2018; ISBN 9780891189688. [Google Scholar]
- Diaz, R.J.; Rosenberg, R. Spreading dead zones and consequences for marine ecosystems. Science (80-) 2008, 321, 926–929. [Google Scholar] [CrossRef] [PubMed]
- Huisman, J.; Matthijs, H.C.P.; Visser, P.M. Harmful Cyanobacteria; Aquatic Ecology Series; Version 3; Springer: Dordrecht, The Netherlands, 2005; ISBN 1402030096. [Google Scholar]
- Townsend, A.R.; Howarth, R.W.; Bazzaz, F.A.; Booth, M.S.; Cleveland, C.C.; Collinge, S.K.; Dobson, A.P.; Epstein, P.R.; Holland, E.A.; Keeney, D.R.; et al. Human health effects of a changing global nitrogen cycle. Front. Ecol. Environ. 2003, 1, 240–246. [Google Scholar] [CrossRef]
- Clifford, E.; Nielsen, M.; Sørensen, K.; Rodgers, M. Nitrogen dynamics and removal in a horizontal flow biofilm reactor for wastewater treatment. Water Res. 2010, 44, 3819–3828. [Google Scholar] [CrossRef] [PubMed]
- Oakley, S.M.; Gold, A.J.; Oczkowski, A.J. Nitrogen control through decentralized wastewater treatment: Process performance and alternative management strategies. Ecol. Eng. 2010, 36, 1520–1531. [Google Scholar] [CrossRef]
- Nemergut, D.R.; Schmidt, S.K.; Fukami, T.; O’Neill, S.P.; Bilinski, T.M.; Stanish, L.F.; Knelman, J.E.; Darcy, J.L.; Lynch, R.C.; Wickey, P.; et al. Patterns and Processes of Microbial Community Assembly. Microbiol. Mol. Biol. Rev. 2013, 77, 342–356. [Google Scholar] [CrossRef] [PubMed]
- USEPA. Management of Small to Medium Sized Municipal Wastewater Treatment Plants; National Service Center for Environmental Publications (NSCEP): Cincinnati, OH, USA, 1979; pp. 1–192.
- USEPA. Clean Watersheds Needs Survey 2012—Report to Congress; United States Environmental Protection Agency: Washington, DC, USA, 2012.
- Brannon, E.Q.; Moseman-Valtierra, S.M.; Lancellotti, B.V.; Wigginton, S.K.; Amador, J.A.; McCaughey, J.C.; Loomis, G.W. Comparison of N2O Emissions and Gene Abundances between Wastewater Nitrogen Removal Systems. J. Environ. Qual. 2017, 46, 931–938. [Google Scholar] [CrossRef] [PubMed]
- USEPA. U.S. Environmental Protection Agency NPDES Permit Writers’ Manual; United States Environmental Protection Agency: Washington, DC, USA, 2010.
- Cydzik-Kwiatkowska, A.; Zielińska, M. Bacterial communities in full-scale wastewater treatment systems. World J. Microbiol. Biotechnol. 2016, 32, 66. [Google Scholar] [CrossRef]
- McLellan, S.L.; Huse, S.M.; Mueller-Spitz, S.R.; Andreishcheva, E.N.; Sogin, M.L. Diversity and population structure of sewage-derived microorganisms in wastewater treatment plant influent. Environ. Microbiol. 2010, 12, 378–392. [Google Scholar] [CrossRef]
- Xia, S.; Duan, L.; Song, Y.; Li, J.; Piceno, Y.M.; Andersen, G.L.; Alvarez-Cohen, L.; Moreno-Andrade, I.; Huang, C.L.; Hermanowicz, S.W. Bacterial community structure in geographically distributed biological wastewater treatment reactors. Environ. Sci. Technol. 2010, 44, 7391–7396. [Google Scholar] [CrossRef]
- Hu, M.; Wang, X.; Wen, X.; Xia, Y. Microbial community structures in different wastewater treatment plants as revealed by 454-pyrosequencing analysis. Bioresour. Technol. 2012, 117, 72–79. [Google Scholar] [CrossRef]
- Wang, X.; Hu, M.; Xia, Y.; Wen, X.; Ding, K. Pyrosequencing analysis of bacterial diversity in 14 wastewater treatment systems in china. Appl. Environ. Microbiol. 2012, 78, 7042–7047. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Shao, M.F.; Ye, L. 454 Pyrosequencing reveals bacterial diversity of activated sludge from 14 sewage treatment plants. ISME J. 2012, 6, 1137–1147. [Google Scholar] [CrossRef]
- Ju, F.; Guo, F.; Ye, L.; Xia, Y.; Zhang, T. Metagenomic analysis on seasonal microbial variations of activated sludge from a full-scale wastewater treatment plant over 4 years. Environ. Microbiol. Rep. 2014, 6, 80–89. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Liu, S.; Zheng, M.; Chen, Q.; Ni, J. Performance Assessment of Full-Scale Wastewater Treatment Plants Based on Seasonal Variability of Microbial Communities via High-Throughput Sequencing. PLoS ONE 2016, 11, e0152998. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Yu, Q.; Yan, G.; Zhu, H.; Xu, X.Y.; Zhu, L. Seasonal bacterial community succession in four typical wastewater treatment plants: Correlations between core microbes and process performance. Sci. Rep. 2018, 8, 4566. [Google Scholar] [CrossRef] [PubMed]
- Ibarbalz, F.M.; Orellana, E.; Figuerola, E.L.M.; Erijman, L. Shotgun metagenomic profiles have a high capacity to discriminate samples of activated sludge according to wastewater type. Appl. Environ. Microbiol. 2016, 82, 5186–5196. [Google Scholar] [CrossRef]
- Ye, L.; Zhang, T. Bacterial communities in different sections of a municipal wastewater treatment plant revealed by 16S rDNA 454 pyrosequencing. Appl. Microbiol. Biotechnol. 2013, 97, 2681–2690. [Google Scholar] [CrossRef]
- Zhang, T.; Ye, L.; Tong, A.H.Y.; Shao, M.F.; Lok, S. Ammonia-oxidizing archaea and ammonia-oxidizing bacteria in six full-scale wastewater treatment bioreactors. Appl. Microbiol. Biotechnol. 2011, 91, 1215–1225. [Google Scholar] [CrossRef]
- Terada, A.; Zhou, S.; Hosomi, M. Presence and detection of anaerobic ammonium-oxidizing (anammox) bacteria and appraisal of anammox process for high-strength nitrogenous wastewater treatment: A review. Clean Technol. Environ. Policy 2011, 13, 759–781. [Google Scholar] [CrossRef]
- Annavajhala, M.K.; Kapoor, V.; Santo-Domingo, J.; Chandran, K. Comammox Functionality Identified in Diverse Engineered Biological Wastewater Treatment Systems. Environ. Sci. Technol. Lett. 2018, 5, 110–116. [Google Scholar] [CrossRef]
- Zhang, W.; Yan, C.; Shen, J.; Wei, R.; Gao, Y.; Miao, A.; Xiao, L.; Yang, L. Characterization of Aerobic Denitrifying Bacterium Pseudomonas mendocina Strain GL6 and Its Potential Application in Wastewater Treatment Plant Effluent. Int. J. Environ. Res. Public Health 2019, 16, 364. [Google Scholar] [CrossRef]
- Che, Y.; Liang, P.; Gong, T.; Cao, X.; Zhao, Y.; Yang, C.; Song, C. Elucidation of major contributors involved in nitrogen removal and transcription level of nitrogen-cycling genes in activated sludge from WWTPs. Sci. Rep. 2017, 7, 44728. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhang, X.X.; Lu, X.; Liu, B.; Li, Y.; Long, C.; Li, A. Abundance and diversity of bacterial nitrifiers and denitrifiers and their functional genes in tannery wastewater treatment plants revealed by high-throughput sequencing. PLoS ONE 2014, 9, e113603. [Google Scholar] [CrossRef] [PubMed]
- Shu, D.; He, Y.; Yue, H.; Wang, Q. Metagenomic and quantitative insights into microbial communities and functional genes of nitrogen and iron cycling in twelve wastewater treatment systems. Chem. Eng. J. 2016, 290, 21–30. [Google Scholar] [CrossRef]
- Yang, B.; Wang, J.; Wang, J.; Xu, H.; Song, X.; Wang, Y.; Li, F.; Liu, Y.; Bai, J. Correlating microbial community structure with operational conditions in biological aerated filter reactor for efficient nitrogen removal of municipal wastewater. Bioresour. Technol. 2018, 250, 374–381. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.Y.; Gao, J.F.; Pan, K.L.; Li, D.C.; Dai, H.H. Temporal dynamics of bacterial communities and predicted nitrogen metabolism genes in a full-scale wastewater treatment plant. RSC Adv. 2017, 7, 56317–56327. [Google Scholar] [CrossRef]
- Johnston, J.; LaPara, T.; Behrens, S. Composition and Dynamics of the Activated Sludge Microbiome during Seasonal Nitrification Failure. Sci. Rep. 2019, 9, 1–15. [Google Scholar] [CrossRef]
- Keegan, M.; Kilroy, K.; Nolan, D.; Dubber, D.; Johnston, P.M.; Misstear, B.D.R.; O’Flaherty, V.; Barrett, M.; Gill, L.W. Assessment of the impact of traditional septic tank soakaway systems on water quality in Ireland. Water Sci. Technol. 2014, 70, 634–641. [Google Scholar] [CrossRef]
- Pussayanavin, T.; Koottatep, T.; Eamrat, R.; Polprasert, C. Enhanced sludge reduction in septic tanks by increasing temperature. J. Environ. Sci. Health Part A Toxic/Hazard. Subst. Environ. Eng. 2015, 50, 81–89. [Google Scholar] [CrossRef]
- Nie, J.Y.; Zhu, N.W.; Zhao, K.; Wu, L.; Hu, Y.H. Analysis of the bacterial community changes in soil for septic tank effluent treatment in response to bio-clogging. Water Sci. Technol. 2011, 63, 1412–1417. [Google Scholar] [CrossRef]
- Robertson, W.D.; Moore, T.A.; Spoelstra, J.; Li, L.; Elgood, R.J.; Clark, I.D.; Schiff, S.L.; Aravena, R.; Neufeld, J.D. Natural Attenuation of Septic System Nitrogen by Anammox. Ground Water 2012, 50, 541–553. [Google Scholar] [CrossRef] [PubMed]
- Tomaras, J.; Sahl, J.W.; Siegrist, R.L.; Spear, J.R. Microbial diversity of septic tank effluent and a soil biomat. Appl. Environ. Microbiol. 2009, 75, 3348–3351. [Google Scholar] [CrossRef] [PubMed]
- Koottatep, T.; Suksiri, P.; Pussayanavin, T.; Polprasert, C. Development of a Novel Multi-soil Layer Constructed Wetland Treating Septic Tank Effluent with Emphasis on Organic and Ammonia Removals. Water Air Soil Pollut. 2018, 229, 258. [Google Scholar] [CrossRef]
- Delgado-Baquerizo, M.; Maestre, F.T.; Reich, P.B.; Jeffries, T.C.; Gaitan, J.J.; Encinar, D.; Berdugo, M.; Campbell, C.D.; Singh, B.K. Microbial diversity drives multifunctionality in terrestrial ecosystems. Nat. Commun. 2016, 7, 10541. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Baca, C.P.; Truhlar, A.M.; Omar, A.E.H.; Rahm, B.G.; Walter, M.T.; Richardson, R.E. Methane and nitrous oxide cycling microbial communities in soils above septic leach fields: Abundances with depth and correlations with net surface emissions. Sci. Total Environ. 2018, 640–641, 429–441. [Google Scholar] [CrossRef] [PubMed]
- Koottatep, T.; Prapasriket, P.; Pussayanavin, T.; Polprasert, C. Performance of up-flow thermophilic septic tank treating blackwater. Environ. Dev. Sustain. 2019, 22, 3691–3700. [Google Scholar] [CrossRef]
- Jin, Z.; Lv, C.; Zhao, M.; Zhang, Y.; Huang, X.; Bei, K.; Kong, H.; Zheng, X. Black water collected from the septic tank treated with a living machine system: HRT effect and microbial community structure. Chemosphere 2018, 210, 745–752. [Google Scholar] [CrossRef]
- Wigginton, S.; Brannon, E.; Kearns, P.J.; Lancellotti, B.; Cox, A.; Loomis, G.W.; Amador, J.A. Nitrifying and denitrifying bacterial communities in advanced nitrogen-removal onsite wastewater treatment systems. J. Environ. Qual. 2018, 47, 1163–1171. [Google Scholar] [CrossRef]
- Lancellotti, B.V.; Loomis, G.W.; Hoyt, K.P.; Avizinis, E.; Amador, J.A. Evaluation of Nitrogen Concentration in Final Effluent of Advanced Nitrogen-Removal Onsite Wastewater Treatment Systems (OWTS). Water Air Soil Pollut. 2017, 228, 383. [Google Scholar] [CrossRef]
- Geets, J.; de Cooman, M.; Wittebolle, L.; Heylen, K.; Vanparys, B.; De Vos, P.; Verstraete, W.; Boon, N. Real-time PCR assay for the simultaneous quantification of nitrifying and denitrifying bacteria in activated sludge. Appl. Microbiol. Biotechnol. 2007, 75, 211–221. [Google Scholar] [CrossRef]
- Junier, P.; Kim, O.-S.; Junier, T.; Ahn, T.-S.; Imhoff, J.F.; Witzel, K.-P. Community analysis of betaproteobacterial ammonia-oxidizing bacteria using the amoCAB operon. Appl. Microbiol. Biotechnol. 2009, 83, 175–188. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Bokulich, N.A.; Subramanian, S.; Faith, J.J.; Gevers, D.; Gordon, J.I.; Knight, R.; Mills, D.A.; Caporaso, J.G. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods 2013, 10, 57–59. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef]
- Mahé, F.; Rognes, T.; Quince, C.; de Vargas, C.; Dunthorn, M. Swarm: Robust and fast clustering method for amplicon-based studies. PeerJ 2014, 2, e593. [Google Scholar] [CrossRef]
- Kearns, P.J.; Angell, J.H.; Feinman, S.G.; Bowen, J.L. Long-term nutrient addition differentially alters community composition and diversity of genes that control nitrous oxide flux from salt marsh sediments. Estuar. Coast. Shelf Sci. 2015, 154, 39–47. [Google Scholar] [CrossRef]
- Pester, M.; Rattei, T.; Flechl, S.; Gröngröft, A.; Richter, A.; Overmann, J.; Reinhold-Hurek, B.; Loy, A.; Wagner, M. amoA-based consensus phylogeny of ammonia-oxidizing archaea and deep sequencing of amoA genes from soils of four different geographic regions. Environ. Microbiol. 2012, 14, 525–539. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Cole, J.R.; Wang, Q.; Fish, J.A.; Chai, B.; McGarrell, D.M.; Sun, Y.; Brown, C.T.; Porras-Alfaro, A.; Kuske, C.R.; Tiedje, J.M. Ribosomal Database Project: Data and tools for high throughput rRNA analysis. Nucleic Acids Res. 2014, 42, D633–D642. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; Stevens, M.H.H.; Wagner, H.; et al. Package ‘vegan’. Community Ecol. Package Vers. 2013, 2, 1–295. [Google Scholar]
- R Core Team. R: A language and environment for statistical computing. Vienna R Found. Stat. Comput. 2012, 1, 12–21. [Google Scholar]
- McMurdie, P.J.; Holmes, S. phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Roy, D.; McEvoy, J.; Blonigen, M.; Amundson, M.; Khan, E. Seasonal variation and ex-situ nitrification activity of ammonia oxidizing archaea in biofilm based wastewater treatment processes. Bioresour. Technol. 2017, 244, 850–859. [Google Scholar] [CrossRef] [PubMed]
- Albright, M.B.N.; Timalsina, B.; Martiny, J.B.H.; Dunbar, J. Comparative Genomics of Nitrogen Cycling Pathways in Bacteria and Archaea. Microb. Ecol. 2018, 77, 597–606. [Google Scholar] [CrossRef] [PubMed]
- Kindaichi, T.; Tsushima, I.; Ogasawara, Y.; Shimokawa, M.; Ozaki, N.; Satoh, H.; Okabe, S. In situ activity and spatial organization of anaerobic ammonium-oxidizing (anammox) bacteria in biofilms. Appl. Environ. Microbiol. 2007, 73, 4931–4939. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, G.A.; Nicholas, D.J. Identification of the sources of nitrous oxide produced by oxidative and reductive processes in Nitrosomonas europaea. Biochem. J. 1972, 126, 1181–1191. [Google Scholar] [CrossRef] [PubMed]
- Valentín-Vargas, A.; Toro-Labrador, G.; Massol-Deyá, A.A. Bacterial Community Dynamics in Full-Scale Activated Sludge Bioreactors: Operational and Ecological Factors Driving Community Assembly and Performance. PLoS ONE 2012, 7, e42524. [Google Scholar] [CrossRef] [PubMed]
- Van Der Gast, C.J.; Jefferson, B.; Reid, E.; Robinson, T.; Bailey, M.J.; Judd, S.J.; Thompson, I.P. Bacterial diversity is determined by volume in membrane bioreactors. Environ. Microbiol. 2006, 8, 1048–1055. [Google Scholar] [CrossRef]
- Horner-Devine, M.C.; Lage, M.; Hughes, J.B.; Bohannan, B.J.M. A taxa–area relationship for bacteria. Nature 2004, 432, 750–753. [Google Scholar] [CrossRef]
- Miyahara, M.; Kim, S.W.; Fushinobu, S.; Takaki, K.; Yamada, T.; Watanabe, A.; Miyauchi, K.; Endo, G.; Wakagi, T.; Shoun, H. Potential of aerobic denitrification by pseudomonas stutzeri TR2 to reduce nitrous oxide emissions from wastewater treatment plants. Appl. Environ. Microbiol. 2010, 76, 4619–4625. [Google Scholar] [CrossRef]
- Su, J.J.; Liu, B.Y.; Liu, C.Y. Comparison of aerobic denitrification under high oxygen atmosphere by Thiosphaera pantotropha ATCC 35512 and Pseudomonas stutzeri SU2 newly isolated from the activated sludge of a piggery wastewater treatment system. J. Appl. Microbiol. 2001, 90, 457–462. [Google Scholar] [CrossRef]
- Zheng, M.; He, D.; Ma, T.; Chen, Q.; Liu, S.; Ahmad, M.; Gui, M.; Ni, J. Reducing NO and N2O emission during aerobic denitrification by newly isolated Pseudomonas stutzeri PCN-1. Bioresour. Technol. 2014, 162, 80–88. [Google Scholar] [CrossRef] [PubMed]
- Ji, B.; Yang, K.; Zhu, L.; Jiang, Y.; Wang, H.; Zhou, J.; Zhang, H. Aerobic denitrification: A review of important advances of the last 30 years. Biotechnol. Bioprocess. Eng. 2015, 20, 643–651. [Google Scholar] [CrossRef]
- Chen, J.; Strous, M. Denitrification and aerobic respiration, hybrid electron transport chains and co-evolution. Biochim. Biophys. Acta Bioenerg. 2013, 1827, 136–144. [Google Scholar] [CrossRef]
- Chen, Y.; Lan, S.; Wang, L.; Dong, S.; Zhou, H.; Tan, Z.; Li, X. A review: Driving factors and regulation strategies of microbial community structure and dynamics in wastewater treatment systems. Chemosphere 2017, 174, 173–182. [Google Scholar] [CrossRef]
- Pruitt, K.D.; Tatusova, T.; Maglott, D.R. NCBI reference sequences (RefSeq): A curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007, 35, 61–65. [Google Scholar] [CrossRef]
- Gao, J.; Luo, X.; Wu, G.; Li, T.; Peng, Y. Abundance and diversity based on amoA genes of ammonia-oxidizing archaea and bacteria in ten wastewater treatment systems. Appl. Microbiol. Biotechnol. 2014, 98, 3339–3354. [Google Scholar] [CrossRef]
- Aigle, A.; Prosser, J.I.; Gubry-Rangin, C. The application of high-throughput sequencing technology to analysis of amoA phylogeny and environmental niche specialisation of terrestrial bacterial ammonia-oxidisers. Environ. Microbiome 2019, 14, 3. [Google Scholar] [CrossRef]
- Land, M.; Hauser, L.; Jun, S.R.; Nookaew, I.; Leuze, M.R.; Ahn, T.H.; Karpinets, T.; Lund, O.; Kora, G.; Wassenaar, T.; et al. Insights from 20 years of bacterial genome sequencing. Funct. Integr. Genom. 2015, 15, 141–161. [Google Scholar] [CrossRef]
- Morris, A.; Meyer, K.; Bohannan, B. Linking microbial communities to ecosystem functions: What we can learn from genotype-phenotype mapping in organisms. Philos. Trans. R. Soc. B Biol. Sci. 2020, 375, 20190244. [Google Scholar] [CrossRef]
- Maron, P.A.; Sarr, A.; Kaisermann, A.; Lévêque, J.; Mathieu, O.; Guigue, J.; Karimi, B.; Bernard, L.; Dequiedt, S.; Terrat, S.; et al. High microbial diversity promotes soil ecosystem functioning. Appl. Environ. Microbiol. 2018, 84, e02738-17. [Google Scholar] [CrossRef]
- Peter, H.; Beier, S.; Bertilsson, S.; Lindström, E.S.; Langenheder, S.; Tranvik, L.J. Function-specific response to depletion of microbial diversity. ISME J. 2011, 5, 351–361. [Google Scholar] [CrossRef] [PubMed]
- Trivedi, C.; Delgado-Baquerizo, M.; Hamonts, K.; Lai, K.; Reich, P.B.; Singh, B.K. Losses in microbial functional diversity reduce the rate of key soil processes. Soil Biol. Biochem. 2019, 135, 267–274. [Google Scholar] [CrossRef]
- Tomás, J.M. The Main Aeromonas Pathogenic Factors. ISRN Microbiol. 2012, 2012, 256261. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Zhang, Z.; Zhang, L.; Li, X. Complete genome sequence of a denitrifying bacterium, Pseudomonas sp. CC6-YY-74, isolated from Arctic Ocean sediment. Mar. Genom. 2017, 35, 47–49. [Google Scholar] [CrossRef]
- Singleton, P.; Sainsbury, D. Dictionary of Microbiology and Molecular Biology, 3rd ed.; John Wiley & Sons, Ltd.: Chichester, UK, 2006; ISBN 9780470056981. [Google Scholar]
- Hiraishi, A.; Hoshino, Y.; Satoh, T. Rhodoferax fermentans gen. nov., sp. nov., a phototrophic purple nonsulfur bacterium previously referred to as the “Rhodocyclus gelatinosus-like” group. Arch. Microbiol. 1991, 155, 330–336. [Google Scholar] [CrossRef]
- Finneran, K.T.; Johnsen, C.V.; Lovley, D.R. Rhodoferax ferrireducens sp. nov., a psychrotolerant, facultatively anaerobic bacterium that oxidizes acetate with the reduction of Fe (III). Int. J. Syst. Evol. Microbiol. 2003, 53, 669–673. [Google Scholar] [CrossRef]
- Mukherjee, S.; Parekh, V. Review of Purification of Industrial Wastewater. Int. J. Eng. Res. 2016, 5, 379–383. [Google Scholar] [CrossRef]
- Bai, Y.; Sun, Q.; Zhao, C.; Wen, D.; Tang, X. Aerobic degradation of pyridine by a new bacterial strain, Shinella zoogloeoides BC026. J. Ind. Microbiol. Biotechnol. 2009, 36, 1391–1400. [Google Scholar] [CrossRef]
| Gene | Genus, Species and Strain of Closest Cultured Specimen | Id. Match (%) | System Type | |||||
|---|---|---|---|---|---|---|---|---|
| Combined | OWTS | WTP | ||||||
| # Samples with Gene | Relative Abundance (%) Mean (SD) | # Samples with Gene | Relative Abundance (%) Mean (SD) | # Samples with Gene | Relative Abundance (%) Mean (SD) | |||
| amoA | Nitrosomonas oligotropha Nm75 | 97 | 45 | 30.2 (24.4) | 25 | 41.1 (27.3) | 20 | 16.5 (8.7) |
| Nitrosomonas sp. Is79A3 | 94 | 43 | 7.8 (5.7) | 23 | 8.3 (6.9) | 20 | 7.2 (4.2) | |
| Nitrosomonas sp. JL21 | 86 | 26 | 0.4 (0.5) | 20 | 0.5 (0.5) | 6 | 0.1 (0.1) | |
| Nitrosomonas sp. JL21 | 97 | 24 | 0.9 (1.0) | 15 | 1.1 (1.0) | 9 | 0.5 (0.6) | |
| Nitrosomonas sp. Nm59 | 85 | 36 | 0.5 (0.9) | 16 | 0.8 (1.1) | 20 | 0.3 (0.3) | |
| Nitrosomonas sp. Nm59 | 92 | 29 | 1.7 (1.2) | 9 | 0.7 (0.6) | 20 | 2.1 (1.2) | |
| Nitrosomonas sp. Nm59 | 88 | 29 | 0.9 (0.7) | 9 | 0.3 (0.3) | 20 | 1.1 (0.6) | |
| Nitrosomonas sp. PY1 | 90 | 27 | 0.4 (0.5) | 11 | 0.6 (0.5) | 16 | 0.2 (0.2) | |
| Nitrosomonas sp. Nm84 | 88 | 28 | 1.0 (0.8) | 8 | 0.8 (0.6) | 20 | 1.1 (0.8) | |
| Nitrosospira sp. L115 | 91 | 31 | 0.7 (0.9) | 20 | 1.0 (1.0) | 11 | 0.1 (0.1) | |
| Nitrosospira sp. Wyke2 | 96 | 39 | 13.3 (18.4) | 24 | 20.9 (20.4) | 15 | 1.2 (1.2) | |
| Nitrosovibrio sp. RY3C | 99 | 26 | 0.5 (0.8) | 19 | 0.6 (0.9) | 7 | 0.1 (0.1) | |
| Unclassified sp. (denovo 1096) | - | 44 | 38.0 (30.2) | 24 | 15.7 (20.2) | 20 | 64.7 (14.6) | |
| nosZ | Aeromonas media WS | 99 | 40 | 12.7 (14.1) | 22 | 12.7 (18.2) | 18 | 12.7 (6.0) |
| Oligotropha carboxidovorans strain C1S131/132.2 | 99 | 29 | 1.7 (1.3) | 12 | 4.1.5 (1.5) | 17 | 1.8 (1.1) | |
| Pseudomonas sp. CC6-YY-74 | 99 | 47 | 23.7 (30.7) | 30 | 36.0 (32.5) | 17 | 2.0 (21.6) | |
| Thauera phenylacetica strain TN9 | 90 | 29 | 5.5 | 16 | 9.4 (17.6) | 13 | 0.8 (0.5) | |
| Thauera phenylacetica strain TN9 | 92 | 37 | 5.0 (9.2) | 19 | 4.6 (12.5) | 18 | 5.4 (2.7) | |
| Unclassified Alphaproteobacteria | 89 | 29 | 3.2 (3.0) | 14 | 4.2 (3.8) | 15 | 2.2 (1.2) | |
| Unclassified Alphaproteobacteria | 85 | 27 | 1.0 (0.7) | 11 | 1.2 (0.8) | 16 | 0.9 (0.5) | |
| Unclassified Alphaproteobacteria | 79 | 26 | 2.5 (1.5) | 8 | 2.3 (2.1) | 18 | 2.7 (1.1) | |
| Unclassified Betaproteobacteria | 81 | 26 | 2.5 (3.3) | 9 | 4.4 (5.0) | 17 | 1.5 (1.0) | |
| Unclassified Betaproteobacteria | 83 | 26 | 2.4 (1.7) | 9 | 1.0 (1.2) | 17 | 3.2 (1.5) | |
| Unclassified Alphaproteobacteria | 78 | 25 | 8.2 (5.6) | 7 | 5.8 (8.8) | 18 | 9.1 (3.1) | |
| Unclassified Betaproteobacteria | 85 | 25 | 3.5 (4.4) | 9 | 0.6 (0.4) | 16 | 5.2 (4.7) | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wigginton, S.K.; Brannon, E.Q.; Kearns, P.J.; Lancellotti, B.V.; Cox, A.; Moseman-Valtierra, S.; Loomis, G.W.; Amador, J.A. Nitrifying and Denitrifying Microbial Communities in Centralized and Decentralized Biological Nitrogen Removing Wastewater Treatment Systems. Water 2020, 12, 1688. https://doi.org/10.3390/w12061688
Wigginton SK, Brannon EQ, Kearns PJ, Lancellotti BV, Cox A, Moseman-Valtierra S, Loomis GW, Amador JA. Nitrifying and Denitrifying Microbial Communities in Centralized and Decentralized Biological Nitrogen Removing Wastewater Treatment Systems. Water. 2020; 12(6):1688. https://doi.org/10.3390/w12061688
Chicago/Turabian StyleWigginton, Sara K., Elizabeth Q. Brannon, Patrick J. Kearns, Brittany V. Lancellotti, Alissa Cox, Serena Moseman-Valtierra, George W. Loomis, and Jose A. Amador. 2020. "Nitrifying and Denitrifying Microbial Communities in Centralized and Decentralized Biological Nitrogen Removing Wastewater Treatment Systems" Water 12, no. 6: 1688. https://doi.org/10.3390/w12061688
APA StyleWigginton, S. K., Brannon, E. Q., Kearns, P. J., Lancellotti, B. V., Cox, A., Moseman-Valtierra, S., Loomis, G. W., & Amador, J. A. (2020). Nitrifying and Denitrifying Microbial Communities in Centralized and Decentralized Biological Nitrogen Removing Wastewater Treatment Systems. Water, 12(6), 1688. https://doi.org/10.3390/w12061688






