Removal of Antibiotic Resistance Genes at Two Conventional Wastewater Treatment Plants of Louisiana, USA
Abstract
:1. Introduction
2. Materials and Methods
2.1. Collection of Wastewater Samples
2.2. Water Quality Parameters
2.3. Bacterial DNA Extraction
2.4. Quantitative Polymerase Chain Reaction (qPCR)
2.5. Data Analysis
3. Results and Discussion
3.1. Occurrence of Total Bacterial 16S rRNA, IntI1, and ARGs in Wastewater Samples
3.2. Reduction of ARGs and IntI1 at WWTPs
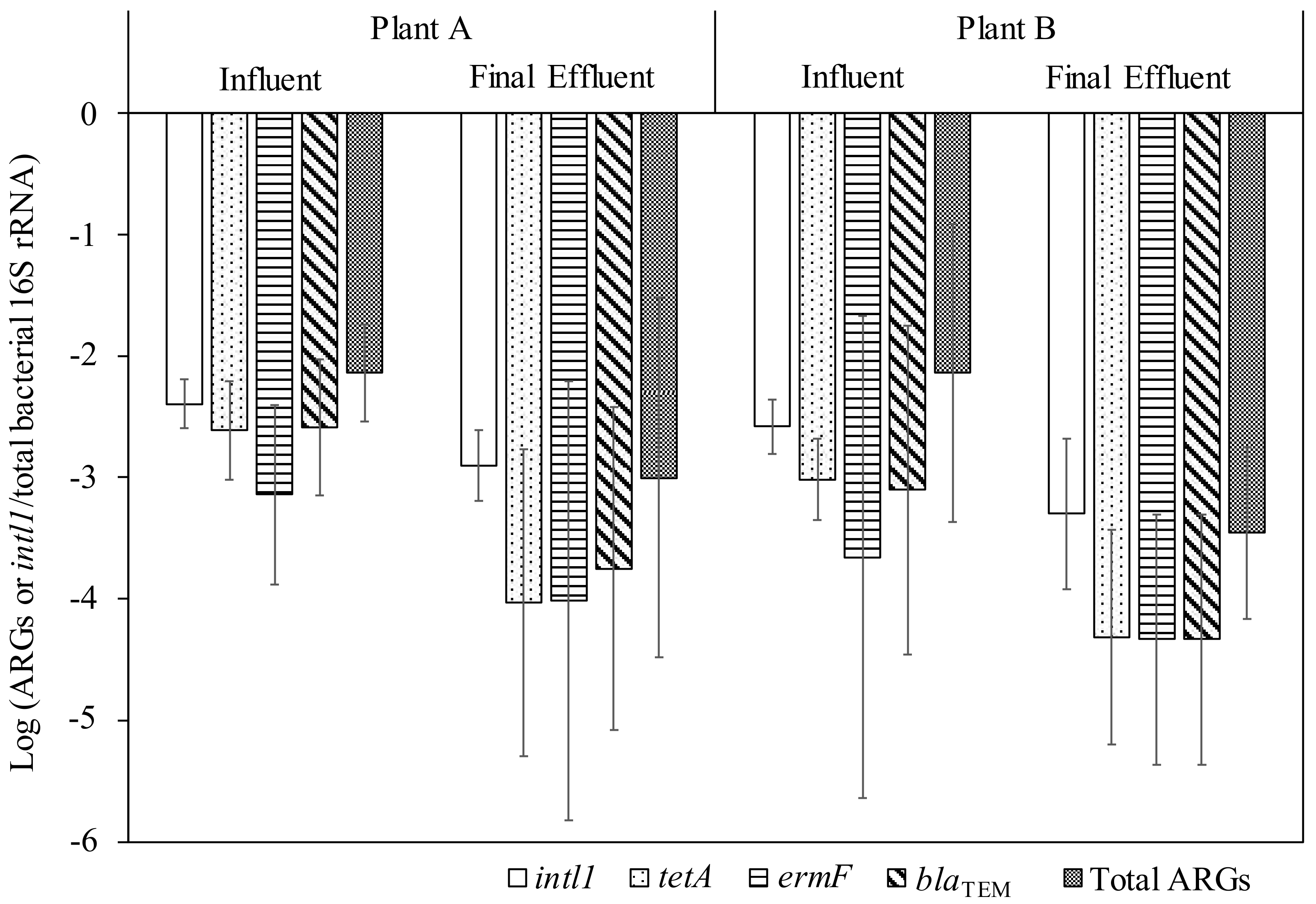
3.3. Relative Abundance of ARGs in Influent and Final Effluent Samples of WWTPs
3.4. Correlation of Individual and Total ARGs with IntI1
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Van Goethem, M.W.; Pierneef, R.; Bezuidt, O.K.I.; Van De Peer, Y.; Cowan, D.A.; Makhalanyane, T.P. A reservoir of “historical” antibiotic resistance genes in remote pristine Antarctic soils. Microbiome 2018, 6, 40. [Google Scholar] [CrossRef] [Green Version]
- Karkman, A.; Pärnänen, K.; Larsson, D.G.J. Fecal pollution can explain antibiotic resistance gene abundances in anthropogenically impacted environments. Nat. Commun. 2019, 10, 80. [Google Scholar] [CrossRef]
- Thakali, O.; Tandukar, S.; Brooks, J.P.; Sherchan, S.P.; Sherchand, J.B.; Haramoto, E. The Occurrence of Antibiotic Resistance Genes in an Urban River in Nepal. Water 2020, 12, 450. [Google Scholar] [CrossRef] [Green Version]
- Levy, S.; Marshall, B. Antibacterial resistance worldwide: Causes, challenges and responses. Nat. Med. 2004, 10, S122–S129. [Google Scholar] [CrossRef]
- Rizzo, L.; Manaia, C.; Merlin, C.; Schwartz, T.; Dagot, C.; Ploy, M.C.; Michael, I.; Fatta-Kassinos, D. Urban wastewater treatment plants as hotspots for antibiotic resistant bacteria and genes spread into the environment: A review. Sci. Total Environ. 2013, 447, 345–360. [Google Scholar] [CrossRef]
- McKinney, C.W.; Pruden, A. Ultraviolet disinfection of antibiotic resistant bacteria and their antibiotic resistance genes in water and wastewater. Environ. Sci. Technol. 2012, 46, 13393–13400. [Google Scholar] [CrossRef]
- Gao, P.; Munir, M.; Xagoraraki, I. Correlation of tetracycline and sulfonamide antibiotics with corresponding resistance genes and resistant bacteria in a conventional municipal wastewater treatment plant. Sci. Total Environ. 2012, 421–422, 173–183. [Google Scholar] [CrossRef]
- Munir, M.; Wong, K.; Xagoraraki, I. Release of antibiotic resistant bacteria and genes in the effluent and biosolids of five wastewater utilities in Michigan. Water Res. 2011, 45, 681–693. [Google Scholar] [CrossRef]
- Lamori, J.G.; Xue, J.; Rachmadi, A.T.; Lopez, G.U.; Kitajima, M.; Gerba, C.P.; Pepper, I.L.; Brooks, J.P.; Sherchan, S. Removal of fecal indicator bacteria and antibiotic resistant genes in constructed wetlands. Environ. Sci. Pollut. Res. 2019, 26, 10188–10197. [Google Scholar] [CrossRef]
- Rafraf, I.D.; Lekunberri, I.; Sànchez-Melsió, A.; Aouni, M.; Borrego, C.M.; Balcázar, J.L. Abundance of antibiotic resistance genes in five municipal wastewater treatment plants in the Monastir Governorate, Tunisia. Environ. Pollut. 2016, 219, 353–358. [Google Scholar] [CrossRef]
- Amarasiri, M.; Sano, D.; Suzuki, S. Understanding human health risks caused by antibiotic resistant bacteria (ARB) and antibiotic resistance genes (ARG) in water environments: Current knowledge and questions to be answered. Crit. Rev. Environ. Sci. Technol. 2019. [Google Scholar] [CrossRef]
- Christgen, B.; Yang, Y.; Ahammad, S.Z.; Li, B.; Rodriquez, D.C.; Zhang, T.; Graham, D.W. Metagenomics shows that low-energy anaerobic-aerobic treatment reactors reduce antibiotic resistance gene levels from domestic wastewater. Environ. Sci. Technol. 2015, 49, 2577–2584. [Google Scholar] [CrossRef]
- Mao, D.; Yu, S.; Rysz, M.; Luo, Y.; Yang, F.; Li, F.; Hou, J.; Mu, Q.; Alvarez, P.J.J. Prevalence and proliferation of antibiotic resistance genes in two municipal wastewater treatment plants. Water Res. 2015, 85, 458–466. [Google Scholar] [CrossRef]
- Hong, P.Y.; Julian, T.R.; Pype, M.L.; Jiang, S.C.; Nelson, K.L.; Graham, D.; Pruden, A.; Manaia, C.M. Reusing treated wastewater: Consideration of the safety aspects associated with antibiotic-resistant bacteria and antibiotic resistance genes. Water 2018, 10, 244. [Google Scholar] [CrossRef] [Green Version]
- Hong, P.Y.; Al-Jassim, N.; Ansari, M.I.; Mackie, R.I. Environmental and public health implications of water reuse: Antibiotics, antibiotic resistant bacteria, and antibiotic resistance genes. Antibiotics 2013, 2, 367–399. [Google Scholar] [CrossRef] [Green Version]
- Volkmann, H.; Schwartz, T.; Bischoff, P.; Kirchen, S.; Obst, U. Detection of clinically relevant antibiotic-resistance genes in municipal wastewater using real-time PCR (TaqMan). J. Microbiol. Methods 2004, 56, 277–286. [Google Scholar] [CrossRef]
- Szczepanowski, R.; Linke, B.; Krahn, I.; Gartemann, K.H.; Gützkow, T.; Eichler, W.; Pühler, A.; Schlüter, A. Detection of 140 clinically relevant antibiotic-resistance genes in the plasmid metagenome of wastewater treatment plant bacteria showing reduced susceptibility to selected antibiotics. Microbiology 2009, 155, 2306–2319. [Google Scholar] [CrossRef] [Green Version]
- Barraud, O.; Baclet, M.C.; Denis, F.; Ploy, M.C. Quantitative multiplex real-time PCR for detecting class 1, 2 and 3 integrons. J. Antimicrob. Chemother. 2010, 65, 1642–1645. [Google Scholar] [CrossRef] [Green Version]
- Narciso-Da-Rocha, C.; Varela, A.R.; Schwartz, T.; Nunes, O.C.; Manaia, C.M. blaTEM and vanA as indicator genes of antibiotic resistance contamination in a hospital-urban wastewater treatment plant system. J. Glob. Antimicrob. Resist. 2014, 2, 309–315. [Google Scholar] [CrossRef] [Green Version]
- Tandukar, S.; Sherchan, S.P.; Haramoto, E. Applicability of crAssphage, pepper mild mottle virus, and tobacco mosaic virus as indicators of reduction of enteric viruses during wastewater treatment. Sci. Rep. 2020, 10, 3616. [Google Scholar] [CrossRef] [Green Version]
- APHA/AWWA/WEF. Standard Methods for the Examination of Water and Wastewater, 20th ed.; Clesceri, L.S., Greenberg, A.E., Eaton, A.D., Eds.; American Public Health Association, American Water Works Association, Water Environment Federation: Washington, DC, USA, 1998. [Google Scholar]
- Nadkarni, M.A.; Martin, F.E.; Jacques, N.A.; Hunter, N. Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology 2002, 148, 257–266. [Google Scholar] [CrossRef] [Green Version]
- Fan, W.; Hamilton, T.; Webster-Sesay, S.; Nikolich, M.P.; Lindler, L.E. Multiplex real-time SYBR Green I PCR assay for detection of tetracycline efflux genes of gram-negative bacteria. Mol. Cell Probes 2007, 21, 245–256. [Google Scholar] [CrossRef]
- Lachmayr, K.L.; Cavanaugh, C.M.; Kerkhof, L.J.; DiRienzo, A.G.; Ford, T.E. Quantifying nonspecific tem β-lactamase (blatem) genes in a wastewater stream. Appl. Environ. Microbiol. 2009, 75, 203–211. [Google Scholar] [CrossRef] [Green Version]
- Sabet, N.S.; Subramaniam, G.; Navaratnam, P.; Sekaran, S.D. Detection of methicillin-and aminoglycoside-resistant genes and simultaneous identification of S. aureus using triplex real-time PCR Taqman assay. J. Microbiol. Methods 2007, 68, 157–162. [Google Scholar] [CrossRef]
- Chen, J.; Yu, Z.; Michel, F.C.; Wittum, T.; Morrison, M. Development and application of real-time PCR assays for quantification of erm genes conferring resistance to macrolides-lincosamides-streptogramin B in livestock manure and manure management systems. Appl. Environ. Microbiol. 2007, 73, 4407–4416. [Google Scholar] [CrossRef] [Green Version]
- McConnell, M.M.; Truelstrup Hansen, L.; Jamieson, R.C.; Neudorf, K.D.; Yost, C.K.; Tong, A. Removal of antibiotic resistance genes in two tertiary level municipal wastewater treatment plants. Sci. Total Environ. 2018, 643, 292–300. [Google Scholar] [CrossRef]
- Schmitz, B.W.; Innes, G.K.; Xue, J.; Gerba, C.P.; Pepper, I.L.; Sherchan, S. Reduction of erythromycin resistance gene erm (F) and class 1 integron-integrase genes in wastewater by Bardenpho treatment. Water Environ. Res. 2020, 1–9. [Google Scholar] [CrossRef]
- Wang, J.; Mao, D.; Mu, Q.; Luo, Y. Fate and proliferation of typical antibiotic resistance genes in five full-scale pharmaceutical wastewater treatment plants. Sci. Total Environ. 2015, 526, 366–373. [Google Scholar] [CrossRef]
- Davies, J.; Davies, D. Origins and Evolution of Antibiotic Resistance. Microbiol. Mol. Biol. Rev. 2010, 74, 417–433. [Google Scholar] [CrossRef] [Green Version]
- Wan, M.T.; Chou, C.C. Spreading of β-lactam resistance gene (mecA) and methicillin-resistant Staphylococcus aureus through municipal and swine slaughterhouse wastewaters. Water Res. 2014, 64, 288–295. [Google Scholar] [CrossRef]
- Börjesson, S.; Melin, S.; Matussek, A.; Lindgren, P.E. A seasonal study of the mecA gene and Staphylococcus aureus including methicillin-resistant S. aureus in a municipal wastewater treatment plant. Water Res. 2009, 43, 925–932. [Google Scholar] [CrossRef] [Green Version]
- Da Silva, M.F.; Tiago, I.; Veríssimo, A.; Boaventura, R.A.; Nunes, O.C.; Manaia, C.M. Antibiotic resistance of enterococci and related bacteria in an urban wastewater treatment plant. FEMS Microbiol. Ecol. 2006, 55, 322–329. [Google Scholar] [CrossRef] [Green Version]
- Sørensen, S.J.; Bailey, M.; Hansen, L.H.; Kroer, N.; Wuertz, S. Studying plasmid horizontal transfer in situ: A critical review. Nat. Rev. Microbiol. 2005, 3, 700–710. [Google Scholar] [CrossRef]
- Yang, Y.; Li, B.; Zou, S.; Fang, H.H.P.; Zhang, T. Fate of antibiotic resistance genes in sewage treatment plant revealed by metagenomic approach. Water Res. 2014, 62, 97–106. [Google Scholar] [CrossRef]
- Wen, Q.; Yang, L.; Duan, R.; Chen, Z. Monitoring and evaluation of antibiotic resistance genes in four municipal wastewater treatment plants in Harbin, Northeast China. Environ. Pollut. 2016, 212, 34–40. [Google Scholar] [CrossRef]
- Anastasi, E.M.; Wohlsen, T.D.; Stratton, H.M.; Katouli, M. Survival of Escherichia coli in two sewage treatment plants using UV irradiation and chlorination for disinfection. Water Res. 2013, 47, 6670–6679. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhuang, Y.; Geng, J.; Ren, H.; Zhang, Y.; Ding, L.; Xu, K. Inactivation of antibiotic resistance genes in municipal wastewater effluent by chlorination and sequential UV/chlorination disinfection. Sci. Total Environ. 2015, 512–513, 125–132. [Google Scholar] [CrossRef]
- Bengtsson-Palme, J.; Hammarén, R.; Pal, C.; Östman, M.; Björlenius, B.; Flach, C.F.; Fick, J.; Kristiansson, E.; Tysklind, M.; Larsson, D.G.J. Elucidating selection processes for antibiotic resistance in sewage treatment plants using metagenomics. Sci. Total Environ. 2016, 572, 697–712. [Google Scholar] [CrossRef]
- Di Cesare, A.; Eckert, E.M.; D’Urso, S.; Bertoni, R.; Gillan, D.C.; Wattiez, R.; Corno, G. Co-occurrence of integrase 1, antibiotic and heavy metal resistance genes in municipal wastewater treatment plants. Water Res. 2016, 94, 208–214. [Google Scholar] [CrossRef]
- Lin, W.; Zhang, M.; Zhang, S.; Yu, X. Can chlorination co-select antibiotic-resistance genes? Chemosphere 2016, 156, 412–419. [Google Scholar] [CrossRef]
- Chen, H.; Zhang, M. Effects of advanced treatment systems on the removal of antibiotic resistance genes in wastewater treatment plants from Hangzhou, China. Environ. Sci. Technol. 2013, 47, 8157–8163. [Google Scholar] [CrossRef]
- Giedraitienė, A.; Vitkauskienė, A.; Naginienė, R.; Pavilonis, A. Antibiotic resistance mechanisms of clinically important bacteria. Medicina (Kaunas) 2011, 47, 37–146. [Google Scholar] [CrossRef]

| WWTP | Tested Gene | Influent | Secondary Effluent | Final Effluent | |||
|---|---|---|---|---|---|---|---|
| No. of Positive Samples (%) | Conc. (Mean ± SD) (log10 copies/mL) | No. of Positive Samples (%) | Conc. (mean ± SD) (log10 copies/mL) | No. of Positive Samples (%) | Conc. (Mean ± SD) (log10 copies/mL) | ||
| Plant A | Total bacterial 16S rRNA | 11 (100) | 7.9 ± 0.7 | 11 (100) | 7.8 ± 0.3 | 11 (100) | 7.4 ± 0.4 |
| tetA | 11 (100) | 5.3 ± 1.0 | 11 (100) | 4.4 ± 0.4 | 9 (82) | 3.3 ± 1.5 | |
| ermF | 11 (100) | 4.8 ± 1.1 | 11 (100) | 4.7 ± 1.8 | 7 (64) | 3.3 ± 1.9 | |
| blaTEM | 11 (100) | 5.3 ± 0.8 | 11 (100) | 4.9 ± 1.3 | 8 (73) | 3.6 ± 1.6 | |
| mecA | 0 (0) | NA | 0 (0) | NA | 0 (0) | NA | |
| intI1 | 11 (100) | 5.5 ± 0.8 | 11 (100) | 5.0 ± 0.4 | 11 (100) | 4.5 ± 0.6 | |
| Plant B | Total bacterial 16S rRNA | 12 (100) | 7.6 ± 1.4 | 12 (100) | 7.1 ± 0.7 | 12 (100) | 5.7 ± 1.0 |
| tetA | 11 (92) | 4.6 ± 1.5 | 12 (100) | 3.6 ± 0.7 | 6 (50) | 1.4 ± 1.2 | |
| ermF | 8 (67) | 4.0 ± 2.5 | 7 (58) | 2.6 ± 1.2 | 0 (0) | NA | |
| blaTEM | 9 (75) | 4.5 ± 2.0 | 5 (42) | 3.1 ± 2.0 | 0 (0) | NA | |
| mecA | 0 (0) | NA | 0 (0) | NA | 0 (0) | NA | |
| intI1 | 12 (100) | 5.0 ± 1.3 | 12 (100) | 4.1 ± 0.9 | 7 (58) | 2.4 ± 1.1 | |
| WWTP | Tested Gene | LRV (Mean ± SD) (No. of Months Used for Calculation) | ||
|---|---|---|---|---|
| Physical–Biological Treatment | Chlorination | Whole Process | ||
| Plant A | Total bacterial 16S rRNA | 0.1 ± 0.6 (11) | 0.4 ± 0.5 * (11) | 0.6 ± 0.8 * (11) |
| tetA | 0.9 ± 1.1 * (11) | 1.1 ± 1.6 # (11) | 2.0 ± 2.1 #* (11) | |
| ermF | 0.1 ± 1.9 (11) | 1.3 ± 1.7 #* (11) | 1.4 ± 2.5 #* (11) | |
| blaTEM | 0.4 ± 1.5 (11) | 1.3 ± 1.6 #* (11) | 1.7 ± 1.8 #* (11) | |
| Total ARGs | 0.2 ± 1.6 (11) | 1.2 ± 1.6 #* (11) | 1.4 ± 2.2 #* (11) | |
| intI1 | 0.5 ± 0.8 (11) | 0.5 ± 0.6 * (11) | 1.1 ± 1.1 * (11) | |
| Plant B | Total bacterial 16S rRNA | 0.5 ± 1.6 (12) | 1.4 ± 1.0 * (12) | 1.9 ± 1.6 * (12) |
| tetA | 1.3 ± 1.2 (11) | 2.2 ± 2.1 #* (12) | 3.6 ± 1.2 #* (11) | |
| ermF | 2.3 ± 2.6 # (8) | 2.1 ± 0.7 #* (7) | 3.9 ± 2.1 #* (8) | |
| blaTEM | 2.7 ± 2.3 # (9) | 4.1 ± 0.6 #* (5) | 4.2 ± 0.9 #* (9) | |
| Total ARGs | 1.5 ± 2.5 (11) | 1.9 ± 1.1 #* (12) | 3.5 ± 1.8 #* (11) | |
| intI1 | 1.2 ± 1.2 (11) | 1.7 ± 1.0 #* (12) | 2.9 ± 1.1 #* (11) | |
| Total coliforms | 0.9 ± 0.1 (12) | 1.6 ± 1.1 #* (12) | 1.7 ± 1.1 #* (12) | |
| E. coli | 0.7 ± 0.6 * (12) | 1.9 ± 0.9 #* (12) | 2.6 ± 1.1 #* (12) | |
| Samples (n = 23) | blaTEM | tetA | ermF | Total ARGs |
|---|---|---|---|---|
| Influent | 0.60 * | 0.90 * | 0.65 * | 0.79 * |
| Secondary effluent | 0.48 * | 0.78 * | 0.64 * | 0.60 * |
| Final effluent | 0.61 * | 0.64 * | 0.53 * | 0.59 * |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thakali, O.; Brooks, J.P.; Shahin, S.; Sherchan, S.P.; Haramoto, E. Removal of Antibiotic Resistance Genes at Two Conventional Wastewater Treatment Plants of Louisiana, USA. Water 2020, 12, 1729. https://doi.org/10.3390/w12061729
Thakali O, Brooks JP, Shahin S, Sherchan SP, Haramoto E. Removal of Antibiotic Resistance Genes at Two Conventional Wastewater Treatment Plants of Louisiana, USA. Water. 2020; 12(6):1729. https://doi.org/10.3390/w12061729
Chicago/Turabian StyleThakali, Ocean, John P. Brooks, Shalina Shahin, Samendra P. Sherchan, and Eiji Haramoto. 2020. "Removal of Antibiotic Resistance Genes at Two Conventional Wastewater Treatment Plants of Louisiana, USA" Water 12, no. 6: 1729. https://doi.org/10.3390/w12061729
APA StyleThakali, O., Brooks, J. P., Shahin, S., Sherchan, S. P., & Haramoto, E. (2020). Removal of Antibiotic Resistance Genes at Two Conventional Wastewater Treatment Plants of Louisiana, USA. Water, 12(6), 1729. https://doi.org/10.3390/w12061729








