Abstract
The proliferation of antibiotic-resistant bacteria (ARB) and the prevalence of antibiotic resistance genes (ARGs) in wastewaters are well-established factors that contribute to the reduced potency of antibiotics used in healthcare worldwide. The human health risk associated with the proliferation of ARB and ARGs need to be understood in order to design mitigation measures to combat their dissemination. Using the PCR analysis of genomic DNA, the prevalence of 41 ARGs active against the commonly used six classes of antibiotics was evaluated in 60 bacterial isolates obtained from pharmaceutical wastewaters in Nigeria. The ARGs most frequently detected from the bacterial isolates in each of the antibiotic classes under study include catA1 (58.3%); sulI (31.7%); tet(E) (30%); aac(3)-IV (28.3%); ermC (20%); blaTEM, blaCTX-M, blaNDM-1 at 18.3% each; which encode for resistance to chloramphenicol, sulfonamides, tetracycline, aminoglycoside, macrolide-lincosamide-streptogramin and β-lactams and penicillins, respectively. Acinetobacter spp., accession number MH396735 expressed the highest number of ARGs of all the bacterial isolates, having at least one gene that encodes for resistance to all the classes of antibiotics in the study. This study highlights wide distribution of ARB and ARGs to the antibiotics tested in the wastewater, making pharmaceutical wastewater reservoirs of ARGs which could potentially be transferred from commensal microorganisms to human pathogens.
1. Introduction
There is a global concern over the use of antibiotics and the development of resistance to the drugs in clinical treatments of infectious diseases [1]. The use of antibiotics in hospitals—particularly with the excretion of their potent forms into the environment—has long rendered clinical sewages as major sources of antibiotic resistance determinants in aquatic environments [2]. Recently, reports show that there are other relevant sources of antibiotic resistance in the environment, such as animal farms [3,4,5], wastewater treatment plants (WWTPs) [6,7,8], etc. Antibiotic resistance genes are now considered as environmental pollutants, there is an obvious need to prevent their further spread [9,10]. To achieve this, there is the need to elucidate their potential reservoirs—especially within the environment. However, the evolution of resistance and the spread of ARGs in wastewater generated from pharmaceutical production processes are less understood.
Pharmaceutical wastewaters contain large traces of antibiotics and other compounds, which at low concentrations—below therapeutic levels—are able to exert selection pressure [11,12]. Studies have shown that wastewaters generated from pharmaceutical production processes are reservoirs of antibiotic-resistant bacteria (ARB) and antibiotic resistance genes (ARGs) [8,13,14] and may accelerate possible horizontal transfer of environmental resistance determinants across endogenous microbial community [11]. This possibility may pose health risks to human and animals [15], with considerably high morbidity and mortality rates, with associated cost implications. Hence, the presence of both ARB and ARGs in environmental samples has been closely associated with the development of antibiotic resistance in treatment of clinical infections [16].
The risks of such clinical infections are potentially more serious in low- and middle-income countries where many hospitals either do not have wastewater treatment plants or they are ineffective. To worsen the situation, in many places—but particularly rural areas—surface water is used for agriculture and domestic purposes or even consumed untreated [2]. Antibiotic resistance genes are disseminated in such waters and have been reported to be more widespread in environmental non-pathogenic microbial populations than was originally believed [17,18,19]. Such resistances spread among bacterial populations have been reported to propagate through two major ways: vertical gene transfer (in the process of proliferation) and horizontal gene transfer, i.e., conjugation, transformation and transduction, promoted by mobile genetic elements (MGEs) [20,21]. However, antibiotic resistance is recognized as a major threat to human public health worldwide, the diversity, distribution and fate of ARGs in urban water systems remain unclear [22].
We investigated the antibiotic resistance gene profiles of bacterial isolates obtained from wastewater samples collected from fourteen pharmaceutical facilities in Lagos and Ogun states, Nigeria (Figure S1). In addition, a central wastewater treatment plant located at Agbara Estate in Ogun State (Figures S2 and S3) that collects both pharmaceutical and household wastewaters within the same industrial settlement was sampled, including the river water that receives the effluent. The selected pharmaceutical facilities are key players in antibiotic production at the secondary and tertiary stage of production in these regions. Pharmaceutical industries in Nigeria produce varying classes of antibiotics and other drug types on a single production plant. In most cases, they lack wastewater treatment before the effluents are released into receiving water bodies [23]. This practice has constituted a potential risk system to the water sources available to end users. This study is aimed at evaluating the degree to which the pharmaceutical wastewater can serve as reservoirs of ARGs which could potentially be transferred to human pathogens.
2. Materials and Methods
2.1. Study Sites and Sample Collection
The study sites and the sample collection procedures were previously described [14]. Briefly, the wastewater samples were collected directly from individual pharmaceutical facilities that play major roles in production and distribution of antibiotics. Most of which were untreated wastewater discharges. The samples were selected from fourteen pharmaceutical companies’ sites in Nigeria over a twenty-six month period. In addition, wastewater samples were collected from a central wastewater treatment plant located in Agbara Industrial Estate of Ogun State. The river that receives the treated effluent was also sampled. Table S1 in the supplementary document show the study sites and sources of samples. A total of 20 samples were collected in duplicates from each site in sterile containers and aseptically transported to the laboratory in 2-L brown glass bottles. The duplicate samples were pooled together to form composite sample and stored at 4 °C until processed for isolation of bacteria between 8 to 24 h after sample collection.
2.2. Bacterial Identification
Bacterial characterization procedures and susceptibility test were as previously described [14]. Bacterial compositions of the water sources were isolated. Wastewater and river water samples were serially diluted and plated on nonselective media, tryptone soya agar (TSA) and plate-count agar (PCA) (Oxoid, Ltd., Basingstoke, Hampshire, United Kingdom) and characterized by the PCR amplification of the 16S rRNA genes, with subsequent grouping using restriction fragment length polymorphism (RFLP) patterns, using BioNumerics version 6.01 (Applied Maths, SintMartens-Latem, Belgium). The 16S rRNA gene amplicons were sequenced (ABI 3730 capillary sequencer (Applied Biosystems)) and classified by construction of phylogenetic trees using the neighbor-joining algorithm with the Ribosomal Database Project II release 9.49 and the GenBank database using the BLAST program.
2.3. DNA Extraction
The genomic DNA was isolated from the sixty bacteria cells using the TIANamp bacteria DNA kit (TIANGEN Biotech Co., Beijing, China) according to the manufacturer’s instructions. The extracted DNA concentrations and quality were checked using the NanoDrop®, (ND-1000, Thermo Scientific, Wilmington, NC, USA.) and agarose gel electrophoresis and stored at −20 °C for further analysis.
2.4. PCR Screening for Antibiotic Resistance Genes and Class 1 Integrons
The screening for specific ARGs encoding resistance was carried out for antibiotics which belong to the classes of tetracycline, aminoglycoside, β-lactams and penicillins, macrolides-lincosamide-streptogramin and chloramphenicol. A total of 16 tetracycline (tet) resistance genes were screened for in all 60 bacterial isolates. The tet genes encoding for tetracycline efflux protein tet(A–E, G, J, Y and Z), ribosomal protection protein tet(BP, M, O, Q, T and W) and inactivating enzyme (tetX), which frequently appear in various environmental compartments, were screened for. In the screening for Aminoglycoside resistance genes, 11 genes conferring resistance to aminoglycoside acetyl transferases (aac(3)-IV, aac(6’)-Ib(aacA4), aac(3)-I, aac(3)-II and aac(3)-III), aminoglycoside phosphotransferases aph(3’)-Ia (aphA1), aph(3’’)-I(strA) and aph(6)-Id(strB) and aminoglycoside nucleotidyltransferases (adenylyltransferases) (ant(3’’)-Ia (aadA), ant(6)-I (aadE) and ant(2’’)-Ia (aadB) were screened for in the bacterial isolates. In the same vein, the presence of 5 clinically important β-lactam resistance genes (bla) encoding β-lactamase TEM, NDM-1, OXA, IMP and CTX-M in the bacterial isolates were screened for. A total number of 5 MLS resistance genes which includes ermA, ermB, ermC and ereA were also investigated. The genes catA1 and cmlA which encode for Chloramphenicol acetyltransferases and specific exporters, respectively were investigated. Also, the presence of 3 sulfonamide resistance genes, sul1, sul2 and sul3) were screened for in the 60 bacterial isolates. The integron 1 (int1) gene was investigated in all the 60 bacterial isolates.
Fifty µL PCR buffer containing 1.5-mM MgCl2, 200 µM of each deoxynucleoside triphosphate, 10 pmol of each primer, 1.25 U of TaKaRa rTaq polymerase and 1 µL of DNA template was used for the screening test (Takara, Dalian, China). The PCR program consisted of initial denaturation at 94 °C for 5 min, followed by 30 cycles of 1 min at 94 °C, 1 min at different annealing temperatures and extension at 72 °C for 1 min and final extension step at 72 °C for 10 min. The specific primers for all the ARGs for each group of antibiotics including the class 1 integrons and their different annealing temperatures for amplification are listed in the supplementary document (Tables S2–S4). Amplified products were separated by 1.5% (wt/vol) agarose gel electrophoresis and visualized under UV light after staining with ethidium bromide.
3. Results
3.1. Tetracycline Resistance Genes
The tetracycline resistance (tet) gene screening detected at least one tet gene in 51.7% of the bacterial isolates that were tested (Table 1). The prevalence of tet genes encoding efflux proteins were clearly more abundant than those encoding ribosomal protection proteins and enzymatic modification in the bacterial isolates (Table 2). The most common tetracycline resistance gene in the bacterial isolates was tet(E), identified in 30.0% of the total bacterial isolates under study and 58% of all bacterial isolates with the tet genes, followed by tet(B), tet(A) and tet(J) (Figure 1). The aforementioned tet genes encode efflux proteins as presented in Table 2. The only ribosomal protection protein determinants, tet(T) was found in 1.7% of the isolates. In addition, tet(X) was the only tetracycline resistance gene encoding for enzymatic modification protein with a prevalence of 5% in the tested bacterial isolates. Table 1 shows a summary of the resistance gene profiles of the bacterial isolates studied and Figure 1 shows their prevalence. It shows that tet genes were not found in all the bacterial isolates that were screened for, but bacterial isolates that carried more than one kind of tet gene were common. Staphylococcus saprophyticus with accession number MH396763 has the highest number of tetracycline resistance genes, the tet genes encountered in this bacterium were tet(A), tet(B), tet(E), tet(J), tet(L) and tet(X).

Table 1.
Phenotypic pattern of antibiotics resistance and resistance gene profile of bacterial isolates.

Table 2.
Prevalence of antibiotic resistance genes and mobile genetic elements from bacterial isolates.
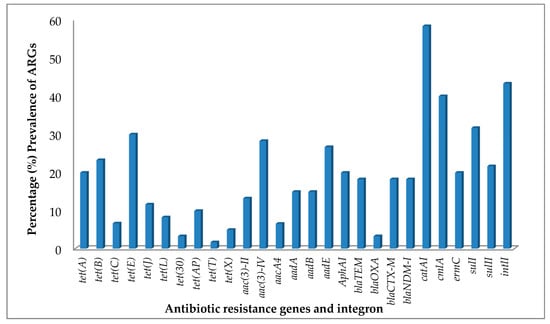
Figure 1.
Percentage prevalence of antibiotic resistance genes (ARGs) and integrons from antibiotic-resistant bacteria (ARB) isolated from pharmaceutical wastewaters.
3.2. Aminoglycoside Resistance Genes
The prevalence of aminoglycoside resistance genes in the bacterial isolates is shown in Table 2. Bacterial isolates that carried at least one kind of aminoglycoside resistance gene were observed in 48.3% of the bacterial isolates. The aminoglycoside resistance genes aac(3)-IV encoding acetyltransferase enzymes were the most abundant aminoglycoside resistance gene in the bacterial isolates (Figure 1), followed by aadE, a nucleotidyltransferase enzymes encoding gene (Figure 1). This group was followed by phosphotransferases enzyme encoding gene, aphA1 and strB both occurring in 20.0% of the bacterial isolates (Table 2). Acinetobacter sp. accession number MH396735 and Bacillus methylotrophicus, accession number MH396733, showed the highest number of aminoglycoside resistance genes (Table 1). The isolates expressed the same eight aminoglycoside resistance genes, aacA4, aac(3)-II, aac(3)-IV, aadA, aadE, aphA1, strA and strB in their genomes, as shown in Table 1.
3.3. β-Lactams and Penicillin Resistance Genes
Sixty bacterial isolates were screened for the β-lactamase encoding gene and 41.7% of the isolates contain at least one bla gene (Table 1). The β-lactamase encoding genes, blaTEM, blaCTX-M and blaNDM-1 showed the highest prevalence in the isolates, with the three occurring in 18.3% of the bacterial isolates; followed by blaOXA which is prevalent in 3.3% (Figure 1) of the isolates. Bacillus sp. and Enterobacter sp. (Table 1) had most the bla genes detected. They both showed expression for the three bla genes, blaTEM, blaCTX-M and blaNDM-1. The β-Lactamase encoding gene, blaIMP, was not found in any of the bacterial isolates screened.
3.4. Macrolide–Lincosamide–Streptogramin and Chloramphenicol Resistance Genes
The MLS resistance gene, ermC was present in 20% of the bacterial isolates (Table 2) and was the only MLS resistance gene expressed. The chloramphenicol resistance genes catA1 and cmlA revealed a high expression of both genes (Table 1) in the bacterial isolates. The genes catA1 and cmlA were present in 56.7% and 40.0% of the bacterial isolates, respectively. The result showed that 38.3% of the bacterial isolates had both catA1 and cmlA genes expressed (Figure 1); whereas 56.7% of the bacterial isolates expressed at least one of the chloramphenicol resistance genes, catA1 and cmlA. The results for the sulfonamide ARGs (sulI and sulII) and integron 1(intl1) have been described previously [14].
4. Discussion
Our results indicate that ARGs are widely distributed among the bacterial isolates. Acinetobacter spp. with accession number MH396735 expressed the highest number of ARGs (50%) of all the bacterial isolates under studied (Table 1). It was isolated from a wastewater treatment plant under study [14]. Acinetobacter sp. was a major bacteria group referred in a review that profiled antibiotic-resistant bacteria that are deemed to be of particular concern in the 21st century [24]. Hence, its relevance in wastewater bacterial communities which agrees with this study, particularly Acinetobacter baumannii, a clinically important antibiotic-resistant bacterium [25,26], reported to be naturally resistant to many antibiotics due to both poor membrane penetration and active efflux pumps [27]. However, the two other Acinetobacter sp. showed no expression for the ARGs under study, except for the one with accession number MH396768 that was tet(B) positive (Table 1). The genes conferring resistance to chloramphenicol was the most frequently detected ARG in the genomes of several species of the isolates. However, reports from other studies show that the total relative abundance of ARGs declined during the wastewater purification process [28,29,30], but the relative abundance of some genes are enriched in the effluent bacterial community [22,31], with accompanying health implications.
Tetracycline-resistant bacteria were reported to emerge in the environments with the introduction of tetracycline [32]. Most environmental tet genes code for transport proteins, which pump the antibiotics out of the bacteria cell and keep the intercellular concentrations low to make ribosomes function normally [33]. In our study, the efflux genes, tet(E), tet(B) and tet(A) were the main tetracycline-resistance genes identified, but tet(E) was the most prevalent of the tet gene, being distributed in 18 genomes of the bacterial isolates. These efflux genes, tet(A–C and E) have been frequently reported in various environmental compartments including activated sludge of sewage treatment plants (STPs) [34] and surface water [35]. The enzyme modification gene tet(X) was the only tet gene encountered in this category in our study. The dissemination of tet(X) resistance gene is of special concern because it confers resistance also against third generation tetracycline tigecycline [36]. However, the use of this antibiotic is strictly regulated; tet(X) has already been observed among pathogenic bacteria [37]. Staphylococcus saprophyticus with accession number MH396763 which showed the highest expression of tetracycline resistance genes, also showed resistance to other groups of ARGs tested, except for the β-lactams and penicillins genes and the MLS genes (Table 1).
Aminoglycoside resistance genes have a major mechanism of resistance, which is the direct deactivation of the antibiotics by enzymatic modification [38]. Aminoglycoside resistance genes encoding for three transferase enzymes, acetyltransferases (AAC), nucleotidyltransferases (ANT) and phosphotransferases (APH), responsible for inactivation of aminoglycoside were detected. In the bacterial isolates, about 50% expressed at least one of the aminoglycoside resistance genes, with the acetyltransferases the most abundant, having aac(3)-IV as the highest occurrence. Reports of several works on the aminoglycoside resistance gene show that the genes of aacC1, C2, C3 and C4, encoding aminoglycoside-3-N-acetyltransferase were often detected in microbial communities or isolates from STPs [39,40,41]. Acinetobacter sp. (MH396735) and Bacillus methylotrophicus (MH396733) expressed 8 out of 9 aminoglycoside resistance genes that tested positive. Coincidentally, both isolates showed expression for the same ARMs genes in this class, despite belonging to Gammaproteobacteria and Firmicutes bacterial groups, respectively.
The β-Lactams are the most widely used antibiotics; resistance to these antibiotics is a severe threat because they have low toxicity and are used to treat a broad range of infections [42]. Resistance usually occurs via hydrolysis of the β-lactam ring, mediated by a wide range of β-lactamases [43] produced by resistant strains that are capable of inactivating beta-lactam drugs. The mechanisms of β-lactam resistance include an inaccessibility of the antibiotics to their target enzymes, modifications of target enzymes and/or direct deactivation of the antibiotics by β-lactamases [44,45]. The bacterial isolates β-lactamase resistance genes, blaNDM-1, blaTEM and blaCTX-M showed the highest prevalence. In line with this, a variety of bla genes have been identified in bacteria derived from STPs [46,47] and surface water [35]. In a similar study of a WWTP of a pharmaceutical industry in China, all of the strains of ESBL-producing Escherichia coli were found to carry at least one β-lactamase gene and the most prevalent ARGs type was blaCTX-M [48]. Their findings were in line with other studies that showed that blaCTX-M was the most dominant ARGs type in WWTPs of industrial origin. These environmental compartments may further serve as reservoirs for β-lactam resistance genes. Enterobacter sp. (Table 1) showed expression for 3 out of the 4 bla genes that tested positive, agreeing with the study which detected bla genes in animal-derived environmental pathogens that includes Enterobacter [47].
Structurally, macrolides, lincosamide and streptogramin are different; they are often investigated simultaneously for microbial resistance since some macrolide resistance genes (erm) encode resistance to two or all three of these compounds [49]. MLS resistance is mostly mediated by rRNA methylases (encoded by erm genes), which methylate the adenine residues to prevent the three antimicrobials from binding to ribosomal protein [33,50]. The erm genes can easily be transferred from one host to another [51], since they are usually acquired and associated with mobile elements, such as transposons [52]. The result shows that ermC was the only MLS resistance gene detected in the isolates. It was also found to be expressed in the genome of Acinetobacter sp. (MH396735). The co-resistance between tetracyclines and MLS antibiotics has been reported to be due to the occurrence of genetic determinants for tetracycline and MLS resistance on very promiscuous transposons that can form genetic linkages. These linkages are capable of increasing the risk for transfer of resistances to pathogenic species that may lead to therapeutic failure with severe consequences [52].
The mechanisms responsible for resistances to chloramphenicol include chloramphenicol acetyltransferases (encoded by cat genes), specific exporters (encoded by cml genes) and multidrug transporters [53]. In the study, bacterial isolates were screened for chloramphenicol resistance catA1 and cmlA. Our results indicate that catA1 and cmlA are widely distributed among the environmental bacterial isolates; with the catA1 gene frequently detected in the genomes of several species. The result shows a very high expression of both genes. The resistance gene catA1 was present in most of the bacterial isolates. In the bacterial isolates there was also a very high expression of cmlA in the genome of most of the bacterial isolates (Table 1). The chloramphenicol resistance gene catA1 shows the highest prevalence of all the resistance gene classes screened for (Table 1), both genes were expressed both in Acinetobacter spp. (MH396735) and Bacillus methylotrophicus (MH396733) genomes.
The location of ARGs on mobile genetic elements, such as integrons makes the horizontal transfer of antibiotic resistance in wastewater possible and easy to achieve among bacteria with same or diverse origins [54]. As presented in the result above (Table 1), integron 1 (Intl1) was detected in almost 50% of the bacterial isolates tested (Table 1). The presence of Intl1 in the genome of the bacterial isolates may indicate a high possibility of horizontal gene transfer of ARGs within the wastewater, hence the high prevalence of ARGs found in the bacterial isolates. Our findings show that sulI is closely associated with the Intl1, this conforms to the report that sulI is a part of class I integron and can be disseminated and transferred horizontally within and between bacterial species in wastewater [40]. This may account for the prevalence of sulI in almost all the compartments sampled. In addition, ARGs to β-lactams and aminoglycoside where found associated with Intl1. Other studies have demonstrated that transposons and integrons carrying more than one type of ARGs often occur in sewage treatment plants [41,46] and surface waters [35]. Integrons were not found in any of the three bacterial isolates from the river water samples (Table 1), this may contribute to the few to none ARGs found in these isolates.
5. Conclusions
This study spread across wastewater obtained from mostly individual pharmaceutical facilities because very few of them have their wastewater passed through a routine/conventional wastewater treatment plant, which was also sampled. The Acinetobacter spp. (MH396735) with the highest expression of ARGs in its genome was isolated from the acclaimed treated water sample from the WWTP. This study clearly demonstrates that pharmaceutical wastewaters are important reservoirs of ARGs for a number of antibiotic classes such as to chloramphenicol, sulfonamides, tetracycline, aminoglycoside, MLS and β-lactams and penicillins. They play a significant role in the emergence of these ARGs. More studies are required to establish the pathways involved in their spread within the environment. Our findings highlight the need for strengthening the active surveillance of the activities of pharmaceutical industries in Nigeria, particularly in the handling and management of wastewater generated from their facilities, with stringent compliance of regulatory authorities in these sectors.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4441/12/7/1897/s1, Table S1: Description of sampled sites, Tables S2–S4: Primers and conditions used to amplify the antibiotic resistance genes ARGs in the study, Figure S1: Map of Nigeria highlighting the study areas: Ogun and Lagos States, Figure S2: A section of the map of Nigeria showing Agbara Industrial Area in Ogun State, Figure S3: Cross-section of the wastewater treatment Plant located in Agbara Industrial Estate, in Ogun State, Nigeria.
Author Contributions
Conceptualization, A.O.; methodology, A.O. and A.M.I.; software, A.M.I.; validation, A.O.; formal analysis, A.O.; investigation, A.O.; resources, A.O. and A.M.I.; data curation, A.O.; writing—original draft preparation, A.O.; writing—Review and Editing, A.O. and A.M.I.; visualization, A.M.I.; supervision, A.M.I.; project administration, A.O. and A.M.I.; funding acquisition, A.O. and A.M.I. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Organization for women in Science for the developing world (OWSD), fund reservation No 3240266476, and The State Key Laboratory of Environmental Aquatic Chemistry, Research Center for Eco-Environmental Sciences, Chinese Academy of Science.
Conflicts of Interest
The authors declare no conflict of interest.
References
- World Health Organization (WHO). Global Antimicrobial Resistance Surveillance System (GLASS) Report: Early Implementation 2017–2018; WHO: Geneva, Switzerland, 2018. [Google Scholar]
- Lien, L.T.; Lan, P.T.; Chuc, N.T.; Hoa, N.Q.; Nhung, P.H.; Thoa, N.T.; Tamhankar, A.J.; Lundborg, C.S. Antibiotic resistance and antibiotic resistance genes in Escherichia coli isolates from hospital wastewater in Vietnam. Int. J. Environ. Res. Public Health 2017, 14, 699. [Google Scholar] [CrossRef]
- Ducey, T.F.; Durso, M.L.; Ibekwe, A.M.; Dungan, R.S.; Jackson, C.R.; Frye, J.G.; Castleberry, B.L.; Rashash, D.M.C.; Rothrock, M.J.; Boykin, D.; et al. A newly developed Escherichia coli isolate panel from a cross section of U.S. animal production systems reveals geographic and commodity-based differences in antibiotic resistance gene carriage. J. Hazard. Mater. 2020, 382, 120991. [Google Scholar] [CrossRef] [PubMed]
- Graham, D.W.; Knapp, C.W.; Christensen, B.T.; McCluskey, S.; Dolfing, J. Appearance of β-lactam resistance genes in agricultural soils and clinical isolates over the 20th century. Sci. Rep. 2016, 6, 21550. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Yuan, Q.; Mathieu, J.; Stadler, L.; Senehi, N.; Sun, R.; Alvarez, P.J.J. Antibiotic resistance genes from livestock waste: Occurrence, dissemination, and treatment. NPJ Clean Water 2020, 3, 4. [Google Scholar] [CrossRef]
- Szczepanowski, R.; Linke, B.; Krahn, I.; Gartemann, K.; Gutzkow, T.; Eichler, W.; Puhler, A.; Schluter, A. Detection of 140 clinically relevant antibiotic-resistance genes in the plasmid metagenome of wastewater treatment plant bacteria showing reduced susceptibility to selected antibiotics. Microbiology 2009, 155, 2306–2319. [Google Scholar] [CrossRef]
- Williams, M.R.; Stedtfeld, R.D.; Guo, X.; Hashsham, S.A. Antimicrobial resistance in the environment. Water Environ. Res. 2016, 88, 1951–1967. [Google Scholar] [CrossRef]
- Hultman, J.; Tamminen, M.; Parnanen, K.; Cairns, J.; Karkman, A.; Virta, M. Host range of antibiotic resistance genes in wastewater treatment plant influent and effluent. FEMS Microbiol. Ecol. 2018, 94, fiy038. [Google Scholar] [CrossRef]
- World Health Organization (WHO). Antimicrobial Resistance: Global Report on Surveillance 2014; WHO: Geneva, Swizerland, 2015. [Google Scholar]
- Roca, I.; Akova, M.; Baquero, F.; Carlet, J.; Cavaleri, M.; Coenen, S.; Cohen, J.; Findlay, D.; Gyssens, I.; Heuer, O.E.; et al. The global threat of antimicrobial resistance: Science for intervention. New Microbes New Infect. 2015, 6, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Karkman, A.; Do, T.T.; Walsh, F.; Virta, M.P.J. Antibiotic-resistance genes in waste water-review. Trends Microbiol. 2017, 26, 220–228. [Google Scholar] [CrossRef] [PubMed]
- Lundborg, C.S.; Tamhankar, A.J. Antibiotic residues in the environment of South East Asia. BMJ 2017, 358. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Yu, T.; Zhang, Y.; Yang, M.; Li, Z.; Liu, M.; Qi, R. Antibiotic Resistance Characteristics of Environmental Bacteria from an Oxytetracycline Production Wastewater Treatment Plant and the Receiving River. Appl. Environ. Microbiol. 2010, 76, 3444–3451. [Google Scholar] [CrossRef] [PubMed]
- Obayiuwana, A.C.; Ogunjobi, A.; Yang, M.; Ibekwe, M. Characterization of Bacterial Communities and their Antibiotic Resistance Profiles in Wastewaters obtained from Pharmaceutical Facilities in Lagos and Ogun States, Nigeria. Int. J. Environ. Res. Public Health 2018, 15, 1365–1378. [Google Scholar] [CrossRef]
- Kemper, N. Veterinary antibiotics in the aquatic and terrestrial environment. Ecol. Indic. 2008, 8, 1–13. [Google Scholar] [CrossRef]
- Iwane, T.; Urase, T.; Yamamoto, K. Possible impact of treated wastewater discharge on incidence of antibiotic resistant bacteria in river water. Water Sci. Technol. 2001, 43, 91–99. [Google Scholar] [CrossRef] [PubMed]
- D’Costa, V.M.; McGram, K.M.; Hughes, D.W.; Wright, G.D. Sampling the antibiotic resistome. Science 2006, 311, 374–377. [Google Scholar] [CrossRef]
- Nesme, J.; Cécillon, S.; Delmont, T.O.; Monier, J.M.; Vogel, T.M.; Simonet, P. Large-scale metagenomic-based study of antibiotic resistance in the environment. Curr. Biol. 2014, 24, 1096–1100. [Google Scholar] [CrossRef]
- Surette, M.D.; Wright, G.D. Lessons from the environmental antibiotic resistome. Annu. Rev. Microbiol. 2017, 71, 309–329. [Google Scholar] [CrossRef]
- Markiewicz, Z.; Kwiatkowski, Z.A. Bakterie, Antybiotyki, Lekooporność; Wydawnictwo Naukowe PWN: Warszawa, Poland, 2008. [Google Scholar]
- Khan, S.; Knapp, C.W.; Beattie, T.K. Antibiotic resistant bacteria found in municipal drinking water. Environ. Process. 2016, 3, 541–552. [Google Scholar] [CrossRef]
- Munck, C.; Albertsen, M.; Telke, A.; Ellabaan, M.; Nielsen, P.H.; Sommer, M.O. Limited dissemination of the wastewater treatment plant core resistome. Nat. Commun. 2015, 6, 8452. [Google Scholar] [CrossRef]
- Ngwuluka, N.C.; Ochekpe, N.A.; Odumosu, P.O. An assessment of pharmaceutical waste management in some Nigerian pharmaceutical industries. Afr. J. Biotechnol. 2011, 10, 11259–11264. [Google Scholar] [CrossRef]
- Fair, R.; Tor, Y. Antibiotics and Bacterial Resistance in the 21st Century. Perspect. Medicin. Chem. 2014, 6, 25–64. [Google Scholar] [CrossRef]
- Lee, C.R.; Lee, J.H.; Park, M.; Park, K.S.; Bae, I.K.; Kim, Y.B.; Cha, C.J.; Jeong, B.C.; Lee, S.H. Biology of Acinetobacter baumannii: Pathogenesis, Antibiotic Resistance Mechanisms, and Prospective Treatment Options. Front. Microbiol. 2017, 7, 55. [Google Scholar] [CrossRef] [PubMed]
- Breijyeh, Z.; Jubeh, B.; Karaman, R. Resistance of Gram-Negative Bacteria to Current Antibacterial Agents and Approaches to Resolve It. Molecules 2020, 25, 1340. [Google Scholar] [CrossRef] [PubMed]
- Vila, J.; Martí, S.; Sánchez-Céspedes, J. Porins, Efflux Pumps and Multidrug Resistance in Acinetobacter baumannii. J. Antimicrob. Chemother. 2007, 59, 1210–1215. [Google Scholar] [CrossRef]
- Yang, Y.; Li, B.; Zou, S.; Fang, H.H.; Zhang, T. Fate of antibiotic resistance genes in sewage treatment plant revealed by metagenomic approach. Water Res. 2014, 62, 97–106. [Google Scholar] [CrossRef] [PubMed]
- Bengtsson-Palme, J.; Hammaren, R.; Pal, C.; Ostman, M.; Bjorlenius, B.; Flach, C.F.; Fick, J.; Kristiansson, E.; Tysklind, M.; Larsson, D.G.J. Elucidating selection processes for antibiotic resistance in sewage treatment plants using metagenomics. Sci. Total Environ. 2016, 572, 697–712. [Google Scholar] [CrossRef]
- Karkman, A.; Johnson, T.A.; Lyra, C.; Stedtfeld, R.D.; Tamminen, M.; Tiedje, J.M.; Virta, M. High-throughput quantification of antibiotic resistance genes from an urban wastewater treatment plant. FEMS Microbiol. Ecol. 2016, 92. [Google Scholar] [CrossRef] [PubMed]
- Mao, D.; Yu, S.; Rysz, M.; Luo, Y.; Yang, F.; Li, F.; Hou, J.; Mu, Q.; Alvarez, P.J. Prevalence and proliferation of antibiotic resistance genes in two municipal wastewater treatment plants. Water Res. 2015, 85, 458–466. [Google Scholar] [CrossRef]
- Dancer, S.J.; Shears, P.; Platt, D.J. Isolation and characterization of coliforms from glacial ice and water in Canada’s high arctic. J. Appl. Microbiol. 1997, 82, 597–609. [Google Scholar] [CrossRef]
- Roberts, M.C. Resistance to tetracycline, macrolide-lincosamide-streptogramin, trimethoprim, and sulfonamide drug classes. Mol. Biotechnol. 2002, 20, 261–283. [Google Scholar] [CrossRef]
- Guillaume, G.; Verbrugge, D.; Chasseur-Libotte, M.L.; Moens, W.; Collard, J.M. PCR typing of tetracycline resistance determinants (Tet A–E) in Salmonella enterica serotype Hadar and in the microbial community of activated sludges from hospital and urban wastewater treatment facilities in Belgium. FEMS Microbiol. Ecol. 2000, 32, 77–85. [Google Scholar]
- Poppe, C.; Martin, L.; Muckle, A.; Archambault, M.; McEwen, S.; Weir, E. Characterization of antimicrobial resistance of Salmonella Newport isolated from animals, the environment, and animal food products in Canada. Can. J. Vet. Res. 2006, 70, 105–114. [Google Scholar] [PubMed]
- Moore, I.F.; Hughes, D.W.; Wright, G.D. Tigecycline is modified by the flavin-dependent monooxygenase tet (x). Biochemistry 2005, 44, 11829–11835. [Google Scholar] [CrossRef] [PubMed]
- Aminov, R.I. Evolution in action: Dissemination of tet (x) into pathogenic microbiota. Front. Microbiol. 2013, 4, 192. [Google Scholar] [CrossRef] [PubMed]
- Shakil, S.; Khan, R.; Zarrilli, R.; Khan, A.U. Aminoglycosides versus bacteria—A description of the action, resistance mechanism, and nosocomial battle ground. J. Biomed. Sci. 2008, 15, 5–14. [Google Scholar] [CrossRef] [PubMed]
- Heuer, H.; Krögerrecklenfort, E.; Wellington, E.M.H.; Egan, S.; van Elsas, J.D.; van Overbeek, L.; Collard, J.M.; Guillaume, G.; Karagouni, A.D.; Nikolakopoulou, T.L.; et al. Gentamicin resistance genes in environmental bacteria: Prevalence and transfer. FEMS Microbiol. Ecol. 2002, 42, 289–302. [Google Scholar] [CrossRef]
- Tennstedt, T.; Szczepanowski, R.; Braun, S.; Pühler, A.; Schlüter, A. Occurrence of integron-associated resistance gene cassettes located on antibiotic resistance plasmids isolated from a wastewater treatment plant. FEMS Microbiol. Ecol. 2003, 45, 239–252. [Google Scholar] [CrossRef]
- Tennstedt, T.; Szczepanowski, R.; Krahn, I.; Pühler, A.; Schlüter, A. Sequence of the 68,869 bp IncP-1a plasmid pTB11 from a wastewater treatment plant reveals a highly conserved backbone, a Tn402-like integron and other transposable elements. Plasmid 2005, 53, 218–238. [Google Scholar] [CrossRef]
- Livermore, D.M. Are all beta-lactams created equal? Scand. J. Infect. Dis. Suppl. 1996, 101, 33–43. [Google Scholar]
- Bush, K.; Bradford, P.A. β-Lactams and β-lactamase inhibitors: An overview. Cold Spring Harbor. Pers. Med. 2016, 6, a025247. [Google Scholar] [CrossRef]
- Walsh, C. Molecular mechanisms that confer antibacterial drug resistance. Nature 2000, 406, 775–778. [Google Scholar] [CrossRef] [PubMed]
- Li, X.Z.; Mehrotra, M.; Ghimire, S.; Adewoye, L. β-Lactam resistance and β-lactamases in bacteria of animal origin. Vet. Microbiol. 2007, 121, 197–214. [Google Scholar] [CrossRef]
- Szczepanowski, R.; Krahn, I.; Linke, B.; Goesmann, A.; Pühler, A.; Schlüter, A. Antibiotic multiresistance plasmid pRSB101 isolated from a wastewater treatment plant is related to plasmids residing in phytopathogenic bacteria and carries eight different resistance determinants including a multidrug transport system. Microbiology 2004, 150, 3613–3630. [Google Scholar] [CrossRef] [PubMed]
- Volkmann, H.; Schwartz, T.; Bischoff, P.; Kirchen, S.; Obst, U. Detection of clinically relevant antibiotic-resistance genes in municipal wastewater using real-time PCR (TaqMan). J. Microbiol. Methods 2004, 56, 277–286. [Google Scholar] [CrossRef]
- Jiang, X.; Cui, X.; Xu, H.; Liu, W.; Tao, F.; Shao, T.; Pan, X.; Zheng, B. Whole gene sequencing of Extended-Spectrum Beta-Lactamase (ESBL)-producing Escherichia coli isolated from a wastewater treatment plant in China. Front. Microbiol. 2019, 10, 1797. [Google Scholar] [CrossRef] [PubMed]
- Roberts, M.C.; Sutcliffe, J.; Courvalin, P.; Jensen, L.B.; Rood, J.; Seppala, H. Nomenclature for macrolide and macrolide-lincosamide-streptogramin B antibiotic resistance determinants. J. Antimicrob. Chemother. 1999, 43, 2823–2830. [Google Scholar] [CrossRef]
- Cetin, E.S.; Gunes, H.; Kaya, S.; Aridogan, B.C.; Demirci, M. Macrolide lincosamide streptogramin B resistance phenotypes in clinical staphylococcal isolates. Int. J. Antimicrob. Agents 2008, 31, 364–368. [Google Scholar] [CrossRef]
- Roberts, M.C. Acquired tetracycline and/or macrolide-lincosamides-streptogramin resistance in anaerobes. Anaerobe 2003, 9, 63–69. [Google Scholar] [CrossRef]
- Marosevic, D.; Kaevska, M.; Jaglic, Z. Resistance to the tetracyclines and macrolide-lincosamide-streptogramin group of antibiotics and its genetic linkage—A review. Ann. Agric. Environ. Med. 2017, 24, 338–344. [Google Scholar] [CrossRef]
- Schwarz, S.; Kehrenberg, C.; Doublet, B.; Cloeckaert, A. Molecular basis of bacterial resistance to chloramphenicol and florfenicol. FEMS Microbiol. Rev. 2004, 28, 519–542. [Google Scholar] [CrossRef]
- Allen, H.K.; Donato, J.; Wang, H.H.; Cloud-Hansen, K.A.; Davies, J.; Handelsman, J. Call of the wild: Antibiotic resistance genes in natural environments. Nat. Rev. Microbiol. 2010, 8, 251–259. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).