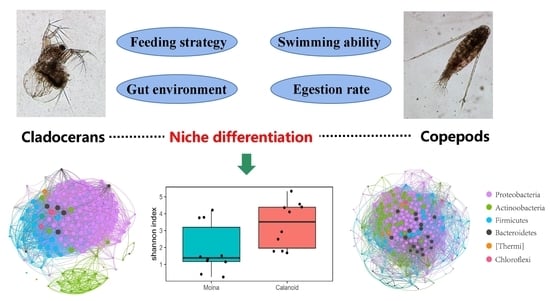
Host Dependence of Zooplankton-Associated Microbes and Their Ecological Implications in Freshwater Lakes
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling Sites, Zooplankton and Water Samples Collection
2.2. Bacterial DNA Extraction and High−Throughput Sequencing of 16S rRNA Gene Amplicons
2.3. Statistical Analysis
3. Results
3.1. Community Structure of Zooplankton−Associated Bacteria
3.2. Significantly Different OTUs between Two Zooplankton−Associated Microbial Groups
3.3. Bacterial Richness and Diversity of Zooplankton Microbiome
3.4. Relationship between Environmental Parameters and Zooplankton Microbiome Composition
3.5. Co−Occurrence Network of Zooplankton−Associated Bacteria
3.6. Biogeochemical Relevant Microbial Groups
4. Discussion
4.1. Community Structures of Zooplankton−Associated Bacteria in Freshwater Lakes
4.2. Zooplankton−Associated Bacteria Exhibited Different Features Depending on the Host Traits
4.3. Potential Ecological Effects of Zooplankton−Associated Bacteria
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Cottenie, K.; Michels, E.; Nuytten, N.; Meester, L.D. Zooplankton metacommunity structure: Regional vs. local processes in highly interconnected ponds. Ecology 2003, 84, 991–1000. [Google Scholar] [CrossRef]
- Kohout, L.; Fott, J. Restoration of zooplankton in a small acidified mountain lake (Plešné Lake, Bohemian Forest) by reintroduction of key species. Biologia 2006, 61, S477–S483. [Google Scholar] [CrossRef]
- Schou, M.O.; Risholt, C.; Lauridsen, T.L.; Søndergaard, M.; Grønkjaer, P.; Jacobsen, L.; Berg, S.; Skov, C.; Brucet, S.; Jeppesen, E.; et al. Restoring lakes by using artificial plant beds: Habitat selection of zooplankton in a clear and a turbid shallow lake. Freshw. Biol. 2009, 54, 1520–1531. [Google Scholar] [CrossRef]
- Hessen, D.O.; Andersen, T. Bacteria as a source of phosphorus for zooplankton. Hydrobiologia 1990, 206, 217–223. [Google Scholar] [CrossRef]
- Steinberg, D.K.; Landry, M.R. Zooplankton and the Ocean Carbon Cycle. Annu. Rev. Mar. Sci. 2017, 9, 413–444. [Google Scholar] [CrossRef]
- Tang, K.; Turk, V.; Grossart, H. Linkage between crustacean zooplankton and aquatic bacteria. Aquat. Microb. Ecol. 2010, 61, 261–277. [Google Scholar] [CrossRef]
- Samad, S.; Lee, H.J.; Cerbin, S.; Meima-Franke, M.; Bodelier, P.L.E. Niche Differentiation of Host-Associated Pelagic Microbes and Their Potential Contribution to Biogeochemical Cycling in Artificially Warmed Lakes. Front. Microbiol. 2020, 11, 582. [Google Scholar] [CrossRef] [PubMed]
- Urban-Rich, J.; McCarty, J.T.; Fernández, J.L.A.; Acuña, J.L. Larvaceans and copepods excrete fluorescent dissolved organic matter (FDOM). J. Exp. Mar. Biol. Ecol. 2006, 332, 96–105. [Google Scholar] [CrossRef]
- Kiko, R.; Hauss, H.; Buchholz, F.; Melzner, F. Ammonium excretion and oxygen respiration of tropical copepods and euphausiids exposed to oxygen minimum zone conditions. Biogeosciences 2016, 13, 2241–2255. [Google Scholar] [CrossRef]
- Valdés, V.; Fernandez, C.; Molina, V.; Escribano, R. Nitrogen excretion by copepods and its effect on ammonia-oxidizing communities from a coastal upwelling zone. Limnol. Oceanogr. 2018, 63, 278–294. [Google Scholar] [CrossRef]
- Bickel, S.L.; Tang, K.W. Zooplankton-associated and free-living bacteria in the York River, Chesapeake Bay: Comparison of seasonal variations and controlling factors. Hydrobiologia 2014, 722, 305–318. [Google Scholar] [CrossRef][Green Version]
- Carman, K.R.; Fred, C.D. Epibiotic microorganisms on copepods and other marine crustaceans. Microsc. Res. Techniq. 1997, 37, 116–135. [Google Scholar] [CrossRef]
- Heidelberg, J.F.; Heidelberg, K.B.; Colwell, R.R. Bacteria of the γ-Subclass Proteobacteria Associated with Zooplankton in Chesapeake Bay. Appl. Environ. Microbiol. 2002, 68, 5296–5303. [Google Scholar] [CrossRef]
- Grossart, H.-P.; Dziallas, C.; Tang, K.W. Bacterial diversity associated with freshwater zooplankton. Environ. Microbiol. Rep. 2010, 1, 50–55. [Google Scholar] [CrossRef] [PubMed]
- Macke, E.; Callens, M.; De Meester, L.; Decaestecker, E. Host-genotype dependent gut microbiota drives zooplankton tolerance to toxic cyanobacteria. Nat. Commun. 2017, 8, 1–13. [Google Scholar] [CrossRef]
- Mushegian, A.A.; Ebert, D. Presence of microbiota reverses the relative performance of Daphnia on two experimental diets. Zoology 2017, 125, 29–31. [Google Scholar] [CrossRef] [PubMed]
- Fong, J.C.; De Guzman, B.E.; Lamborg, C.H.; Sison-Mangus, M.P. The Mercury-Tolerant Microbiota of the Zooplankton Daphnia Aids in Host Survival and Maintains Fecundity under Mercury Stress. Environ. Sci. Technol. 2019, 53, 14688–14699. [Google Scholar] [CrossRef]
- Sison-Mangus, M.P.; Mushegian, A.A.; Ebert, D. Water fleas require microbiota for survival, growth and reproduction. ISME J. 2015, 9, 59–67. [Google Scholar] [CrossRef] [PubMed]
- Eckert, E.; Pernthaler, J. Bacterial epibionts of Daphnia: A potential route for the transfer of dissolved organic carbon in freshwater food webs. ISME J. 2014, 8, 1808–1819. [Google Scholar] [CrossRef]
- De Angelis, M.A.; Lee, C. Methane production during zooplankton grazing on marine phytoplankton. Limnol. Oceanogr. 1994, 39, 1298–1308. [Google Scholar] [CrossRef]
- Scavotto, R.E.; Claudia, D.; Mikkel, B.T.; Lasse, R.; Pia, H.M. 2015. Nitrogen-fixing bacteria associated with copepods in coastal waters of the North Atlantic Ocean. Env. Microbiol. 2015, 17, 3754–3765. [Google Scholar] [CrossRef]
- Shoemaker, K.M.; McCliment, E.A.; Moisander, P.H. Copepod-Associated Gammaproteobacterial Alkaline Phosphatases in the North Atlantic Subtropical Gyre. Front. Microbiol. 2020, 11, 1033. [Google Scholar] [CrossRef]
- Moisander, P.H.; Shoemaker, K.M.; Daley, M.C.; McCliment, E.; Larkum, J.; Altabet, M.A. Copepod-Associated Gammaproteobacteria Respire Nitrate in the Open Ocean Surface Layers. Front. Microbiol. 2018, 9, 2390. [Google Scholar] [CrossRef] [PubMed]
- Greenberg, A.E.; Trussell, R.R.; Lenore, S.C.; American Water Works Association. Standard methods for the examination of water and wastewater: Supplement to the sixteenth edition. Am. J. Public Health Nations Health 2005, 56, 387. [Google Scholar] [CrossRef]
- Jespersen, A.M.; Christoffersen, K. Measurements of chlorophyll-a from phytoplankton using ethanol as extraction solvent. Arch. Hydrobiol. 1987, 109, 445–454. [Google Scholar]
- Quince, C.; Lanzén, A.; Curtis, T.; Davenport, R.J.; Hall, N.; Head, I.; Read, L.F.; Sloan, W.T. Accurate determination of microbial diversity from 454 pyrosequencing data. Nat. Methods 2009, 6, 639–641. [Google Scholar] [CrossRef] [PubMed]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef]
- Chao, A.; Shen, T.-J. Nonparametric estimation of Shannon’s index of diversity when there are unseen species in sample. Environ. Ecol. Stat. 2003, 10, 429–443. [Google Scholar] [CrossRef]
- Mouillot, D. A comparison of species diversity estimators. Popul. Ecol. 1999, 41, 203–215. [Google Scholar] [CrossRef]
- Kilbane, J.J.; Ranganathan, R.; Cleveland, L.; Kayser, K.J.; Ribiero, C.; Linhares, M.M. Selective Removal of Nitrogen from Quinoline and Petroleum by Pseudomonas ayucida IGTN9m. Appl. Environ. Microbiol. 2000, 66, 688–693. [Google Scholar] [CrossRef]
- Delille, D.; Razouls, S. Community structures of heterotrophic bacteria of copepod fecal pellets. J. Plankton Res. 1994, 16, 603–615. [Google Scholar] [CrossRef]
- Hansen, B.; Bech, G. Bacteria associated with a marine planktonic copepod in culture. I. Bacterial genera in seawater, body surface, intestines and fecal pellets and succession during fecal pellet degradation. J. Plankton Res. 1996, 18, 257–273. [Google Scholar] [CrossRef]
- Shoemaker, K.M.; Moisander, P.H. Seasonal variation in the copepod gut microbiome in the subtropical North Atlantic Ocean. Environ. Microbiol. 2017, 19, 3087–3097. [Google Scholar] [CrossRef]
- Tang, K.; Dziallas, C.; Hutalle-Schmelzer, K.; Grossart, H.-P. Effects of food on bacterial community composition associated with the copepod Acartia tonsa Dana. Biol. Lett. 2009, 5, 549–553. [Google Scholar] [CrossRef]
- Mathews, L.; Faithfull, C.L.; Lenz, P.H.; Nelson, C.E. The effects of food stoichiometry and temperature on copepods are mediated by ontogeny. Oecologia 2018, 188, 75–84. [Google Scholar] [CrossRef]
- Frankel-Bricker, J.; Song, M.; Benner, M.J.; Schaack, S. Variation in the Microbiota Associated with Daphnia magna Across Genotypes, Populations, and Temperature. Microb. Ecol. 2019, 79, 731–742. [Google Scholar] [CrossRef] [PubMed]
- Sullam, K.E.; Pichon, S.; Schaer, T.M.M.; Ebert, D. The Combined Effect of Temperature and Host Clonal Line on the Microbiota of a Planktonic Crustacean. Microb. Ecol. 2017, 76, 506–517. [Google Scholar] [CrossRef] [PubMed]
- Eckert, E.M.; Anicic, N.; Fontaneto, D. Freshwater zooplankton microbiome composition is highly flexible and strongly influenced by the environment. Mol. Ecol. 2021, 30, 1545–1558. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Cao, X.; Zeng, J.; Li, H.; Zhao, D.; Wu, Q.L. Distinct shifts in bacterioplankton community composition and functional gene structure between macrophyte- and phytoplankton-dominated regimes in a large shallow lake. Limnol. Oceanogr. 2020, 65, 11373. [Google Scholar] [CrossRef]
- Wu, Q.L.; Zwart, G.; Wu, J.; Agterveld, M.P.K.-V.; Liu, S.; Hahn, M.W. Submersed macrophytes play a key role in structuring bacterioplankton community composition in the large, shallow, subtropical Taihu Lake, China. Environ. Microbiol. 2010, 9, 2765–2774. [Google Scholar] [CrossRef]
- Meunier, C.L.; Boersma, M.; Wiltshire, K.; Malzahn, A. Zooplankton eat what they need: Copepod selective feeding and potential consequences for marine systems. Oikos 2016, 125, 50–58. [Google Scholar] [CrossRef]
- Malkiel, E.; Sheng, J.; Katz, J.; Strickler, J.R. The three-dimensional flow field generated by a feeding calanoid copepod measured using digital holography. J. Exp. Biol. 2003, 206, 3657–3666. [Google Scholar] [CrossRef]
- Smirnov, N.N. Excretion, Physiology of the Cladocera, 2nd ed.; Academic Press: Cambridge, CA, USA, 2017; Chapter 7; pp. 113–119. ISBN 9780128051948. [Google Scholar] [CrossRef]
- Shoemaker, K.M.; Duhamel, S.; Moisander, P.H. Copepods promote bacterial community changes in surrounding seawater through farming and nutrient enrichment. Environ. Microbiol. 2019, 21, 3737–3750. [Google Scholar] [CrossRef]
- He, X.; Wang, W.-X. Relative importance of inefficient feeding and consumer excretion to organic carbon flux from Daphnia. Freshw. Biol. 2006, 51, 1911–1923. [Google Scholar] [CrossRef]
- Seuront, L.; Yamazaki, H.; Souissi, S. Hydrodynamic disturbance and zooplankton swimming behavior. Zool. Stud. 2004, 43, 376–387. [Google Scholar]
- Peerakietkhajorn, S.; Kato, Y.; Kasalický, V.; Matsuura, T.; Watanabe, H. BetaproteobacteriaLimnohabitansstrains increase fecundity in the crustacean Daphnia magna: Symbiotic relationship between major bacterioplankton and zooplankton in freshwater ecosystem. Environ. Microbiol. 2015, 18, 2366–2374. [Google Scholar] [CrossRef]
- Perga, M.-E.; Kainz, M.; Matthews, B.; Mazumder, A. Carbon pathways to zooplankton: Insights from the combined use of stable isotope and fatty acid biomarkers. Freshw. Biol. 2006, 51, 2041–2051. [Google Scholar] [CrossRef]
- Kainz, M.J.; Perga, M.E.; Arts, M.T.; Mazumder, A. Essential fatty acid concentrations of different seston sizes and zooplankton: A field study of monomictic coastal lakes. J. Plankton Res. 2009, 6, 635–645. [Google Scholar] [CrossRef]
- De Corte, D.; Srivastava, A.; Koski, M.; Garcia, J.A.L.; Takaki, Y.; Yokokawa, T.; Nunoura, T.; Elisabeth, N.H.; Sintes, E.; Herndl, G.J. Metagenomic insights into zooplankton-associated bacterial communities. Environ. Microbiol. 2017, 20, 492–505. [Google Scholar] [CrossRef] [PubMed]
- Loreau, M. Linking biodiversity and ecosystems: Towards a unifying ecological theory. Philos. Trans. R. Soc. B: Biol. Sci. 2010, 365, 49–60. [Google Scholar] [CrossRef]
- Bruno, J.F.; Stachowicz, J.J.; Bertness, M.D. Inclusion of facilitation into ecological theory. Trends Ecol. Evol. 2003, 18, 119–125. [Google Scholar] [CrossRef]
- Becker, J.; Eisenhauer, N.; Scheu, S.; Jousset, A. Increasing antagonistic interactions cause bacterial communities to collapse at high diversity. Ecol. Lett. 2012, 15, 468–474. [Google Scholar] [CrossRef] [PubMed]
- Koeppel, A.; Wu, M. Species matter: The role of competition in the assembly of congeneric bacteria. ISME J. 2014, 8, 531–540. [Google Scholar] [CrossRef]
- Zhou, J.; Qin, B.; Zhu, G.; Zhang, Y.; Gao, G. Long-term variation of zooplankton communities in a large, heterogenous lake: Implications for future environmental change scenarios. Environ. Res. 2020, 187, 109704. [Google Scholar] [CrossRef]
- Braun, S.; Proctor, L.; Zani, S.; Mellon, M.; Zehr, J. Molecular evidence for zooplankton-associated nitrogen-fixing anaerobes based on amplification of the nifH gene. FEMS Microbiol. Ecol. 1999, 28, 273–279. [Google Scholar] [CrossRef][Green Version]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Q.; Hao, Z.; Ding, R.; Li, H.; Tang, X.; Chen, F. Host Dependence of Zooplankton-Associated Microbes and Their Ecological Implications in Freshwater Lakes. Water 2021, 13, 2949. https://doi.org/10.3390/w13212949
Wang Q, Hao Z, Ding R, Li H, Tang X, Chen F. Host Dependence of Zooplankton-Associated Microbes and Their Ecological Implications in Freshwater Lakes. Water. 2021; 13(21):2949. https://doi.org/10.3390/w13212949
Chicago/Turabian StyleWang, Qianhong, Zheng Hao, Ruirui Ding, Huabing Li, Xiangming Tang, and Feizhou Chen. 2021. "Host Dependence of Zooplankton-Associated Microbes and Their Ecological Implications in Freshwater Lakes" Water 13, no. 21: 2949. https://doi.org/10.3390/w13212949
APA StyleWang, Q., Hao, Z., Ding, R., Li, H., Tang, X., & Chen, F. (2021). Host Dependence of Zooplankton-Associated Microbes and Their Ecological Implications in Freshwater Lakes. Water, 13(21), 2949. https://doi.org/10.3390/w13212949