Towards the Optimization of eDNA/eRNA Sampling Technologies for Marine Biosecurity Surveillance
Abstract
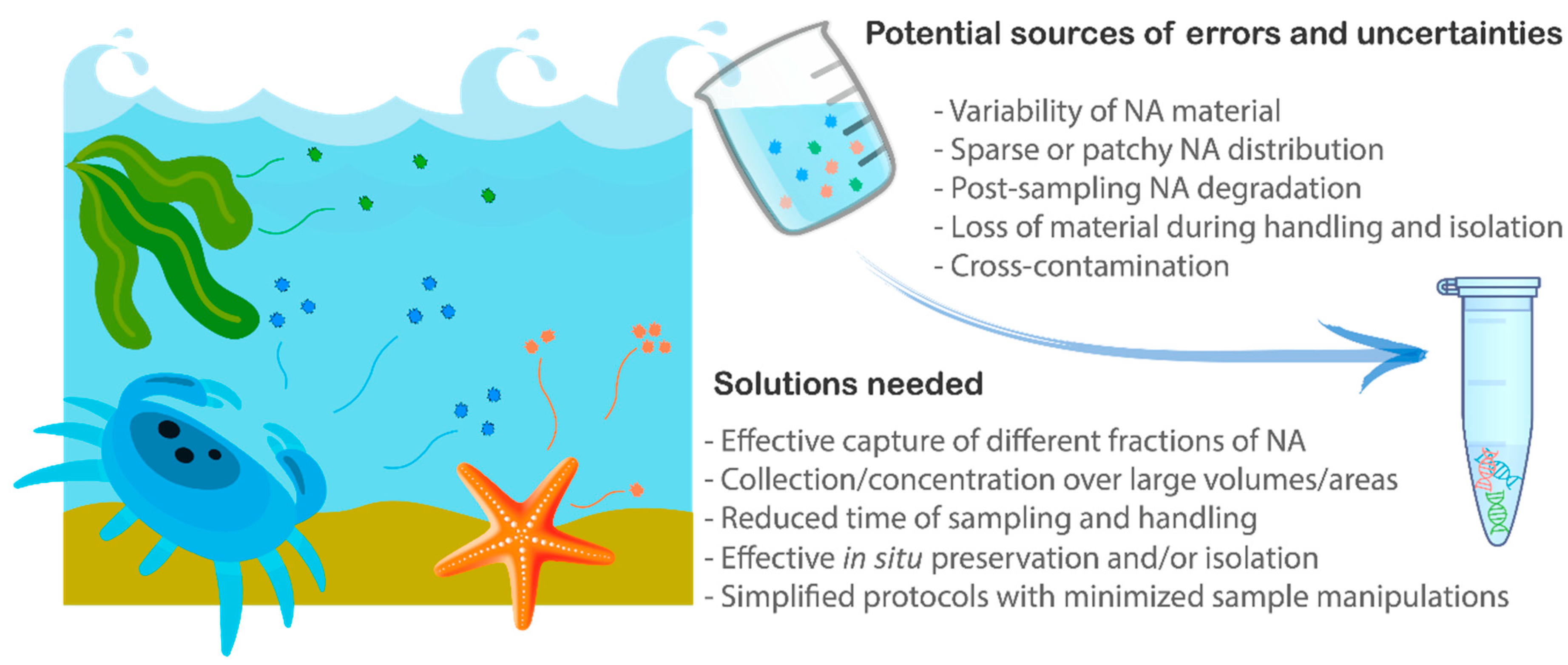
:1. Introduction
2. Applications of Molecular Tools for Marine Biosecurity Surveillance
3. Sample Collection
4. Post-Sampling Capture and Concentration of eDNA
5. Preservation
6. Extraction of Nucleic Acids
7. Outlook
8. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ojaveer, H.; Olenin, S.; Narščius, A.; Florin, A.-B.; Ezhova, E.; Gollasch, S.; Jensen, K.R.; Lehtiniemi, M.; Minchin, D.; Normant-Saremba, M.; et al. Dynamics of Biological Invasions and Pathways over Time: A Case Study of a Temperate Coastal Sea. Biol. Invasions 2017, 19, 799–813. [Google Scholar] [CrossRef]
- Cohen, A.N.; Carlton, J.T. Accelerating Invasion Rate in a Highly Invaded Estuary. Science 1998, 279, 555–558. [Google Scholar] [CrossRef] [Green Version]
- Seebens, H.; Blackburn, T.M.; Dyer, E.E.; Genovesi, P.; Hulme, P.E.; Jeschke, J.M.; Pagad, S.; Pyšek, P.; Winter, M.; Arianoutsou, M.; et al. No Saturation in the Accumulation of Alien Species Worldwide. Nat. Commun. 2017, 8, 14435. [Google Scholar] [CrossRef]
- Seebens, H.; Schwartz, N.; Schupp, P.J.; Blasius, B. Predicting the Spread of Marine Species Introduced by Global Shipping. Proc. Natl. Acad. Sci. USA 2016, 113, 5646–5651. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ojaveer, H.; Galil, B.S.; Carlton, J.T.; Alleway, H.; Goulletquer, P.; Lehtiniemi, M.; Marchini, A.; Miller, W.; Occhipinti-Ambrogi, A.; Peharda, M.; et al. Historical Baselines in Marine Bioinvasions: Implications for Policy and Management. PLos ONE 2018, 13, e0202383. [Google Scholar] [CrossRef] [Green Version]
- Wonham, M.; Walton, W.; Ruiz, G.; Frese, A.; Galil, B. Going to the Source: Role of the Invasion Pathway in Determining Potential Invaders. Mar. Ecol. Prog. Ser. 2001, 215, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Leppäkoski, E.; Gollasch, S.; Olenin, S. Invasive Aquatic Species of Europe. Distribution, Impacts and Management; Springer: Dordrecht, The Netherlands, 2002; ISBN 978-94-015-9956-6. [Google Scholar]
- Sardain, A.; Sardain, E.; Leung, B. Global Forecasts of Shipping Traffic and Biological Invasions to 2050. Nat. Sustain. 2019, 2, 274–282. [Google Scholar] [CrossRef]
- Larson, E.R.; Graham, B.M.; Achury, R.; Coon, J.J.; Daniels, M.K.; Gambrell, D.K.; Jonasen, K.L.; King, G.D.; LaRacuente, N.; Perrin-Stowe, T.I.; et al. From EDNA to Citizen Science: Emerging Tools for the Early Detection of Invasive Species. Front. Ecol. Environ. 2020, 18, 194–202. [Google Scholar] [CrossRef]
- Hebert, P.D.N.; Cywinska, A.; Ball, S.L.; deWaard, J.R. Biological Identifications through DNA Barcodes. Proc. R. Soc. Lond. B 2003, 270, 313–321. [Google Scholar] [CrossRef] [Green Version]
- Hajibabaei, M.; Singer, G.A.C.; Hebert, P.D.N.; Hickey, D.A. DNA Barcoding: How It Complements Taxonomy, Molecular Phylogenetics and Population Genetics. Trends Genet. 2007, 23, 167–172. [Google Scholar] [CrossRef]
- Wood, S.A.; Biessy, L.; Latchford, J.L.; Zaiko, A.; von Ammon, U.; Audrezet, F.; Cristescu, M.E.; Pochon, X. Release and Degradation of Environmental DNA and RNA in a Marine System. Sci. Total Environ. 2020, 704, 135314. [Google Scholar] [CrossRef]
- Baird, D.J.; Hajibabaei, M. Biomonitoring 2.0: A New Paradigm in Ecosystem Assessment Made Possible by next-Generation DNA Sequencing: NEWS AND VIEWS: OPINION. Mol. Ecol. 2012, 21, 2039–2044. [Google Scholar] [CrossRef]
- Pawlowski, J.; Kelly-Quinn, M.; Altermatt, F.; Apothéloz-Perret-Gentil, L.; Beja, P.; Boggero, A.; Borja, A.; Bouchez, A.; Cordier, T.; Domaizon, I.; et al. The Future of Biotic Indices in the Ecogenomic Era: Integrating (e)DNA Metabarcoding in Biological Assessment of Aquatic Ecosystems. Sci. Total Environ. 2018, 637–638, 1295–1310. [Google Scholar] [CrossRef]
- Ansorge, W.J. Next-Generation DNA Sequencing Techniques. New Biotechnol. 2009, 25, 195–203. [Google Scholar] [CrossRef]
- Zhou, X.; Li, Y.; Liu, S.; Yang, Q.; Su, X.; Zhou, L.; Tang, M.; Fu, R.; Li, J.; Huang, Q. Ultra-Deep Sequencing Enables High-Fidelity Recovery of Biodiversity for Bulk Arthropod Samples without PCR Amplification. GigaSci 2013, 2, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keeley, N.; Wood, S.A.; Pochon, X. Development and Preliminary Validation of a Multi-Trophic Metabarcoding Biotic Index for Monitoring Benthic Organic Enrichment. Ecol. Indic. 2018, 85, 1044–1057. [Google Scholar] [CrossRef]
- Sepulveda, A.J.; Nelson, N.M.; Jerde, C.L.; Luikart, G. Are Environmental DNA Methods Ready for Aquatic Invasive Species Management? Trends Ecol. Evol. 2020, 35, 668–678. [Google Scholar] [CrossRef]
- van der Heyde, M.; Bunce, M.; Wardell-Johnson, G.; Fernandes, K.; White, N.E.; Nevill, P. Testing Multiple Substrates for Terrestrial Biodiversity Monitoring Using Environmental DNA Metabarcoding. Mol. Ecol. Resour 2020, 20, 732–745. [Google Scholar] [CrossRef] [PubMed]
- Vander Zanden, M.J.; Hansen, G.J.A.; Higgins, S.N.; Kornis, M.S. A Pound of Prevention, plus a Pound of Cure: Early Detection and Eradication of Invasive Species in the Laurentian Great Lakes. J. Great Lakes Res. 2010, 36, 199–205. [Google Scholar] [CrossRef]
- Trebitz, A.S.; Hoffman, J.C.; Darling, J.A.; Pilgrim, E.M.; Kelly, J.R.; Brown, E.A.; Chadderton, W.L.; Egan, S.P.; Grey, E.K.; Hashsham, S.A.; et al. Early Detection Monitoring for Aquatic Non-Indigenous Species: Optimizing Surveillance, Incorporating Advanced Technologies, and Identifying Research Needs. J. Environ. Manag. 2017, 202, 299–310. [Google Scholar] [CrossRef]
- Lyal, C.H.C.; Miller, S.E. Capacity of United States Federal Government and Its Partners to Rapidly and Accurately Report the Identity (Taxonomy) of Non-Native Organisms Intercepted in Early Detection Programs. Biol. Invasions 2020, 22, 101–127. [Google Scholar] [CrossRef] [Green Version]
- Chain, F.J.J.; Brown, E.A.; MacIsaac, H.J.; Cristescu, M.E. Metabarcoding Reveals Strong Spatial Structure and Temporal Turnover of Zooplankton Communities among Marine and Freshwater Ports. Divers. Distrib. 2016, 22, 493–504. [Google Scholar] [CrossRef] [Green Version]
- von Ammon, U.; Wood, S.A.; Laroche, O.; Zaiko, A.; Tait, L.; Lavery, S.; Inglis, G.; Pochon, X. The Impact of Artificial Surfaces on Marine Bacterial and Eukaryotic Biofouling Assemblages: A High-Throughput Sequencing Analysis. Mar. Environ. Res. 2018, 133, 57–66. [Google Scholar] [CrossRef] [PubMed]
- Brys, R.; Halfmaerten, D.; Neyrinck, S.; Mauvisseau, Q.; Auwerx, J.; Sweet, M.; Mergeay, J. Reliable EDNA Detection and Quantification of the European Weather Loach ( Misgurnus Fossilis ). J. Fish. Biol. 2020, jfb.14315. [Google Scholar] [CrossRef]
- Witzel, N.A.; Taheri, A.; Miller, B.T.; Hardman, R.H.; Withers, D.I.; Spear, S.F.; Sutton, W.B. Validation of an Environmental DNA Protocol to Detect a Stream-breeding Amphibian, the Streamside Salamander (Ambystoma Barbouri). Environ. DNA 2020, 2, 554–564. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.; Aw, T.G.; Teal, T.K.; Rose, J.B. Metagenomic Investigation of Viral Communities in Ballast Water. Environ. Sci. Technol. 2015, 49, 8396–8407. [Google Scholar] [CrossRef] [Green Version]
- Darling, J.A.; Martinson, J.; Gong, Y.; Okum, S.; Pilgrim, E.; Lohan, K.M.P.; Carney, K.J.; Ruiz, G.M. Ballast Water Exchange and Invasion Risk Posed by Intracoastal Vessel Traffic: An Evaluation Using High Throughput Sequencing. Environ. Sci. Technol. 2018, 52, 9926–9936. [Google Scholar] [CrossRef] [Green Version]
- Deiner, K.; Walser, J.-C.; Mächler, E.; Altermatt, F. Choice of Capture and Extraction Methods Affect Detection of Freshwater Biodiversity from Environmental DNA. Biol. Conserv. 2015, 183, 53–63. [Google Scholar] [CrossRef]
- Wood, S.A.; Pochon, X.; Ming, W.; von Ammon, U.; Woods, C.; Carter, M.; Smith, M.; Inglis, G.; Zaiko, A. Considerations for Incorporating Real-Time PCR Assays into Routine Marine Biosecurity Surveillance Programmes: A Case Study Targeting the Mediterranean Fanworm ( Sabella Spallanzanii ) and Club Tunicate ( Styela Clava ). Genome 2019, 62, 137–146. [Google Scholar] [CrossRef] [Green Version]
- Darling, J.A.; Mahon, A.R. From Molecules to Management: Adopting DNA-Based Methods for Monitoring Biological Invasions in Aquatic Environments. Environ. Res. 2011, 111, 978–988. [Google Scholar] [CrossRef] [PubMed]
- Murray, D.C.; Coghlan, M.L.; Bunce, M. From Benchtop to Desktop: Important Considerations When Designing Amplicon Sequencing Workflows. PLos ONE 2015, 10, e0124671. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Furlan, E.M.; Davis, J.; Duncan, R.P. Identifying Error and Accurately Interpreting Environmental DNA Metabarcoding Results: A Case Study to Detect Vertebrates at Arid Zone Waterholes. Mol. Ecol. Resour. 2020, 20, 1259–1276. [Google Scholar] [CrossRef]
- Thalinger, B.; Deiner, K.; Harper, L.R.; Rees, H.C.; Blackman, R.C.; Sint, D.; Traugott, M.; Goldberg, C.S.; Bruce, K. A Validation Scale to Determine the Readiness of Environmental DNA Assays for Routine Species Monitoring; Molecular Biology: Hoboken, NJ, USA, 2020. [Google Scholar]
- Darling, J.A.; Pochon, X.; Abbott, C.L.; Inglis, G.J.; Zaiko, A. The Risks of Using Molecular Biodiversity Data for Incidental Detection of Species of Concern. Divers. Distrib. 2020, 26, 1116–1121. [Google Scholar] [CrossRef]
- Kumar, G.; Eble, J.E.; Gaither, M.R. A Practical Guide to Sample Preservation and Pre-PCR Processing of Aquatic Environmental DNA. Mol. Ecol. Resour. 2020, 20, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Tsuri, K.; Ikeda, S.; Hirohara, T.; Shimada, Y.; Minamoto, T.; Yamanaka, H. Messenger RNA Typing of Environmental RNA (ERNA): A Case Study on Zebrafish Tank Water with Perspectives for the Future Development of ERNA Analysis on Aquatic Vertebrates. Environ. DNA 2021, 3, 14–21. [Google Scholar] [CrossRef]
- Bott, N.J.; Ophel-Keller, K.M.; Sierp, M.T.; Herdina; Rowling, K.P.; McKay, A.C.; Loo, M.G.K.; Tanner, J.E.; Deveney, M.R. Toward Routine, DNA-Based Detection Methods for Marine Pests. Biotechnol. Adv. 2010, 28, 706–714. [Google Scholar] [CrossRef]
- Wood, S.; Smith, K.; Banks, J.; Tremblay, L.; Rhodes, L.; Mountfort, D.; Cary, S.; Pochon, X. Molecular Genetic Tools for Environmental Monitoring of New Zealand’s Aquatic Habitats, Past, Present and the Future. New Zealand J. Mar. Freshw. Res. 2013, 47, 90–119. [Google Scholar] [CrossRef] [Green Version]
- Rius, M.; Bourne, S.; Hornsby, H.G.; Chapman, M.A. Applications of Next-Generation Sequencing to the Study of Biological Invasions. Curr. Zool. 2015, 61, 488–504. [Google Scholar] [CrossRef] [Green Version]
- Zaiko, A.; Pochon, X.; Garcia-Vazquez, E.; Olenin, S.; Wood, S.A. Advantages and Limitations of Environmental DNA/RNA Tools for Marine Biosecurity: Management and Surveillance of Non-Indigenous Species. Front. Mar. Sci. 2018, 5, 322. [Google Scholar] [CrossRef] [Green Version]
- Richardson, M.F.; Sherman, C.D.H.; Lee, R.S.; Bott, N.J.; Hirst, A.J. Multiple Dispersal Vectors Drive Range Expansion in an Invasive Marine Species. Mol. Ecol. 2016, 25, 5001–5014. [Google Scholar] [CrossRef] [PubMed]
- Beng, K.C.; Corlett, R.T. Applications of Environmental DNA (EDNA) in Ecology and Conservation: Opportunities, Challenges and Prospects. Biodivers. Conserv. 2020, 29, 2089–2121. [Google Scholar] [CrossRef]
- Furlan, E.M.; Gleeson, D. Improving Reliability in Environmental DNA Detection Surveys through Enhanced Quality Control. Mar. Freshw. Res. 2017, 68, 388. [Google Scholar] [CrossRef]
- Foote, A.D.; Thomsen, P.F.; Sveegaard, S.; Wahlberg, M.; Kielgast, J.; Kyhn, L.A.; Salling, A.B.; Galatius, A.; Orlando, L.; Gilbert, M.T.P. Investigating the Potential Use of Environmental DNA (EDNA) for Genetic Monitoring of Marine Mammals. PLos ONE 2012, 7, e41781. [Google Scholar] [CrossRef]
- Valentini, A.; Taberlet, P.; Miaud, C.; Civade, R.; Herder, J.; Thomsen, P.F.; Bellemain, E.; Besnard, A.; Coissac, E.; Boyer, F.; et al. Next-Generation Monitoring of Aquatic Biodiversity Using Environmental DNA Metabarcoding. Mol. Ecol. 2016, 25, 929–942. [Google Scholar] [CrossRef] [Green Version]
- Mathieu, C.; Hermans, S.M.; Lear, G.; Buckley, T.R.; Lee, K.C.; Buckley, H.L. A Systematic Review of Sources of Variability and Uncertainty in EDNA Data for Environmental Monitoring. Front. Ecol. Evol. 2020, 8, 135. [Google Scholar] [CrossRef]
- Taberlet, P.; Coissac, E.; Hajibabaei, M.; Rieseberg, L.H. Environmental DNA: ENVIRONMENTAL DNA. Mol. Ecol. 2012, 21, 1789–1793. [Google Scholar] [CrossRef] [PubMed]
- Pochon, X.; Zaiko, A.; Fletcher, L.M.; Laroche, O.; Wood, S.A. Wanted Dead or Alive? Using Metabarcoding of Environmental DNA and RNA to Distinguish Living Assemblages for Biosecurity Applications. PLos ONE 2017, 12, e0187636. [Google Scholar] [CrossRef] [PubMed]
- Cristescu, M.E. Can Environmental RNA Revolutionize Biodiversity Science? Trends Ecol. Evol. 2019, 34, 694–697. [Google Scholar] [CrossRef]
- Kitahashi, T.; Sugime, S.; Inomata, K.; Nishijima, M.; Kato, S.; Yamamoto, H. Meiofaunal Diversity at a Seamount in the Pacific Ocean: A Comprehensive Study Using Environmental DNA and RNA. Deep Sea Res. Part. I: Oceanogr. Res. Pap. 2020, 160, 103253. [Google Scholar] [CrossRef]
- Brandt, M.I.; Trouche, B.; Henry, N.; Liautard-Haag, C.; Maignien, L.; de Vargas, C.; Wincker, P.; Poulain, J.; Zeppilli, D.; Arnaud-Haond, S. An Assessment of Environmental Metabarcoding Protocols Aiming at Favoring Contemporary Biodiversity in Inventories of Deep-Sea Communities. Front. Mar. Sci. 2020, 7, 234. [Google Scholar] [CrossRef]
- Zaiko, A.; Wood, S.A.; Pochon, X.; Biessy, L.; Laroche, O.; Croot, P.; Garcia-Vazquez, E. Elucidating Biodiversity Shifts in Ballast Water Tanks during a Cross-Latitudinal Transfer: Complementary Insights from Molecular Analyses. Environ. Sci. Technol. 2020, 54, 8443–8454. [Google Scholar] [CrossRef]
- Dell’Anno, A.; Danovaro, R. Extracellular DNA Plays a Key Role in Deep-Sea Ecosystem Functioning. Science 2005, 309, 2179. [Google Scholar] [CrossRef]
- Corinaldesi, C.; Beolchini, F.; Dell’Anno, A. Damage and Degradation Rates of Extracellular DNA in Marine Sediments: Implications for the Preservation of Gene Sequences. Mol. Ecol. 2008, 17, 3939–3951. [Google Scholar] [CrossRef]
- Ficetola, G.F.; Poulenard, J.; Sabatier, P.; Messager, E.; Gielly, L.; Leloup, A.; Etienne, D.; Bakke, J.; Malet, E.; Fanget, B.; et al. DNA from Lake Sediments Reveals Long-Term Ecosystem Changes after a Biological Invasion. Sci. Adv. 2018, 4, eaar4292. [Google Scholar] [CrossRef] [Green Version]
- Andruszkiewicz, E.A.; Sassoubre, L.M.; Boehm, A.B. Persistence of Marine Fish Environmental DNA and the Influence of Sunlight. PLos ONE 2017, 12, e0185043. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pinfield, R.; Dillane, E.; Runge, A.K.W.; Evans, A.; Mirimin, L.; Niemann, J.; Reed, T.E.; Reid, D.G.; Rogan, E.; Samarra, F.I.P.; et al. False-negative Detections from Environmental DNA Collected in the Presence of Large Numbers of Killer Whales ( Orcinus Orca ). Environ. DNA 2019, 1, 316–328. [Google Scholar] [CrossRef] [Green Version]
- Murakami, H.; Yoon, S.; Kasai, A.; Minamoto, T.; Yamamoto, S.; Sakata, M.K.; Horiuchi, T.; Sawada, H.; Kondoh, M.; Yamashita, Y.; et al. Dispersion and Degradation of Environmental DNA from Caged Fish in a Marine Environment. Fish. Sci. 2019, 85, 327–337. [Google Scholar] [CrossRef]
- Barnes, M.A.; Turner, C.R.; Jerde, C.L.; Renshaw, M.A.; Chadderton, W.L.; Lodge, D.M. Environmental Conditions Influence EDNA Persistence in Aquatic Systems. Environ. Sci. Technol. 2014, 48, 1819–1827. [Google Scholar] [CrossRef]
- Strickler, K.M.; Fremier, A.K.; Goldberg, C.S. Quantifying Effects of UV-B, Temperature, and PH on EDNA Degradation in Aquatic Microcosms. Biol. Conserv. 2015, 183, 85–92. [Google Scholar] [CrossRef]
- Harrison, J.B.; Sunday, J.M.; Rogers, S.M. Predicting the Fate of EDNA in the Environment and Implications for Studying Biodiversity. Proc. R. Soc. B. 2019, 286, 20191409. [Google Scholar] [CrossRef] [PubMed]
- Jo, T.; Murakami, H.; Yamamoto, S.; Masuda, R.; Minamoto, T. Effect of Water Temperature and Fish Biomass on Environmental DNA Shedding, Degradation, and Size Distribution. Ecol. Evol 2019, 9, 1135–1146. [Google Scholar] [CrossRef] [Green Version]
- Adrian-Kalchhauser, I.; Burkhardt-Holm, P. An EDNA Assay to Monitor a Globally Invasive Fish Species from Flowing Freshwater. PLos ONE 2016, 11, e0147558. [Google Scholar] [CrossRef] [Green Version]
- Kasai, A.; Takada, S.; Yamazaki, A.; Masuda, R.; Yamanaka, H. The Effect of Temperature on Environmental DNA Degradation of Japanese Eel. Fish. Sci. 2020, 86, 465–471. [Google Scholar] [CrossRef]
- Rey, A.; Basurko, O.C.; Rodriguez-Ezpeleta, N. Considerations for Metabarcoding-based Port Biological Baseline Surveys Aimed at Marine Nonindigenous Species Monitoring and Risk Assessments. Ecol. Evol. 2020, 10, 2452–2465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bouchet, P.; Phillips, C.; Huang, Z.; Meeuwig, J.; Foster, S.; Przeslawski, R. Comparative Assessment of Pelagic Sampling Methods Used in Marine Monitoring. Report to the National Environmental Science Programme; Marine Biodiversity Hub, 2018; p. 149. Available online: https://www.researchgate.net/profile/Phil-Bouchet/publication/327237111_Comparative_assessment_of_pelagic_sampling_methods_used_in_marine_monitoring/links/5b836fce4585151fd134f890/Comparative-assessment-of-pelagic-sampling-methods-used-in-marine-monitoring.pdf (accessed on 16 April 2021).
- Koziol, A.; Stat, M.; Simpson, T.; Jarman, S.; DiBattista, J.D.; Harvey, E.S.; Marnane, M.; McDonald, J.; Bunce, M. Environmental DNA Metabarcoding Studies Are Critically Affected by Substrate Selection. Mol. Ecol. Resour. 2019, 19, 366–376. [Google Scholar] [CrossRef]
- von Ammon, U.; Wood, S.A.; Laroche, O.; Zaiko, A.; Lavery, S.D.; Inglis, G.J.; Pochon, X. Linking Environmental DNA and RNA for Improved Detection of the Marine Invasive Fanworm Sabella Spallanzanii. Front. Mar. Sci. 2019, 6, 621. [Google Scholar] [CrossRef]
- Schabacker, J.C.; Amish, S.J.; Ellis, B.K.; Gardner, B.; Miller, D.L.; Rutledge, E.A.; Sepulveda, A.J.; Luikart, G. Increased EDNA Detection Sensitivity Using a Novel High-volume Water Sampling Method. Environ. DNA 2020, 2, 244–251. [Google Scholar] [CrossRef] [Green Version]
- Lacoursière-Roussel, A.; Howland, K.; Normandeau, E.; Grey, E.K.; Archambault, P.; Deiner, K.; Lodge, D.M.; Hernandez, C.; Leduc, N.; Bernatchez, L. eDNA Metabarcoding as a New Surveillance Approach for Coastal Arctic Biodiversity. Ecol. Evol. 2018, 8, 7763–7777. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holman, L.E.; de Bruyn, M.; Creer, S.; Carvalho, G.; Robidart, J.; Rius, M. Detection of Introduced and Resident Marine Species Using Environmental DNA Metabarcoding of Sediment and Water. Sci. Rep. 2019, 9, 11559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Antich, A.; Palacín, C.; Cebrian, E.; Golo, R.; Wangensteen, O.S.; Turon, X. Marine Biomonitoring with EDNA: Can Metabarcoding of Water Samples Cut It as a Tool for Surveying Benthic Communities? Mol. Ecol. 2020, mec.15641. [Google Scholar] [CrossRef]
- Outinen, O.; Forsström, T.; Yli-Rosti, J.; Vesakoski, O.; Lehtiniemi, M. Monitoring of Sessile and Mobile Epifauna—Considerations for Non-Indigenous Species. Mar. Pollut. Bull. 2019, 141, 332–342. [Google Scholar] [CrossRef]
- Pearman, J.K.; Ammon, U.; Laroche, O.; Zaiko, A.; Wood, S.A.; Zubia, M.; Planes, S.; Pochon, X. Metabarcoding as a Tool to Enhance Marine Surveillance of Nonindigenous Species in Tropical Harbors: A Case Study in Tahiti. Environ. DNA 2021, 3, 173–189. [Google Scholar] [CrossRef]
- Grey, E.K.; Bernatchez, L.; Cassey, P.; Deiner, K.; Deveney, M.; Howland, K.L.; Lacoursière-Roussel, A.; Leong, S.C.Y.; Li, Y.; Olds, B.; et al. Effects of Sampling Effort on Biodiversity Patterns Estimated from Environmental DNA Metabarcoding Surveys. Sci. Rep. 2018, 8, 8843. [Google Scholar] [CrossRef] [Green Version]
- Aylagas, E.; Borja, Á.; Muxika, I.; Rodríguez-Ezpeleta, N. Adapting Metabarcoding-Based Benthic Biomonitoring into Routine Marine Ecological Status Assessment Networks. Ecol. Indic. 2018, 95, 194–202. [Google Scholar] [CrossRef]
- Pilliod, D.S.; Laramie, M.B.; MacCoy, D.; Maclean, S. Integration of EDNA-Based Biological Monitoring within the U.S. Geological Survey’s National Streamgage Network. J. Am. Water Resour Assoc. 2019, 55, 1505–1518. [Google Scholar] [CrossRef] [Green Version]
- von Ammon, U.; Jeffs, A.; Zaiko, A.; van der Rees, A.; Goodwin, D.; Beckley, L.E.; Malpot, E.; Pochon, X. A Portable Cruising Speed Net: Expanding Global Collection of Sea Surface Plankton Data. Frontiers in Marine Science. in press.
- Bessey, C.; Jarman, S.N.; Berry, O.; Olsen, Y.S.; Bunce, M.; Simpson, T.; Power, M.; McLaughlin, J.; Edgar, G.J.; Keesing, J. Maximizing Fish Detection with EDNA Metabarcoding. Environ. DNA 2020, 2, 493–504. [Google Scholar] [CrossRef]
- Carim, K.J.; Christianson, K.R.; McKelvey, K.M.; Pate, W.M.; Silver, D.B.; Johnson, B.M.; Galloway, B.T.; Young, M.K.; Schwartz, M.K. Environmental DNA Marker Development with Sparse Biological Information: A Case Study on Opossum Shrimp (Mysis Diluviana). PLos ONE 2016, 11, e0161664. [Google Scholar] [CrossRef] [Green Version]
- Djurhuus, A.; Port, J.; Closek, C.J.; Yamahara, K.M.; Romero-Maraccini, O.; Walz, K.R.; Goldsmith, D.B.; Michisaki, R.; Breitbart, M.; Boehm, A.B.; et al. Evaluation of Filtration and DNA Extraction Methods for Environmental DNA Biodiversity Assessments across Multiple Trophic Levels. Front. Mar. Sci. 2017, 4, 314. [Google Scholar] [CrossRef] [Green Version]
- Spens, J.; Evans, A.R.; Halfmaerten, D.; Knudsen, S.W.; Sengupta, M.E.; Mak, S.S.T.; Sigsgaard, E.E.; Hellström, M. Comparison of Capture and Storage Methods for Aqueous Macrobial eDNA Using an Optimized Extraction Protocol: Advantage of Enclosed Filter. Methods Ecol. Evol. 2017, 8, 635–645. [Google Scholar] [CrossRef] [Green Version]
- Jeunen, G.; Knapp, M.; Spencer, H.G.; Taylor, H.R.; Lamare, M.D.; Stat, M.; Bunce, M.; Gemmell, N.J. Species-level Biodiversity Assessment Using Marine Environmental DNA Metabarcoding Requires Protocol Optimization and Standardization. Ecol. Evol. 2019, 9, 1323–1335. [Google Scholar] [CrossRef] [Green Version]
- Collins, R.A.; Wangensteen, O.S.; O’Gorman, E.J.; Mariani, S.; Sims, D.W.; Genner, M.J. Persistence of Environmental DNA in Marine Systems. Commun. Biol. 2018, 1, 185. [Google Scholar] [CrossRef] [Green Version]
- Sassoubre, L.M.; Yamahara, K.M.; Gardner, L.D.; Block, B.A.; Boehm, A.B. Quantification of Environmental DNA (EDNA) Shedding and Decay Rates for Three Marine Fish. Environ. Sci. Technol. 2016, 50, 10456–10464. [Google Scholar] [CrossRef] [PubMed]
- Salter, I. Seasonal Variability in the Persistence of Dissolved Environmental DNA (EDNA) in a Marine System: The Role of Microbial Nutrient Limitation. PLos ONE 2018, 13, e0192409. [Google Scholar] [CrossRef] [Green Version]
- Thomas, A.C.; Nguyen, P.L.; Howard, J.; Goldberg, C.S. A Self-preserving, Partially Biodegradable EDNA Filter. Methods Ecol. Evol. 2019, 10, 1136–1141. [Google Scholar] [CrossRef] [Green Version]
- Yoerger, D.R.; Curran, M.; Fujii, J.; German, C.R.; Gomez-Ibanez, D.; Govindarajan, A.F.; Howland, J.C.; Llopiz, J.K.; Wiebe, P.H.; Hobson, B.W.; et al. Mesobot: An Autonomous Underwater Vehicle for Tracking and Sampling Midwater Targets. In Proceedings of the 2018 IEEE/OES Autonomous Underwater Vehicle Workshop (AUV), Porto, Portugal, 6–9 November 2018; pp. 1–7. [Google Scholar]
- Yamahara, K.M.; Preston, C.M.; Birch, J.; Walz, K.; Marin, R.; Jensen, S.; Pargett, D.; Roman, B.; Ussler, W.; Zhang, Y.; et al. In Situ Autonomous Acquisition and Preservation of Marine Environmental DNA Using an Autonomous Underwater Vehicle. Front. Mar. Sci. 2019, 6, 373. [Google Scholar] [CrossRef] [Green Version]
- Kirtane, A.; Atkinson, J.D.; Sassoubre, L. Design and Validation of Passive Environmental DNA Samplers Using Granular Activated Carbon and Montmorillonite Clay. Environ. Sci. Technol. 2020, 54, 11961–11970. [Google Scholar] [CrossRef] [PubMed]
- Leray, M.; Knowlton, N. DNA Barcoding and Metabarcoding of Standardized Samples Reveal Patterns of Marine Benthic Diversity. Proc. Natl. Acad. Sci. USA 2015, 112, 2076–2081. [Google Scholar] [CrossRef] [Green Version]
- Zaiko, A.; Schimanski, K.; Pochon, X.; Hopkins, G.A.; Goldstien, S.; Floerl, O.; Wood, S.A. Metabarcoding Improves Detection of Eukaryotes from Early Biofouling Communities: Implications for Pest Monitoring and Pathway Management. Biofouling 2016, 32, 671–684. [Google Scholar] [CrossRef] [PubMed]
- Turner, C.R.; Barnes, M.A.; Xu, C.C.Y.; Jones, S.E.; Jerde, C.L.; Lodge, D.M. Particle Size Distribution and Optimal Capture of Aqueous Macrobial eDNA. Methods Ecol. Evol. 2014, 5, 676–684. [Google Scholar] [CrossRef] [Green Version]
- Renshaw, M.A.; Olds, B.P.; Jerde, C.L.; McVeigh, M.M.; Lodge, D.M. The Room Temperature Preservation of Filtered Environmental DNA Samples and Assimilation into a Phenol–Chloroform–Isoamyl Alcohol DNA Extraction. Mol. Ecol. Resour 2015, 15, 168–176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eichmiller, J.J.; Best, S.E.; Sorensen, P.W. Effects of Temperature and Trophic State on Degradation of Environmental DNA in Lake Water. Environ. Sci. Technol. 2016, 50, 1859–1867. [Google Scholar] [CrossRef]
- Minamoto, T.; Fukuda, M.; Katsuhara, K.R.; Fujiwara, A.; Hidaka, S.; Yamamoto, S.; Takahashi, K.; Masuda, R. Environmental DNA Reflects Spatial and Temporal Jellyfish Distribution. PLos ONE 2017, 12, e0173073. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deiner, K.; Lopez, J.; Bourne, S.; Holman, L.; Seymour, M.; Grey, E.K.; Lacoursière, A.; Li, Y.; Renshaw, M.A.; Pfrender, M.E.; et al. Optimising the Detection of Marine Taxonomic Richness Using Environmental DNA Metabarcoding: The Effects of Filter Material, Pore Size and Extraction Method. MBMG 2018, 2, e28963. [Google Scholar] [CrossRef]
- Capo, E.; Spong, G.; Königsson, H.; Byström, P. Effects of Filtration Methods and Water Volume on the Quantification of Brown Trout ( Salmo Trutta ) and Arctic Char ( Salvelinus Alpinus ) EDNA Concentrations via Droplet Digital PCR. Environ. DNA 2020, 2, 152–160. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Wikfors, G.H.; Rose, J.M.; McBride, R.S.; Milke, L.M.; Mercaldo-Allen, R. Application of Environmental DNA Metabarcoding to Spatiotemporal Finfish Community Assessment in a Temperate Embayment. Front. Mar. Sci. 2019, 6, 674. [Google Scholar] [CrossRef] [Green Version]
- Kelly, R.P.; Port, J.A.; Yamahara, K.M.; Crowder, L.B. Using Environmental DNA to Census Marine Fishes in a Large Mesocosm. PLos ONE 2014, 9, e86175. [Google Scholar] [CrossRef] [Green Version]
- Egan, S.P.; Barnes, M.A.; Hwang, C.-T.; Mahon, A.R.; Feder, J.L.; Ruggiero, S.T.; Tanner, C.E.; Lodge, D.M. Rapid Invasive Species Detection by Combining Environmental DNA with Light Transmission Spectroscopy: EDNA-LTS Species Detection. Conserv. Lett. 2013, 6, 402–409. [Google Scholar] [CrossRef]
- Barnes, M.A.; Chadderton, W.L.; Jerde, C.L.; Mahon, A.R.; Turner, C.R.; Lodge, D.M. Environmental Conditions Influence EDNA Particle Size Distribution in Aquatic Systems. Environ. DNA 2020, edn3.160. [Google Scholar] [CrossRef]
- Jane, S.F.; Wilcox, T.M.; McKelvey, K.S.; Young, M.K.; Schwartz, M.K.; Lowe, W.H.; Letcher, B.H.; Whiteley, A.R. Distance, Flow and PCR Inhibition: EDNA Dynamics in Two Headwater Streams. Mol. Ecol. Resour. 2015, 15, 216–227. [Google Scholar] [CrossRef]
- Hunter, M.E.; Ferrante, J.A.; Meigs-Friend, G.; Ulmer, A. Improving EDNA Yield and Inhibitor Reduction through Increased Water Volumes and Multi-Filter Isolation Techniques. Sci. Rep. 2019, 9, 5259. [Google Scholar] [CrossRef]
- Sepulveda, A.J.; Schabacker, J.; Smith, S.; Al-Chokhachy, R.; Luikart, G.; Amish, S.J. Improved Detection of Rare, Endangered and Invasive Trout in Using a New Large-volume Sampling Method for EDNA Capture. Environ. DNA 2019, 1, 227–237. [Google Scholar] [CrossRef]
- Wittwer, C.; Nowak, C.; Strand, D.A.; Vrålstad, T.; Thines, M.; Stoll, S. Comparison of Two Water Sampling Approaches for EDNA-Based Crayfish Plague Detection. Limnologica 2018, 70, 1–9. [Google Scholar] [CrossRef]
- Not, F.; del Campo, J.; Balagué, V.; de Vargas, C.; Massana, R. New Insights into the Diversity of Marine Picoeukaryotes. PLos ONE 2009, 4, e7143. [Google Scholar] [CrossRef] [Green Version]
- de Vargas, C.; Audic, S.; Henry, N.; Decelle, J.; Mahe, F.; Logares, R.; Lara, E.; Berney, C.; Le Bescot, N.; Probert, I.; et al. Eukaryotic Plankton Diversity in the Sunlit Ocean. Science 2015, 348, 1261605. [Google Scholar] [CrossRef] [Green Version]
- Moushomi, R.; Wilgar, G.; Carvalho, G.; Creer, S.; Seymour, M. Environmental DNA Size Sorting and Degradation Experiment Indicates the State of Daphnia Magna Mitochondrial and Nuclear EDNA Is Subcellular. Sci. Rep. 2019, 9, 12500. [Google Scholar] [CrossRef]
- Majaneva, M.; Diserud, O.H.; Eagle, S.H.C.; Boström, E.; Hajibabaei, M.; Ekrem, T. Environmental DNA Filtration Techniques Affect Recovered Biodiversity. Sci. Rep. 2018, 8, 4682. [Google Scholar] [CrossRef] [PubMed]
- Mahon, A.R.; Jerde, C.L.; Galaska, M.; Bergner, J.L.; Chadderton, W.L.; Lodge, D.M.; Hunter, M.E.; Nico, L.G. Validation of EDNA Surveillance Sensitivity for Detection of Asian Carps in Controlled and Field Experiments. PLos ONE 2013, 8, e58316. [Google Scholar] [CrossRef] [Green Version]
- Takahara, T.; Minamoto, T.; Doi, H. Using Environmental DNA to Estimate the Distribution of an Invasive Fish Species in Ponds. PLos ONE 2013, 8, e56584. [Google Scholar] [CrossRef] [Green Version]
- Jerde, C.L.; Mahon, A.R.; Chadderton, W.L.; Lodge, D.M. “Sight-Unseen” Detection of Rare Aquatic Species Using Environmental DNA: EDNA Surveillance of Rare Aquatic Species. Conserv. Lett. 2011, 4, 150–157. [Google Scholar] [CrossRef]
- Wilcox, T.M.; Carim, K.J.; Young, M.K.; McKelvey, K.S.; Franklin, T.W.; Schwartz, M.K. Comment: The Importance of Sound Methodology in Environmental DNA Sampling. North. Am. J. Fish. Manag. 2018, 38, 592–596. [Google Scholar] [CrossRef]
- Sales, N.G.; Wangensteen, O.S.; Carvalho, D.C.; Mariani, S. Influence of Preservation Methods, Sample Medium and Sampling Time on EDNA Recovery in a Neotropical River. Environ. DNA 2019, 1, 119–130. [Google Scholar] [CrossRef] [Green Version]
- Doi, H.; Uchii, K.; Matsuhashi, S.; Takahara, T.; Yamanaka, H.; Minamoto, T. Isopropanol Precipitation Method for Collecting Fish Environmental DNA. Limnol. Oceanogr. Methods 2017, 15, 212–218. [Google Scholar] [CrossRef] [Green Version]
- Goldberg, C.S.; Sepulveda, A.; Ray, A.; Baumgardt, J.; Waits, L.P. Environmental DNA as a New Method for Early Detection of New Zealand Mudsnails ( Potamopyrgus Antipodarum ). Freshw. Sci. 2013, 32, 792–800. [Google Scholar] [CrossRef] [Green Version]
- Pilliod, D.S.; Goldberg, C.S.; Arkle, R.S.; Waits, L.P. Estimating Occupancy and Abundance of Stream Amphibians Using Environmental DNA from Filtered Water Samples. Can. J. Fish. Aquat. Sci. 2013, 70, 1123–1130. [Google Scholar] [CrossRef]
- Hajibabaei, M.; Spall, J.L.; Shokralla, S.; van Konynenburg, S. Assessing Biodiversity of a Freshwater Benthic Macroinvertebrate Community through Non-Destructive Environmental Barcoding of DNA from Preservative Ethanol. BMC Ecol. 2012, 12, 28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamanaka, H.; Minamoto, T.; Matsuura, J.; Sakurai, S.; Tsuji, S.; Motozawa, H.; Hongo, M.; Sogo, Y.; Kakimi, N.; Teramura, I.; et al. A Simple Method for Preserving Environmental DNA in Water Samples at Ambient Temperature by Addition of Cationic Surfactant. Limnology 2017, 18, 233–241. [Google Scholar] [CrossRef] [Green Version]
- Seutin, G.; White, B.N.; Boag, P.T. Preservation of Avian Blood and Tissue Samples for DNA Analyses. Can. J. Zool. 1991, 69, 82–90. [Google Scholar] [CrossRef]
- Longmire, J.; Maltbie, M.; Baker, R. Use of ‘Lysis Buffer’ in DNA Isolation and Its Implications for Museum Collections. Mus. Tex. Tech. Univ. 1997, 163, 1–3. [Google Scholar]
- van der Loos, L.M.; Nijland, R. Biases in Bulk: DNA Metabarcoding of Marine Communities and the Methodology Involved. Mol. Ecol. 2020, mec.15592. [Google Scholar] [CrossRef]
- Mäki, A.; Salmi, P.; Mikkonen, A.; Kremp, A.; Tiirola, M. Sample Preservation, DNA or RNA Extraction and Data Analysis for High-Throughput Phytoplankton Community Sequencing. Front. Microbiol. 2017, 8, 1848. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gray, M.A.; Pratte, Z.A.; Kellogg, C.A. Comparison of DNA Preservation Methods for Environmental Bacterial Community Samples. FEMS MicroBiol. Ecol. 2013, 83, 468–477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wegleitner, B.J.; Jerde, C.L.; Tucker, A.; Chadderton, W.L.; Mahon, A.R. Long Duration, Room Temperature Preservation of Filtered EDNA Samples. Conserv. Genet. Resour. 2015, 7, 789–791. [Google Scholar] [CrossRef]
- Williams, K.E.; Huyvaert, K.P.; Piaggio, A.J. No Filters, No Fridges: A Method for Preservation of Water Samples for EDNA Analysis. BMC Res. Notes 2016, 9, 298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goldberg, C.S.; Turner, C.R.; Deiner, K.; Klymus, K.E.; Thomsen, P.F.; Murphy, M.A.; Spear, S.F.; McKee, A.; Oyler-McCance, S.J.; Cornman, R.S.; et al. Critical Considerations for the Application of Environmental DNA Methods to Detect Aquatic Species. Methods Ecol. Evol. 2016, 7, 1299–1307. [Google Scholar] [CrossRef]
- Bachoon, D.S.; Chen, F.; Hodson, R.E. RNA Recovery and Detection of MRNA by RT-PCR from Preserved Prokaryotic Samples. Fems Microbiol. Lett. 2001, 201, 127–132. [Google Scholar] [CrossRef]
- van Eijsden, R.G.E.; Stassen, C.; Daenen, L.; Van Mulders, S.E.; Bapat, P.M.; Siewers, V.; Goossens, K.V.Y.; Nielsen, J.; Delvaux, F.R.; Van Hummelen, P.; et al. A Universal Fixation Method Based on Quaternary Ammonium Salts (RNAlater) for Omics-Technologies: Saccharomyces Cerevisiae as a Case Study. Biotechnol. Lett. 2013, 35, 891–900. [Google Scholar] [CrossRef] [PubMed]
- Rissanen, A.J.; Kurhela, E.; Aho, T.; Oittinen, T.; Tiirola, M. Storage of Environmental Samples for Guaranteeing Nucleic Acid Yields for Molecular Microbiological Studies. Appl. MicroBiol. Biotechnol. 2010, 88, 977–984. [Google Scholar] [CrossRef] [PubMed]
- Dahm, R. Discovering DNA: Friedrich Miescher and the Early Years of Nucleic Acid Research. Hum. Genet. 2008, 122, 565–581. [Google Scholar] [CrossRef]
- Goldenberger, D.; Perschil, I.; Ritzler, M.; Altwegg, M. A Simple “Universal” DNA Extraction Procedure Using SDS and Proteinase K Is Compatible with Direct PCR Amplification. Genome Res. 1995, 4, 368–370. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Bruns, M.A.; Tiedje, J.M. DNA Recovery from Soils of Diverse Composition. Appl. Environ. Microbiol. 1996, 62, 316–322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ogram, A.; Sayler, G.S.; Barkay, T. The Extraction and Purification of Microbial DNA from Sediments. J. Microbiol. Methods 1987, 7, 57–66. [Google Scholar] [CrossRef]
- Tsai, Y.L.; Olson, B.H. Rapid Method for Direct Extraction of DNA from Soil and Sediments. Appl. Environ. Microbiol. 1991, 57, 1070–1074. [Google Scholar] [CrossRef] [Green Version]
- Natarajan, V.P.; Zhang, X.; Morono, Y.; Inagaki, F.; Wang, F. A Modified SDS-Based DNA Extraction Method for High Quality Environmental DNA from Seafloor Environments. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tkach, V.; Pawlowski, J.W. A New Method of DNA Extraction from the Ethanol-Fixed Parasitic Worms. Acta Parasitol. 1999, 44, 147–148. [Google Scholar]
- Porebski, S.; Bailey, L.G.; Baum, B.R. Modification of a CTAB DNA Extraction Protocol for Plants Containing High Polysaccharide and Polyphenol Components. Plant. Mol. Biol. Rep. 1997, 15, 8–15. [Google Scholar] [CrossRef]
- Sambrook, J.; Russell, D. Purification of Nucleic Acids by Extraction with Phenol: Chloroform. Cold Spring Harb. Protoc. 2006. [Google Scholar] [CrossRef]
- Porteous, L.A.; Seidler, R.J.; Watrud, L.S. An Improved Method for Purifying DNA from Soil for Polymerase Chain Reaction Amplification and Molecular Ecology Applications. Mol. Ecol. 1997, 6, 787–791. [Google Scholar] [CrossRef]
- Manoli, F.; Dalas, E. Spontaneous Precipitation of Calcium Carbonate in the Presence of Ethanol, Isopropanol and Diethylene Glycol. J. Cryst. Growth 2000, 218, 359–364. [Google Scholar] [CrossRef]
- Deiner, K.; Altermatt, F. Transport Distance of Invertebrate Environmental DNA in a Natural River. PLos ONE 2014, 9, e88786. [Google Scholar] [CrossRef] [Green Version]
- Lever, M.A.; Torti, A.; Eickenbusch, P.; Michaud, A.B.; Šantl-Temkiv, T.; Jørgensen, B.B. A Modular Method for the Extraction of DNA and RNA, and the Separation of DNA Pools from Diverse Environmental Sample Types. Front. Microbiol. 2015, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geerts, A.N.; Boets, P.; Van den Heede, S.; Goethals, P.; Van der heyden, C. A Search for Standardized Protocols to Detect Alien Invasive Crayfish Based on Environmental DNA (EDNA): A Lab and Field Evaluation. Ecol. Indic. 2018, 84, 564–572. [Google Scholar] [CrossRef]
- Pearman, J.K.; Keeley, N.B.; Wood, S.A.; Laroche, O.; Zaiko, A.; Thomson-Laing, G.; Biessy, L.; Atalah, J.; Pochon, X. Comparing Sediment DNA Extraction Methods for Assessing Organic Enrichment Associated with Marine Aquaculture. PeerJ 2020, 8, e10231. [Google Scholar] [CrossRef] [PubMed]
- Lear, G.; Dickie, I.; Banks, J.; Boyer, S.; Buckley, H.; Buckley, T.; Cruickshank, R.; Dopheide, A.; Handley, K.; Hermans, S.; et al. Methods for the Extraction, Storage, Amplification and Sequencing of DNA from Environmental Samples. NZ 2018, 42, 1A–50A. [Google Scholar] [CrossRef] [Green Version]
- Tsuji, S.; Takahara, T.; Doi, H.; Shibata, N.; Yamanaka, H. The Detection of Aquatic Macroorganisms Using Environmental DNA Analysis—A Review of Methods for Collection, Extraction, and Detection. Environ. DNA 2019, 1, 99–108. [Google Scholar] [CrossRef] [Green Version]
- Hinlo, R.; Gleeson, D.; Lintermans, M.; Furlan, E. Methods to Maximise Recovery of Environmental DNA from Water Samples. PLos ONE 2017, 12, e0179251. [Google Scholar] [CrossRef] [PubMed]
- Sidstedt, M.; Jansson, L.; Nilsson, E.; Noppa, L.; Forsman, M.; Rådström, P.; Hedman, J. Humic Substances Cause Fluorescence Inhibition in Real-Time Polymerase Chain Reaction. Anal. Biochem. 2015, 487, 30–37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sidstedt, M.; Rådström, P.; Hedman, J. PCR Inhibition in QPCR, DPCR and MPS—Mechanisms and Solutions. Anal. Bioanal. Chem. 2020, 412, 2009–2023. [Google Scholar] [CrossRef] [Green Version]
- Uchii, K.; Doi, H.; Okahashi, T.; Katano, I.; Yamanaka, H.; Sakata, M.K.; Minamoto, T. Comparison of Inhibition Resistance among PCR Reagents for Detection and Quantification of Environmental DNA. Environ. DNA 2019, 1, 359–367. [Google Scholar] [CrossRef] [Green Version]
- Lance, R.F.; Guan, X. Variation in Inhibitor Effects on QPCR Assays and Implications for EDNA Surveys. Can. J. Fish. Aquat. Sci. 2020, 77, 23–33. [Google Scholar] [CrossRef]
- McKee, A.M.; Spear, S.F.; Pierson, T.W. The Effect of Dilution and the Use of a Post-Extraction Nucleic Acid Purification Column on the Accuracy, Precision, and Inhibition of Environmental DNA Samples. Biol. Conserv. 2015, 183, 70–76. [Google Scholar] [CrossRef]
- Yuan, Q.-B.; Huang, Y.-M.; Wu, W.-B.; Zuo, P.; Hu, N.; Zhou, Y.-Z.; Alvarez, P.J.J. Redistribution of Intracellular and Extracellular Free & Adsorbed Antibiotic Resistance Genes through a Wastewater Treatment Plant by an Enhanced Extracellular DNA Extraction Method with Magnetic Beads. Environ. Int. 2019, 131, 104986. [Google Scholar] [CrossRef]
- Sanches, T.M.; Schreier, A.D. Optimizing an EDNA Protocol for Estuarine Environments: Balancing Sensitivity, Cost and Time. PLos ONE 2020, 15, e0233522. [Google Scholar] [CrossRef]
- Green, H.C.; Field, K.G. Sensitive Detection of Sample Interference in Environmental QPCR. Water Res. 2012, 46, 3251–3260. [Google Scholar] [CrossRef]
- Turner, C.R.; Uy, K.L.; Everhart, R.C. Fish Environmental DNA Is More Concentrated in Aquatic Sediments than Surface Water. Biol. Conserv. 2015, 183, 93–102. [Google Scholar] [CrossRef] [Green Version]
- Marshall, N.T.; Vanderploeg, H.A.; Chaganti, S.R. Environmental (e)RNA Advances the Reliability of EDNA by Predicting Its Age. Sci. Rep. 2021, 11, 2769. [Google Scholar] [CrossRef]
- Munk, B.; Lebuhn, M. Process Diagnosis Using Methanogenic Archaea in Maize-Fed, Trace Element Depleted Fermenters. Anaerobe 2014, 29, 22–28. [Google Scholar] [CrossRef] [PubMed]
- Lebuhn, M.; Derenkó, J.; Rademacher, A.; Helbig, S.; Munk, B.; Pechtl, A.; Stolze, Y.; Prowe, S.; Schwarz, W.; Schlüter, A.; et al. DNA and RNA Extraction and Quantitative Real-Time PCR-Based Assays for Biogas Biocenoses in an Interlaboratory Comparison. Bioengineering 2016, 3, 7. [Google Scholar] [CrossRef] [PubMed]
- Leese, F.; Bouchez, A.; Abarenkov, K.; Altermatt, F.; Borja, Á.; Bruce, K.; Ekrem, T.; Čiampor, F.; Čiamporová-Zaťovičová, Z.; Costa, F.O.; et al. Why We Need Sustainable Networks Bridging Countries, Disciplines, Cultures and Generations for Aquatic Biomonitoring 2.0: A Perspective Derived From the DNAqua-Net COST Action. In Advances in Ecological Research; Elsevier: Amsterdam, The Netherlands, 2018; Volume 58, pp. 63–99. ISBN 978-0-12-813949-3. [Google Scholar]
- Peckys, D.B.; Melechko, A.V.; Simpson, M.L.; McKnight, T.E. Immobilization and Release Strategies for DNA Delivery Using Carbon Nanofiber Arrays and Self-Assembled Monolayers. Nanotechnology 2009, 20, 145304. [Google Scholar] [CrossRef] [PubMed]
- Liu, J. Adsorption of DNA onto Gold Nanoparticles and Graphene Oxide: Surface Science and Applications. Phys. Chem. Chem. Phys. 2012, 14, 10485. [Google Scholar] [CrossRef] [Green Version]
- Matthew, R.A.; Gopi, M.M.; Menon, P.; Jayakumar, R.; Vijayachandran, L.S. Synthesis of Electrospun Silica Nanofibers for Protein/DNA Binding. Mater. Lett. 2016, 184, 5–8. [Google Scholar] [CrossRef]
- Wood, S.A.; Holland, P.T.; MacKenzie, L. Development of Solid Phase Adsorption Toxin Tracking (SPATT) for Monitoring Anatoxin-a and Homoanatoxin-a in River Water. Chemosphere 2011, 82, 888–894. [Google Scholar] [CrossRef]
- Hale, S.E.; Oen, A.M.P.; Cornelissen, G.; Jonker, M.T.O.; Waarum, I.-K.; Eek, E. The Role of Passive Sampling in Monitoring the Environmental Impacts of Produced Water Discharges from the Norwegian Oil and Gas Industry. Mar. Pollut. Bull. 2016, 111, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Gallori, E.; Bazzicalupo, M.; Dal Canto, L.; Fani, R.; Nannipieri, P.; Vettori, C.; Stotzky, G. Transformation of Bacillus Subtilis by DNA Bound on Clay in Non-Sterile Soil. FEMS Microbiol. Ecol. 1994, 15, 119–126. [Google Scholar] [CrossRef]
- Vandeventer, P.E.; Lin, J.S.; Zwang, T.J.; Nadim, A.; Johal, M.S.; Niemz, A. Multiphasic DNA Adsorption to Silica Surfaces under Varying Buffer, PH, and Ionic Strength Conditions. J. Phys. Chem. B 2012, 116, 5661–5670. [Google Scholar] [CrossRef] [Green Version]
- Lozano, H.J.; Busto, N.; Lari, M.; Leal, J.M.; García, B. Binding of Aluminium/Cacodylate Complexes with DNA and RNA. Experimental and “ in Silico ”Study. New J. Chem. 2018, 42, 8137–8144. [Google Scholar] [CrossRef]
- Sheng, X.; Qin, C.; Yang, B.; Hu, X.; Liu, C.; Waigi, M.G.; Li, X.; Ling, W. Metal Cation Saturation on Montmorillonites Facilitates the Adsorption of DNA via Cation Bridging. Chemosphere 2019, 235, 670–678. [Google Scholar] [CrossRef] [PubMed]
- Mali, P.; Bhattacharjee, N.; Searson, P.C. Electrochemically Programmed Release of Biomolecules and Nanoparticles. Nano Lett. 2006, 6, 1250–1253. [Google Scholar] [CrossRef] [PubMed]
- Lomas, H.; Canton, I.; MacNeil, S.; Du, J.; Armes, S.P.; Ryan, A.J.; Lewis, A.L.; Battaglia, G. Biomimetic PH Sensitive Polymersomes for Efficient DNA Encapsulation and Delivery. Adv. Mater. 2007, 19, 4238–4243. [Google Scholar] [CrossRef]
- Mariani, S.; Baillie, C.; Colosimo, G.; Riesgo, A. Sponges as Natural Environmental DNA Samplers. Curr. Biol. 2019, 29, R401–R402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, J.; Mukherjee, P.; Gao, H.; Luan, Q.; Papautsky, I. Label-Free Microfluidic Sorting of Microparticles. APL Bioeng. 2019, 3, 041504. [Google Scholar] [CrossRef] [Green Version]
- Watanabe, M.; Kenmotsu, H.; Ko, R.; Wakuda, K.; Ono, A.; Imai, H.; Taira, T.; Naito, T.; Murakami, H.; Abe, M.; et al. Isolation and Molecular Analysis of Circulating Tumor Cells from Lung Cancer Patients Using a Microfluidic Chip Type Cell Sorter. Cancer Sci. 2018, 109, 2539–2548. [Google Scholar] [CrossRef] [Green Version]
- Shakeel Syed, M.; Rafeie, M.; Henderson, R.; Vandamme, D.; Asadnia, M.; Ebrahimi Warkiani, M. A 3D-Printed Mini-Hydrocyclone for High Throughput Particle Separation: Application to Primary Harvesting of Microalgae. Lab. Chip 2017, 17, 2459–2469. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Bai, X.; Wu, C.; Li, W.; Liu, K.; Kiani, A. Recent Advances in the Beneficiation of Ultrafine Coal Particles. Fuel Process. Technol. 2018, 178, 104–125. [Google Scholar] [CrossRef]
- Nielsen, P.E.; Egholm, M. An Introduction to Peptide Nucleic Acid. Curr Issues Mol. Biol. 1999, 1, 89–104. [Google Scholar]
- Metaferia, B.; Wei, J.S.; Song, Y.K.; Evangelista, J.; Aschenbach, K.; Johansson, P.; Wen, X.; Chen, Q.; Lee, A.; Hempel, H.; et al. Development of Peptide Nucleic Acid Probes for Detection of the HER2 Oncogene. PLos ONE 2013, 8, e58870. [Google Scholar] [CrossRef] [Green Version]
- Tutkus, M.; Rakickas, T.; Kopūstas, A.; Ivanovaitė, Š.; Venckus, O.; Navikas, V.; Zaremba, M.; Manakova, E.; Valiokas, R. Fixed DNA Molecule Arrays for High-Throughput Single DNA–Protein Interaction Studies. Langmuir 2019, 35, 5921–5930. [Google Scholar] [CrossRef]
- Dowle, E.; Pochon, X.; Keeley, N.; Wood, S.A. Assessing the Effects of Salmon Farming Seabed Enrichment Using Bacterial Community Diversity and High-Throughput Sequencing. FEMS Microbiol. Ecol. 2015, 91, fiv089. [Google Scholar] [CrossRef]
- Slon, V.; Hopfe, C.; Weiß, C.L.; Mafessoni, F.; de la Rasilla, M.; Lalueza-Fox, C.; Rosas, A.; Soressi, M.; Knul, M.V.; Miller, R.; et al. Neandertal and Denisovan DNA from Pleistocene Sediments. Science 2017, 356, 605–608. [Google Scholar] [CrossRef] [Green Version]
- Notomi, T. Loop-Mediated Isothermal Amplification of DNA. Nucleic Acids Res. 2000, 28, e63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- James, A.; Macdonald, J. Recombinase Polymerase Amplification: Emergence as a Critical Molecular Technology for Rapid, Low-Resource Diagnostics. Expert Rev. Mol. Diagn. 2015, 15, 1475–1489. [Google Scholar] [CrossRef] [Green Version]
- Lobato, I.M.; O’Sullivan, C.K. Recombinase Polymerase Amplification: Basics, Applications and Recent Advances. TrAC Trends Anal. Chem. 2018, 98, 19–35. [Google Scholar] [CrossRef] [PubMed]
- Gootenberg, J.S.; Abudayyeh, O.O.; Kellner, M.J.; Joung, J.; Collins, J.J.; Zhang, F. Multiplexed and Portable Nucleic Acid Detection Platform with Cas13, Cas12a, and Csm6. Science 2018, 360, 439–444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kellner, M.J.; Koob, J.G.; Gootenberg, J.S.; Abudayyeh, O.O.; Zhang, F. SHERLOCK: Nucleic Acid Detection with CRISPR Nucleases. Nat. Protoc. 2019, 14, 2986–3012. [Google Scholar] [CrossRef] [PubMed]
- Posthuma-Trumpie, G.A.; Korf, J.; van Amerongen, A. Lateral Flow (Immuno)Assay: Its Strengths, Weaknesses, Opportunities and Threats. A Literature Survey. Anal. Bioanal. Chem. 2009, 393, 569–582. [Google Scholar] [CrossRef] [Green Version]
- Williams, M.; O’Grady, J.; Ball, B.; Carlsson, J.; Eyto, E.; McGinnity, P.; Jennings, E.; Regan, F.; Parle-McDermott, A. The Application of CRISPR-Cas for Single Species Identification from Environmental DNA. Mol. Ecol. Resour. 2019, 19, 1106–1114. [Google Scholar] [CrossRef] [PubMed]
- Ravindran, V.B.; Khallaf, B.; Surapaneni, A.; Crosbie, N.D.; Soni, S.K.; Ball, A.S. Detection of Helminth Ova in Wastewater Using Recombinase Polymerase Amplification Coupled to Lateral Flow Strips. Water 2020, 12, 691. [Google Scholar] [CrossRef] [Green Version]
- Hansen, S.; Abd El Wahed, A. Point-Of-Care or Point-Of-Need Diagnostic Tests: Time to Change Outbreak Investigation and Pathogen Detection. Trop. Med. 2020, 5, 151. [Google Scholar] [CrossRef]
- Chowdhury, R.; Ghosh, P.; Khan, M.d.A.A.; Hossain, F.; Faisal, K.; Nath, R.; Baker, J.; Wahed, A.A.E.; Maruf, S.; Nath, P.; et al. Evaluation of Rapid Extraction Methods Coupled with a Recombinase Polymerase Amplification Assay for Point-of-Need Diagnosis of Post-Kala-Azar Dermal Leishmaniasis. Trop. Med. 2020, 5, 95. [Google Scholar] [CrossRef]
- Mondal, D.; Ghosh, P.; Khan, M.A.A.; Hossain, F.; Böhlken-Fascher, S.; Matlashewski, G.; Kroeger, A.; Olliaro, P.; Abd El Wahed, A. Mobile Suitcase Laboratory for Rapid Detection of Leishmania Donovani Using Recombinase Polymerase Amplification Assay. Parasites Vectors 2016, 9, 281. [Google Scholar] [CrossRef] [Green Version]
- Faye, O.; Faye, O.; Soropogui, B.; Patel, P.; El Wahed, A.A.; Loucoubar, C.; Fall, G.; Kiory, D.; Magassouba, N.; Keita, S.; et al. Development and Deployment of a Rapid Recombinase Polymerase Amplification Ebola Virus Detection Assay in Guinea in 2015. Euro Surveill. 2015, 20, 30053. [Google Scholar] [CrossRef] [PubMed]
- Vereecke, N.; Bokma, J.; Haesebrouck, F.; Nauwynck, H.; Boyen, F.; Pardon, B.; Theuns, S. High Quality Genome Assemblies of Mycoplasma Bovis Using a Taxon-Specific Bonito Basecaller for MinION and Flongle Long-Read Nanopore Sequencing. BMC Bioinform. 2020, 21, 517. [Google Scholar] [CrossRef] [PubMed]
- Chan, W.S.; Au, C.H.; Lam, H.Y.; Wang, C.L.N.; Ho, D.N.-Y.; Lam, Y.M.; Chu, D.K.W.; Poon, L.L.M.; Chan, T.L.; Zee, J.S.-T.; et al. Evaluation on the Use of Nanopore Sequencing for Direct Characterization of Coronaviruses from Respiratory Specimens, and a Study on Emerging Missense Mutations in Partial RdRP Gene of SARS-CoV-2. Virol. J. 2020, 17, 183. [Google Scholar] [CrossRef] [PubMed]

| Term | Definition |
|---|---|
| Acid washing | A common practice for decontaminating surfaces and apparatus from organismal material that may contaminate various steps in the pipeline (from sample collection through molecular analyses). |
| Bead-beating | Mechanical way to disrupt cells; filters are placed into a tube containing beads and lysis buffer, and placed on a shaking bead beater for a fixed amount of time. |
| Bioinformatics | Suite of software tools used to analyze genetic data. |
| Capture efficiency | How well a method retains genetic material—for example, material or pore size can affect utility of a filter. |
| cDNA | The DNA strand that is complementary to RNA; part of the intermediate step between genomic DNA and protein, used as a measure of gene activity. |
| Concentrate | Through filtration, the contents of a sample are distilled into a smaller volume, thereby increasing the chances of capturing rare or low-abundance organisms. |
| Cross-contamination | When genetic material from target or non-target species contributes inaccurately to molecular analyses, due to inadequate decontamination of surfaces and apparatus. |
| Decontamination | Sterilization of surfaces and apparatus from organismal material that may contaminate various steps in the pipeline (from sample collection through molecular analyses). |
| Degradation | The breaking down of genetic material (DNA/RNA) through enzymatic action (DNA/RNAses) or abiotic factors (e.g., temperature, UV light). |
| Diluted bleach | A common practice (10–50% for >10 min) for decontaminating surfaces and apparatus from organismal material that may contaminate various steps in the pipeline (from sample collection through molecular analyses). |
| Dissolved eDNA/RNA | Free-floating, naked nucleic acid (DNA/RNA) in the water column (i.e., not contained within or adsorbed to any particles). |
| DNA | Deoxyribonucleic acid; central storage of genetic information for organisms (except RNA viruses). In eDNA, analyses targeted for gene presence of species (single or multiple species). |
| DNAse | Deoxyribonuclease; group of enzymes that can degrade DNA, thereby affecting quality and quantity. |
| False positive | An instance where a sample should have been negative, but the result was positive; contamination from improper sample handling can lead to a false positive. |
| False negative | An instance where a sample should have been positive, but the result was negative; inhibitors can lead to a false negative. |
| "Fit-for-purpose” | The concept that a pipeline (sample collection through data analysis) needs to be formulated for each particular sampling context. In contrast to “one-size-fits-all”. |
| Inhibitors | A variety of substances of known (e.g., tannins, humics) and unknown type that can be co-extracted with nucleic acids and hinder the performance of downstream enzymatic reactions (e.g., the amplification steps in quantitative polymerase chain reaction (qPCR) and metabarcoding). |
| Metabarcoding | A genetic method that amplifies homologous gene(s) across species in order to gain perspective into the taxonomic constituents of a community. |
| Molecular signal | Results derived from any number of assays (e.g., qPCR, metabarcoding) that detect and possibly quantify genetic material. |
| Niskin bottle | A columnar sampling bottle that can be triggered to capture a whole water sample from a desired depth in the water column. |
| “one-size-fits-all” | The concept that one pipeline (sample collection through data analysis) can be formulated and used in all field and experimental contexts. In contrast to “fit-for-purpose”. |
| Particle size | Refers to an array of particle types that may be encountered in the water column: whole cells, broken/damaged cell pieces, and naked nucleic acids from lysed cells. Any of these forms can be free-floating or adsorbed to other (non-)organic material. |
| Plankton tow | Vertical or horizontal pull of a specialized net to filter and concentrate water column contents into a smaller volume, thereby increasing the chances of capturing rare or low-abundance organisms. |
| Precipitation | Concentration and purification of nucleic acids (DNA/RNA) through chemical means. |
| Preservation | Near-immediate immobilization of a sample (through a combination of buffers/freezing and transport/storage conditions) to maintain integrity of genetic material. |
| qPCR | Quantitative polymerase chain reaction; also called real-time PCR, because the amplification of a genetic target can be monitored during the reaction, and a determination of copy numbers of that target can be made. This is in contrast to PCR, which cannot be monitored in real time and produces a qualitative (positive/negative) result. |
| RNA | Ribonucleic acid; intermediate step between genomic DNA and protein, used as a measure of gene activity. |
| RNAse | Ribonuclease; group of enzymes that can degrade RNA, thereby affecting quality and quantity. |
| Settlement plates | Artificial structures (e.g., plastic polymer material) deployed in aquatic environments for passive sampling of marine biofouling; can be used to study recruitment of sessile taxa and non-indigenous species surveillance. |
| Snap-freeze | A method to immediately preserve genetic material after sample filtration; the filter is housed in a tube and submerged in liquid nitrogen or on dry ice. |
| Sterility | Maintaining a clean/aseptic environment throughout the entire pipeline to eliminate cross-contamination of organismal genetic material between samples at all stages (sample collection through processing and molecular manipulations). |
| Total eDNA/eRNA | Environmental DNA/RNA in all forms (whole and partial cells, free NAs in solution (dissolved) or adsorbed to particles). |
| Van Dorn | Large chamber water sampler that allows for sampling from one depth or a composite of several depths. |
| Field Workflow Considerations | Large Volume/Area Coverage | Lower Inhibitor Effect | Possibility of Visual Pre-Screening of Biodiversity | Selectivity for Invasive Taxa | Affinity of Signal to Source Location | Homogeneity of Material (Effective Replication) | Non-Disruptive to Ecosystem | Non-Hazardous (H&S Wise) | Capture of all NA Fractions | Suitable for Varying Depth/Locations | Lab Workflow Considerations | Easy/Inexpensive | No Specialized Equipment/ Infrastructure Required | Possible in Field | Lower Risk of Compromising Integrity of NAs | Time-Eefficient | Low-Waste | Non-Hazardous | Efficient Cross-Contamination Control |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Soft sediment | Post-sampling concentration (water samples) | ||||||||||||||||||
| Benthic samplers | Centrifugation | ||||||||||||||||||
| Scuba diving | Precipitation (chemical) | ||||||||||||||||||
| Hard bottom | Filtration | ||||||||||||||||||
| Scuba diving | Preservation | ||||||||||||||||||
| Settlement plates | Snap-freezing | ||||||||||||||||||
| Biofouling on artificial structures | Preservation buffers | ||||||||||||||||||
| Scuba diving | Desiccation | ||||||||||||||||||
| Settlement plates | |||||||||||||||||||
| Water column | Color key | ||||||||||||||||||
| Whole water samples | Yes | ||||||||||||||||||
| In-situ filtration | No | ||||||||||||||||||
| Plankton nets | Neutral/context dependent | ||||||||||||||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bowers, H.A.; Pochon, X.; von Ammon, U.; Gemmell, N.; Stanton, J.-A.L.; Jeunen, G.-J.; Sherman, C.D.H.; Zaiko, A. Towards the Optimization of eDNA/eRNA Sampling Technologies for Marine Biosecurity Surveillance. Water 2021, 13, 1113. https://doi.org/10.3390/w13081113
Bowers HA, Pochon X, von Ammon U, Gemmell N, Stanton J-AL, Jeunen G-J, Sherman CDH, Zaiko A. Towards the Optimization of eDNA/eRNA Sampling Technologies for Marine Biosecurity Surveillance. Water. 2021; 13(8):1113. https://doi.org/10.3390/w13081113
Chicago/Turabian StyleBowers, Holly A., Xavier Pochon, Ulla von Ammon, Neil Gemmell, Jo-Ann L. Stanton, Gert-Jan Jeunen, Craig D. H. Sherman, and Anastasija Zaiko. 2021. "Towards the Optimization of eDNA/eRNA Sampling Technologies for Marine Biosecurity Surveillance" Water 13, no. 8: 1113. https://doi.org/10.3390/w13081113







