A Digital Light Microscopic Method for Diatom Surveys Using Embedded Acid-Cleaned Samples
Abstract
:1. Introduction
2. Materials and Methods
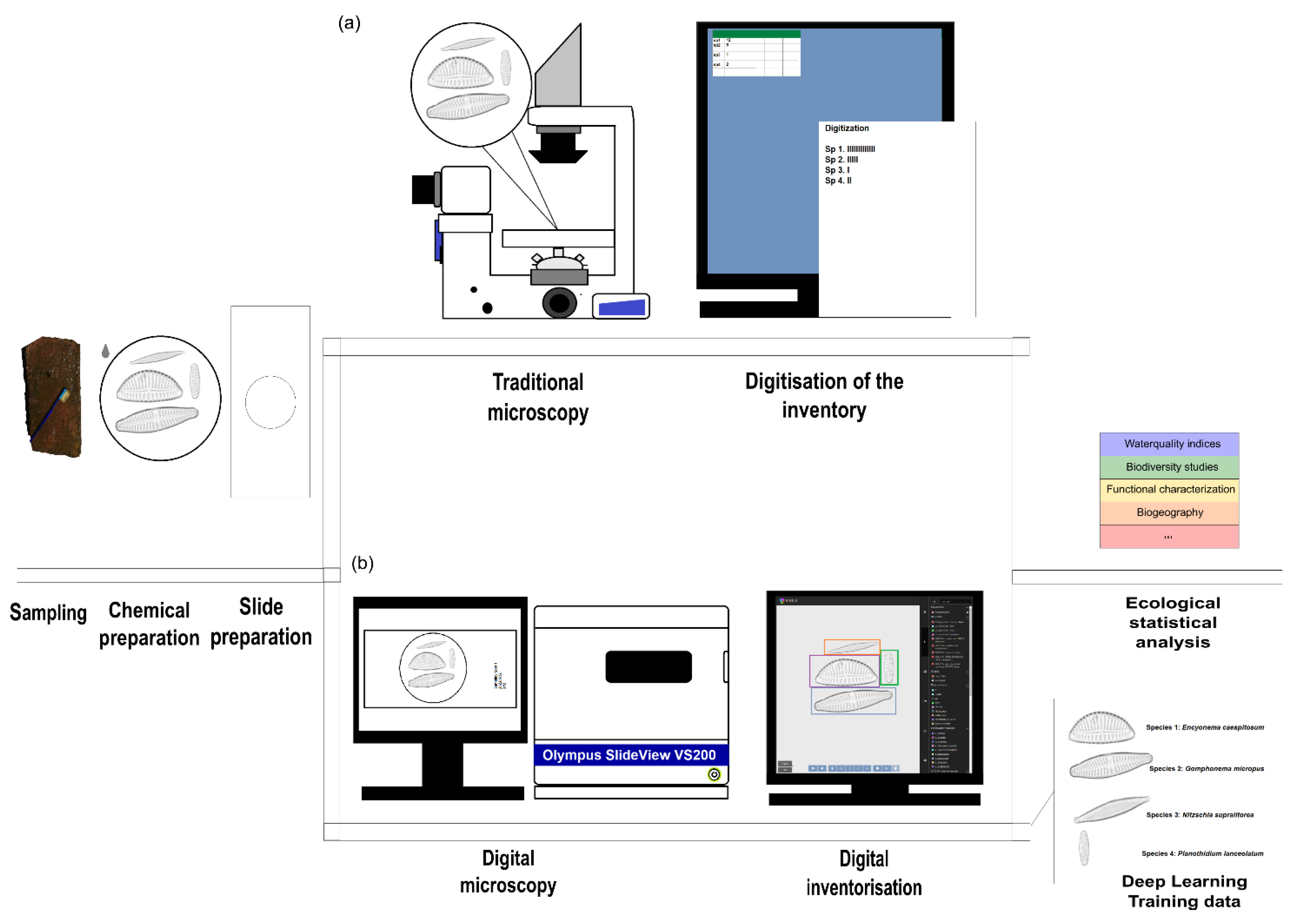
2.1. Diatom Collection, Preparation and Processing
2.2. “Traditional” Diatom Identification Workflow
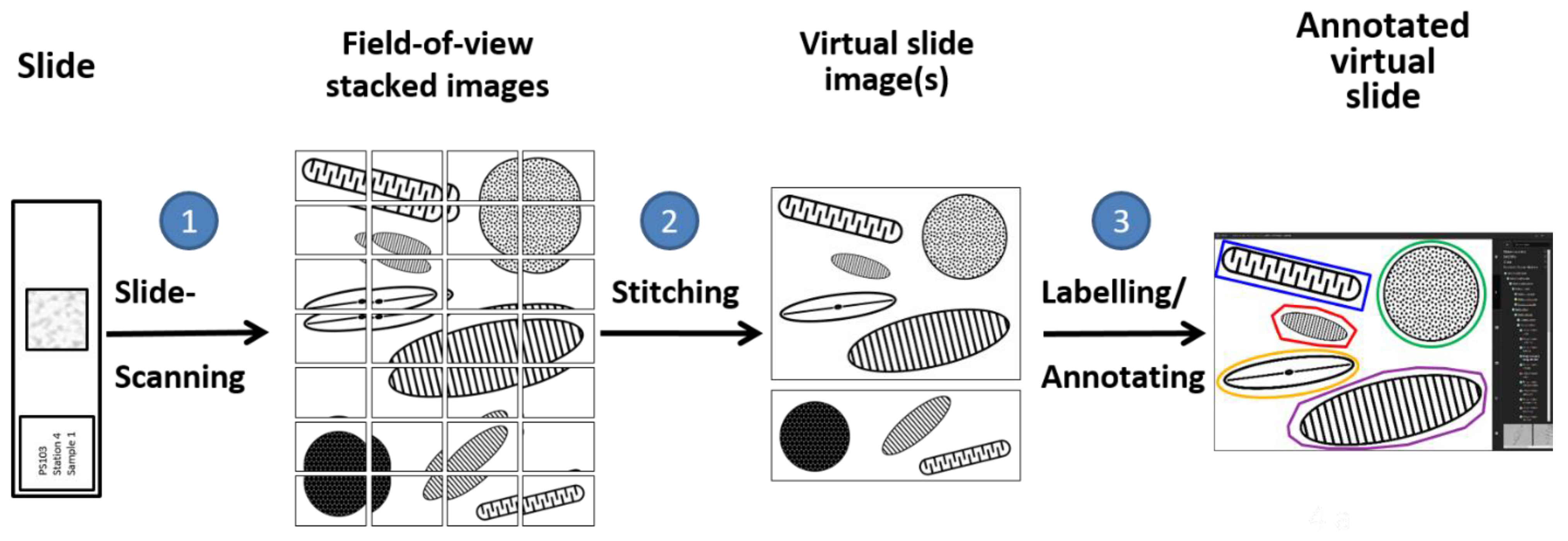
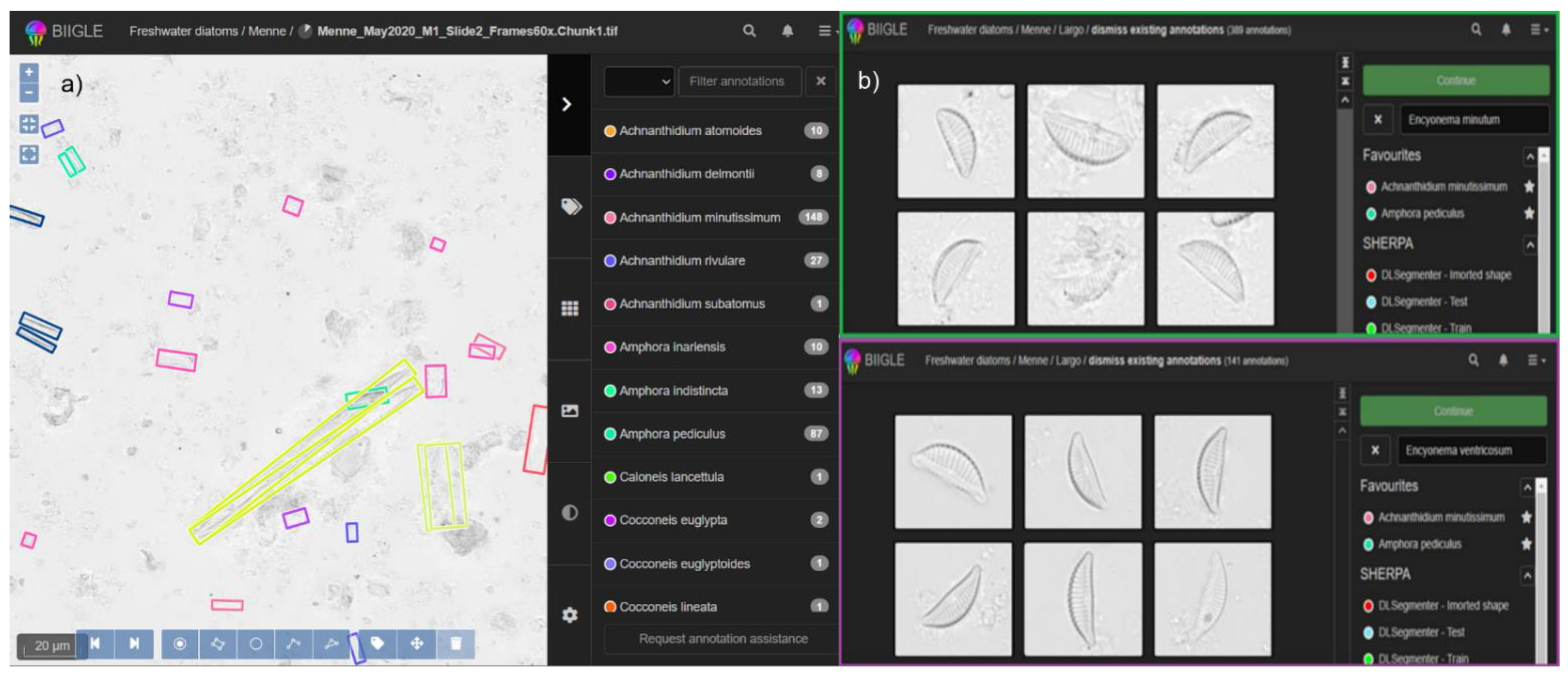
2.3. Digital Diatom Identification Workflow
2.4. Calculation of Biotic Indices
2.5. Statistical Analysis
3. Results & Discussion
3.1. Time Requirement
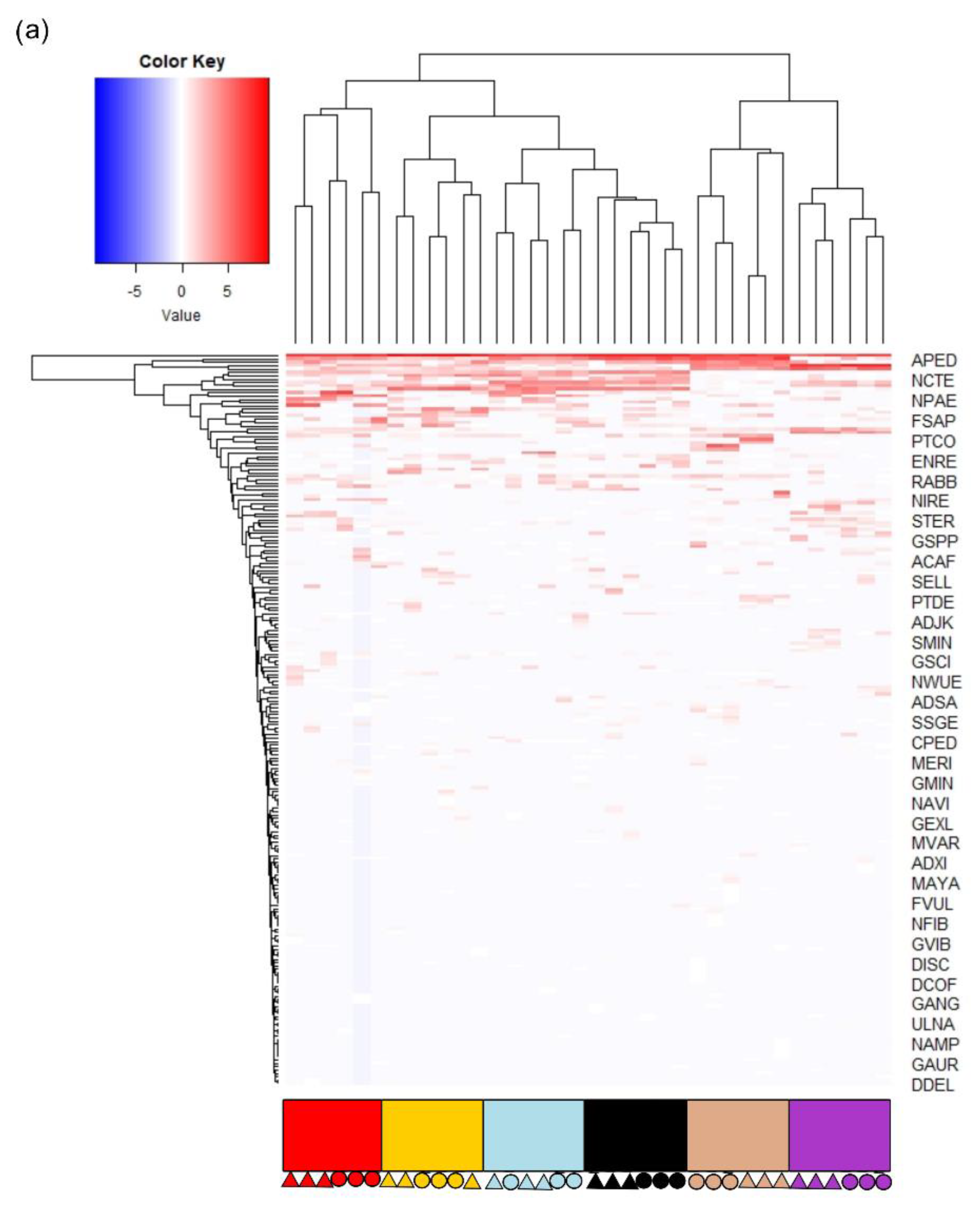
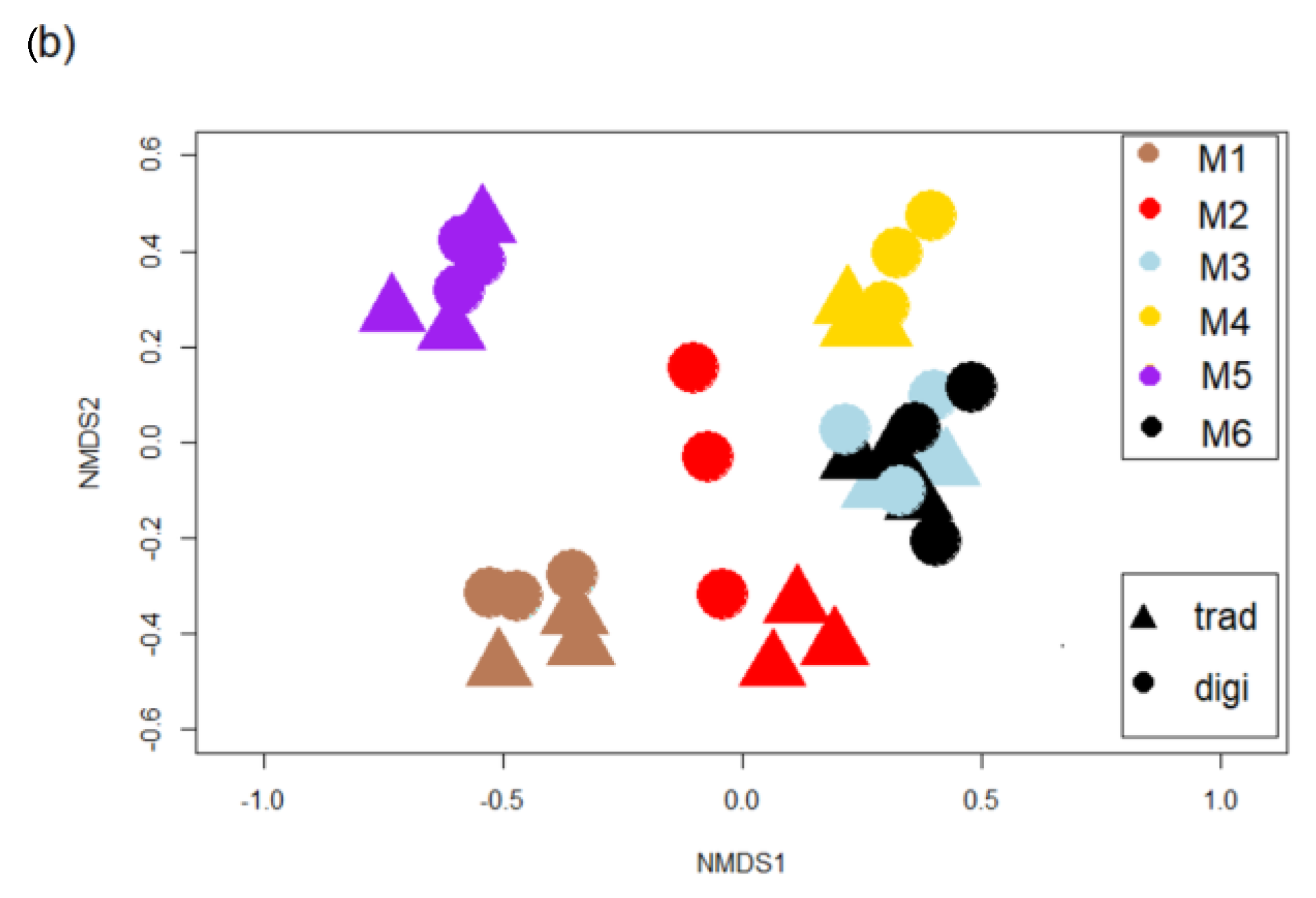
3.2. Diatom Communities
3.3. Testing the Effect of Difficult Species Complexes
3.4. Biotic Indices
3.5. Overall Observations
3.5.1. The Digital Workflow Provides Comparable Results with the “Traditional” One
3.5.2. The Digital Workflow Is Slightly More Time Consuming, but Enables Better Scientific Practice
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lama, G.F.C.; Errico, A.; Pasquino, V.; Mirzaei, S.; Preti, F.; Chirico, G.B. Velocity uncertainty quantification based on Riparian vegetation indices in open channels colonized by Phragmites australis. J. Ecohydraulics 2022, 7, 71–76. [Google Scholar] [CrossRef]
- Jalonen, J.; Järvelä, J.; Koivusalo, H.; Hyyppä, H. Deriving Floodplain Topography and Vegetation Characteristics for Hydraulic Engineering Applications by Means of Terrestrial Laser Scanning. J. Hydraul. Eng. 2014, 140, 4014056. [Google Scholar] [CrossRef]
- Lowe, R.L. Environmental Requirements and Pollution Tolerance of Freshwater Diatoms EPA-670/4-74-005; National Environmental Research Center Office of Research and Development, U.S. Environmental Protection Agency: Cincinnati, OH, USA, 1974. [Google Scholar]
- Patrick, R. The Importance of Monitoring Change. In Biological Monitoring of Water and Effluent Quality; Cairns, J., Dikinson, K.L., Westlake, G., Eds.; American Society for Testing and Materials: West Conshohocken, PA, USA, 1976; pp. 157–190. [Google Scholar]
- Falasco, E.; Bona, F.; Monauni, C.; Zeni, A.; Piano, E. Environmental and spatial factors drive diatom species distribution in Alpine streams: Implications for biomonitoring. Ecol. Indic. 2019, 106, 105441. [Google Scholar] [CrossRef] [Green Version]
- Ehrenberg, C.G. Die Infusionsthierchen als Vollkommene Organismen; Leopold Voss: Leipzig, Germany, 1838. [Google Scholar]
- Lobo, E.A.; Heinrich, C.G.; Schuch, M.; Wetzel, C.E.; Ector, L. Chapter 11. Diatoms as Bioindicators in Rivers Eduardo A. Lobo, Carla Giselda Heinrich, Marilia Schuch, Carlos Eduardo Wetzel, and Luc Ector. In River Algae; Necchi, O., Ed.; Springer: Cham, Switzerland, 2016; ISBN 978-3-319-31983-4. [Google Scholar]
- Smol, J.P.; Stoermer, E.F. (Eds.) The Diatoms: Applications for the Environmental and Earth Sciences, 2nd ed.; Cambridge University Press: Cambridge, UK, 2012; ISBN 978-1-107-56496-1. [Google Scholar]
- Morales, E.A.; Siver, P.A.; Trainor, F.R. Identification of diatoms (Bacillariophyceae) during ecological assessments: Comparison between Light Microscopy and Scanning Electron Microscopy techniques. Analysis 2001, 151, 95–103. [Google Scholar] [CrossRef]
- Pérez-Burillo, J.; Trobajo, R.; Vasselon, V.; Rimet, F.; Bouchez, A.; Mann, D.G. Evaluation and sensitivity analysis of diatom DNA metabarcoding for WFD bioassessment of Mediterranean rivers. Sci. Total Environ. 2020, 727, 138445. [Google Scholar] [CrossRef]
- Bailet, B.; Bouchez, A.; Franc, A.; Frigerio, J.-M.; Keck, F.; Karjalainen, S.-M.; Rimet, F.; Schneider, S.; Kahlert, M. Molecular versus morphological data for benthic diatoms biomonitoring in Northern Europe freshwater and consequences for ecological status. Metabarcoding Metagenomics 2019, 3, e34002. [Google Scholar] [CrossRef] [Green Version]
- Borrego-Ramos, M.; Bécares, E.; García, P.; Nistal, A.; Blanco, S. Epiphytic Diatom-Based Biomonitoring in Mediterranean Ponds: Traditional Microscopy versus Metabarcoding Approaches. Water 2021, 13, 1351. [Google Scholar] [CrossRef]
- Kahlert, M.; Ács, É.; Almeida, S.F.P.; Blanco, S.; Dressler, M.; Ector, L.; Karjalainen, S.M.; Liess, A.; Mertens, A.; van der Wal, J.; et al. Quality assurance of diatom counts in Europe: Towards harmonized datasets. Hydrobiologia 2016, 772, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Beszteri, B.; Allen, C.; Almandoz, G.O.; Armand, L.; Bárcena, M.A.; Cantzler, H.; Crosta, X.; Esper, O.; Jordan, R.W.; Kauer, G.; et al. Quantitative comparison of taxa and taxon concepts in the diatom genus Fragilariopsis a case study on using slide scanning multiexpert image annotation and image analysis in taxonomy. J. Phycol. 2018, 54, 705–719. [Google Scholar] [CrossRef] [Green Version]
- Tyree, M.A.; Bishop, I.W.; Hawkins, C.P.; Mitchell, R.; Spaulding, S.A. Reduction of taxonomic bias in diatom species data. Limnol. Oceanogr. Methods 2020, 18, 271–279. [Google Scholar] [CrossRef]
- Kloster, M.; Esper, O.; Kauer, G.; Beszteri, B. Large-Scale Permanent Slide Imaging and Image Analysis for Diatom Morphometrics. Appl. Sci. 2017, 7, 330. [Google Scholar] [CrossRef] [Green Version]
- Cristóbal, G.; Blanco, S.; Bueno, G. (Eds.) Modern Trends in Diatom Identification; Springer International Publishing: Cham, Switzerland, 2020; ISBN 978-3-030-39211-6. [Google Scholar]
- Du Buf, J.M.H.; Bayer, M.M. (Eds.) Automatic Diatom Identification (ADIAC); World Scientific Publishing Co. Pte. Ltd.: London, UK, 2002; ISBN 978-85-7811-079-6. [Google Scholar]
- Pinto, R.; Vilarinho, R.; Carvalho, A.P.; Moreira, J.A.; Guimarães, L.; Oliva-Teles, L. Novel Approach to Freshwater Diatom Profiling and Identification Using Raman Spectroscopy and Chemometric Analysis. Water 2022, 14, 2116. [Google Scholar] [CrossRef]
- Burfeid-Castellanos, A.M.; Kloster, M.; Cambra, J.; Beszteri, B. Both hydrology and physicochemistry influence diatom morphometry. Diatom Res. 2020, 35, 1–12. [Google Scholar] [CrossRef]
- Burfeid Castellanos, A.M.; Martín-Martín, R.P.; Kloster, M.; Angulo-Preckler, C.; Avila, C.; Beszteri, B.; Burfeid-Castellanos, A.M.; Kloster, M.; Angulo-Preckler, C.; Beszteri, B. Epiphytic diatom community structure and richness is determined by macroalgal host and location in the South Shetland Islands (Antarctica). PLoS ONE 2021, 16, e0250629. [Google Scholar] [CrossRef]
- Langenkämper, D.; Zurowietz, M.; Schoening, T.; Nattkemper, T.W. BIIGLE 2.0—Browsing and Annotating Large Marine Image Collections. Front. Mar. Sci. 2017, 4, 83. [Google Scholar] [CrossRef] [Green Version]
- Zurowietz, M.; Nattkemper, T.W. Current Trends and Future Directions of Large Scale Image and Video Annotation: Observations From Four Years of BIIGLE 2.0. Front. Mar. Sci. 2021, 8, 1752. [Google Scholar] [CrossRef]
- CEN. UNE-EN 13946:2014; Water Quality—Guidance for the Routine Sampling and Preparation of Benthic Diatoms from Rivers and Lakes. CEN-CENELEC Management Centre: Brussels, Belgium, 2014.
- CEN. UNE-EN 14407:2014; Water Quality—Guidance Standard for the Identification, Enumeration and Interpretation of Benthic Diatom Samples from rivers and lakes. CEN-CENELEC Management Centre: Brussels, Belgium, 2014.
- Taylor, J.C.; Harding, W.R.; Archibald, C.G.M. A Methods Manual for the Collection, Preparation and Analysis of Diatom Samples Version 1.0; TT 281/07; Water Research Commission: Pretoria, South Africa, 2007; ISBN 1-77005-483-9. [Google Scholar]
- Bey, M.-Y.; Ector, L. Atlas des Diatomées des Cours d’eau de la Région Rhône-Alpes; Tomes 1–6; Direction régionale de l'environnement, de l'aménagement et du logement, Préfet de la Région Auvergne-Rhône-Alpes: Lyon, France, 2013; ISBN 978-2-11-129817-0. [Google Scholar]
- Cantonati, M.; Hoffmann, G.; Kelly, M.G.; Lange-Bertalot, H.; Werum, M. Freshwater Benthic Diatoms of Central Europe: Over 800 Common Species Used in Ecological Assessment; Koeltz Botanical Books: Schmitten-Oberreifenberg, Germany, 2017; ISBN 978-3-946583-06-6. [Google Scholar]
- Bahls, L.L.; Edlund, M.B.; Kociolek, J.P.; Lowe, R.L.; Potapova, M.G.; Spaulding, S.A.; Bishop, I.; Burge, D.R.L.; English, J.; Furey, P.; et al. Diatoms of North America. Available online: https://diatoms.org/ (accessed on 30 June 2020).
- Levkov, Z.; Metzeltin, D.; Pavlov, A. Luticola and Luticolopsis. In Diatoms of the European Inland Waters and Comparable Habitats, 1st ed.; Bertalot, H.L., Ed.; Koeltz Botanical Books: Oberreifenberg, Germany, 2013; p. 698. ISBN 978-3-87429-439-3. [Google Scholar]
- Trobajo, R.; Rovira, L.; Ector, L.; Wetzel, C.E.; Kelly, M.G.; Mann, D.G. Morphology and identity of some ecologically important small Nitzschia species. Diatom Res. 2013, 28, 37–59. [Google Scholar] [CrossRef]
- Helicon Focus; Helicon Soft Ltd.: Kharkiv, Ukraine, 2020.
- Kloster, M.; Langenkämper, D.; Zurowietz, M.; Beszteri, B.; Nattkemper, T.W. Deep learning-based diatom taxonomy on virtual slides. Sci. Rep. 2020, 10, 14416. [Google Scholar] [CrossRef]
- Schoening, T.; Osterloff, J.; Nattkemper, T.W. RecoMIA—Recommendations for Marine Image Annotation: Lessons Learned and Future Directions. Front. Mar. Sci. 2016, 3, 59. [Google Scholar] [CrossRef] [Green Version]
- Bailet, B.; Apothéloz-Perret-Gentil, L.; Baričević, A.; Chonova, T.; Franc, A.; Frigerio, J.-M.; Kelly, M.; Mora, D.; Pfannkuchen, M.; Proft, S.; et al. Diatom DNA metabarcoding for ecological assessment: Comparison among bioinformatics pipelines used in six European countries reveals the need for standardization. Sci. Total Environ. 2020, 745, 140948. [Google Scholar] [CrossRef]
- Lecointe, C.; Coste, M.; Prygiel, J. “Omnidia”: Software for taxonomy, calculation of diatom indices and inventories management. Hydrobiologia 1993, 269, 509–513. [Google Scholar] [CrossRef]
- Lecointe, C.; Coste, M. Omnidia. 2015. Available online: https://omnidia.fr/en/ (accessed on 11 May 2020).
- Rott, E. Indikationslisten für Aufwuchsalgen in Österreichischen Fließgewässern. 2. Trophieindikation sowie geochemische Präferenz: Taxonomische und Toxikologische Anmerkungen; Bundesministerium f. Land- u. Forstwirtschaft: Wien, Austria, 1999. [Google Scholar]
- Descy, J.P.; Coste, M. Utilisation des diatomeés benthiques pour la mesure de la qualité des eaux du bassin Artois-Picardie: Bilan et perspectives. Ann. De Limnol. Int. J. Limnol. 1990, 29, 255–267. [Google Scholar]
- Prygiel, J.; Leveque, L.; Iserentant, R. Un nouvel indice Diatomique Pratique por l’évaluation de la qualité des eaux en réseau de surveillance. Rev. Des Sci. De L’Eau 1996, 1, 97–113. [Google Scholar]
- Prygiel, J.; Carpentier, P.; Almeida, S.F.P.; Coste, M.; Druart, J.-C.; Ector, L.; Guillard, D.; Honoré, M.-A.; Iserentant, R.; Ledeganck, P.; et al. Determination of the biological diatom index (IBD NF T 90–354): Results of an intercomparison exercise. J. Appl. Phycol. 2002, 14, 27–40. [Google Scholar] [CrossRef]
- Corse, P.B. Cemagref. Etude des Méthodes Biologiques Quantitatives D’appréciation de la Qualité des Eaux. Rapport Division Qualité des Eaux Lyon. 1982. Available online: https://www.documentation.eauetbiodiversite.fr/notice/00000000015def9cd65cdb328423f073 (accessed on 2 January 2019).
- Kelly, M.G.; Whitton, B.A. The trophic diatom index: A new index for monitoring eutrophication in rivers. J. Appl. Phycol. 1995, 7, 433–444. [Google Scholar] [CrossRef]
- Rott, E.; Hofmann, P.G.; Pall, K.; Pfister, P.; Pipp, E. Indikationslisten für Aufwuchsalgen in Österreichischen Fliessgewässern, Teil 1: Saprobielle Indikation; Bundesministerium f. Land- u. Forstwirtschaft: Wien, Austria, 1997. [Google Scholar]
- Sládeček, V. Diatoms as indicators of organic pollution. Acta Hydrochim. Et Hydrobiol. 1986, 14, 555–566. [Google Scholar] [CrossRef]
- R Development Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2008; ISBN 3-900051-07-0. [Google Scholar]
- RStudio Team. RStudio: Integrated Development for R; RStudio, Inc.: Boston, MA, USA, 2015. [Google Scholar]
- Oksanen, J. Multivariate Analysis of Ecological Communities in R. Ph.D. Thesis, University of Oulu, Oulu, Finland, 2013. [Google Scholar]
- Martinez Arbizu, P. pairwiseAdonis: Pairwise Multilevel Comparison Using Adonis. 2020. Available online: https://github.com/pmartinezarbizu/pairwiseAdonis (accessed on 1 March 2020).
- Warnes, G.R.; Bolker, B.; Bonebakker, L.; Huber, W.; Liaw, A.; Lumley, T.; Magnusson, A.; Moeller, S.; Schwartz, M. Package ‘gplots’. 2015. Available online: https://cran.r-project.org/web/packages/gplots/gplots.pdf (accessed on 20 May 2018).
- Wojtal, A.Z.; Ector, L.; van de Vijver, B.; Morales, E.A.; Blanco, S.; Piatek, J.; Smieja, A. The Achnanthidium minutissimum complex (Bacillariophyceae ) in southern Poland. Algol. Stud. 2011, 136–137, 211–238. [Google Scholar] [CrossRef]
- Jahn, R.; Kusber, W.-H.; Romero, O.E. Cocconeis pediculus Ehrenberg and C. placentula Ehrenberg var. placentula (Bacillariophyta): Typification and taxonomy. Fottea 2009, 9, 275–288. [Google Scholar] [CrossRef]
- Zgrundo, A.; Lemke, P.; Pniewski, F.; Cox, E.J.; Latała, A. Morphological and molecular phylogenetic studies on Fistulifera saprophila. Diatom Res. 2013, 28, 431–443. [Google Scholar] [CrossRef]
- Trobajo, R.; Clavero, E.; Chepurnov, V.A.; Sabbe, K.; Mann, D.G.; Ishihara, S.; Cox, E.J. Morphological, genetic and mating diversity within the widespread bioindicator Nitzschia palea (Bacillariophyceae). Phycologia 2009, 48, 443–459. [Google Scholar] [CrossRef]
- Williams, D.M. Synedra, Ulnaria: Definitions and descriptions—A partial resolution. Diatom Res. 2011, 26, 149–153. [Google Scholar] [CrossRef]
- Novais, M.H.; Jüttner, I.; de van Vijver, B.; Morais, M.M.; Hoffmann, L.; Ector, L. Morphological variability within the Achnanthidium minutissimum species complex (Bacillariophyta): Comparison between the type material of Achnanthes minutissima and related taxa, and new freshwater Achnanthidium species from Portugal. Phytotaxa 2015, 224, 101–139. [Google Scholar] [CrossRef]
- Pinseel, E.; Vanormelingen, P.; Hamilton, P.B.; Vyverman, W.; de van Vijver, B.; Kopalova, K. Molecular and morphological characterization of the Achnanthidium minutissimum complex (Bacillariophyta) in Petuniabukta (Spitsbergen, High Arctic) including the description of A. digitatum sp. nov. Eur. J. Phycol. 2017, 52, 264–280. [Google Scholar] [CrossRef]
- Wetzel, C.E.; Jüttner, I.; Gurung, S.; Ector, L. Analysis of the type material of Achnanthes minutissima var. macrocephala (Bacillariophyta) and description of two new small capitate Achnanthidium species from Europe and the Himalaya. Plant Ecol. Evol. 2019, 152, 340–350. [Google Scholar] [CrossRef]
- Kelly, M.G. Use of similarity measures for quality control of benthic diatom samples. Water Res. 2001, 35, 2784–2788. [Google Scholar] [CrossRef]
- ter Braak, C.J.F.; van Dam, H. Inferring pH from diatoms: A comparison of old and new calibration methods. Hydrobiologia 1989, 178, 209–223. [Google Scholar] [CrossRef]
- Hamsher, S.E.; LeGresley, M.M.; Martin, J.L.; Saunders, G.W. A comparison of morphological and molecular-based surveys to estimate the species richness of Chaetoceros and Thalassiosira (bacillariophyta), in the Bay of Fundy. PLoS ONE 2013, 8, e73521. [Google Scholar] [CrossRef] [Green Version]
- Lowndes, J.S.S.; Best, B.D.; Scarborough, C.; Afflerbach, J.C.; Frazier, M.R.; O'Hara, C.C.; Jiang, N.; Halpern, B.S. Our path to better science in less time using open data science tools. Nat. Ecol. Evol. 2017, 1, 160. [Google Scholar] [CrossRef] [Green Version]
- Kahlert, M.; Kelly, M.G.; Albert, R.-L.; Almeida, S.F.P.; Bešta, T.; Blanco, S.; Coste, M.; Denys, L.; Ector, L.; Fránková, M.; et al. Identification versus counting protocols as sources of uncertainty in diatom-based ecological status assessments. Hydrobiologia 2012, 695, 109–124. [Google Scholar] [CrossRef]
- Kahlert, M.; Kelly, M.G.; Mann, D.G.; Rimet, F.; Sato, S.; Bouchez, A.; Keck, F. Connecting the morphological and molecular species concepts to facilitate species identification within the genus Fragilaria (Bacillariophyta). J. Phycol. 2019, 55, 948–970. [Google Scholar] [CrossRef]
- Bishop, I.W.; Esposito, R.M.; Tyree, M.; Spaulding, S.A.; BISHOP, I.W. A diatom voucher flora from selected southeast rivers (USA). Phytotaxa 2017, 332, 101. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.S.; Bishop, I.W.; Spaulding, S.A.; Mitchell, R.M.; Yuan, L.L. Taxonomic harmonization may reveal a stronger association between diatom assemblages and total phosphorus in large datasets. Ecol. Indic. 2019, 102, 166–174. [Google Scholar] [CrossRef]
- Kloster, M.; Kauer, G.; Beszteri, B. SHERPA: An image segmentation and outline feature extraction tool for diatoms and other objects. BMC Bioinform. 2014, 15, 218. [Google Scholar] [CrossRef]
- Rojas Camacho, O.; Forero, M.G.; Menéndez, J.M.; Forero, M.; Menéndez, J. A Tuning Method for Diatom Segmentation Techniques. Appl. Sci. 2017, 7, 762. [Google Scholar] [CrossRef]
- Libreros, J.; Bueno, G.; Trujillo, M.; Ospina, M. Diatom Segmentation in Water Resources. In Advances in Computing, Proceedings of the 13th Colombian conference, CCC 2018, Cartagena, Colombia, 26–28 September 2018; Jairo, E., Serrano, C., Martínez-Santos, J.C., Eds.; Springer: Cham, Switzerland, 2018; pp. 83–97. ISBN 978-3-319-98998-3. [Google Scholar]
- Ivanova, M.V.; Vuntesmeri, Y.V.; Bukhtiiarova, L.M. Automated Detection and Classification of the Diatom Microscopic Photo Images. Mìkrosist. Elektron. Akust. 2019, 24, 18–25. [Google Scholar] [CrossRef]
- Fischer, S.; Shahbazkia, H.; Bunke, H. Contour Extraction. In Automatic Diatom Identification (ADIAC); Du Buf, J.M.H., Bayer, M.M., Eds.; World Scientific Publishing Co Pte Ltd.: London, UK, 2002; pp. 93–108. ISBN 978-85-7811-079-6. [Google Scholar]
- Westenberg, M.A.; Roerdink, J. Mixed-Method Identifications. In Automatic Diatom Identification (ADIAC); Du Buf, J.M.H., Bayer, M.M., Eds.; World Scientific Publishing Co Pte Ltd.: London, UK, 2002; pp. 245–258. ISBN 978-85-7811-079-6. [Google Scholar]
- Spaulding, S.A.; Jewson, D.H.; Bixby, R.J.; Nelson, H.; McKnight, D.M. Automated measurement of diatom size. Limnol. Oceanogr. Methods 2012, 10, 882–890. [Google Scholar] [CrossRef]
- Glemser, B.; Kloster, M.; Esper, O.; Eggers, S.L.; Kauer, G.; Beszteri, B. Biogeographic differentiation between two morphotypes of the Southern Ocean diatom Fragilariopsis kerguelensis. Polar Biol. 2019, 42, 1369–1376. [Google Scholar] [CrossRef] [Green Version]
- Jalba, A.C.; Wilkinson, M.H.F.; Roerdink, J.B.T.M. Automatic segmentation of diatom images for classification. Microsc. Res. Tech. 2004, 65, 72–85. [Google Scholar] [CrossRef] [Green Version]
- Bueno, G.; Deniz, O.; Pedraza, A.; Ruiz-Santaquiteria, J.; Salido, J.; Cristóbal, G.; Borrego-Ramos, M.; Blanco, S. Automated Diatom Classification (Part A): Handcrafted Feature Approaches. Appl. Sci. 2017, 7, 753. [Google Scholar] [CrossRef] [Green Version]
- Pedraza, A.; Bueno, G.; Deniz, O.; Cristóbal, G.; Blanco, S.; Borrego-Ramos, M. Automated Diatom Classification (Part B): A Deep Learning Approach. Appl. Sci. 2017, 7, 460. [Google Scholar] [CrossRef] [Green Version]
- Abdullah; Ali, S.; Khan, Z.; Hussain, A.; Athar, A.; Kim, H.-C. Computer Vision Based Deep Learning Approach for the Detection and Classification of Algae Species Using Microscopic Images. Water 2022, 14, 2219. [Google Scholar] [CrossRef]
| Combined Name/Complex | Species in Complete Dataset | Rationale |
|---|---|---|
| Achnanthidium minutissimum (Kützing) Czarnecki complex | Achnanthidium alteragracillimum Round & Bukhtiyarova | Relative similarity of the valves may bring over-specification of diatoms and enhance error [51] |
| Achnanthidium affine (Grunow) Czarnecki | ||
| Achnanthidium minussimum (Kützing) Czarnecki | ||
| Achnanthidium minutissimum var. jackii (Rabenhorst) Lange-Bertalot | ||
| Achnanthidium saprophilum (Kobayashi & Mayama) Round & Bukhtiyarova | ||
| Cocconeis placentula Ehrenberg | Cocconeis placentula var. euglypta Ehrenberg | Following diatoms.org, we selected this as a complex [52] |
| Cocconeis placentula var. euglyptoides (Geitler) Lange-Bertalot | ||
| Cocconeis placentula Ehrenberg | ||
| Cocconeis lineata Ehrenberg | ||
| Fistulifera saprophila (Lange-Bertalot & Bonik) Lange-Bertalot | Fistulifera saprophila (Lange-Bertalot & Bonik) Lange-Bertalot | Taxa difficult to differentiate with the “traditional” method [53], pooled into F. saprophila. |
| Fistulifera pellucida (Kützing) Lange-Bertalot | ||
| Luticola mutica (Kützing) D.G. Mann | Luticola goeppertiana (Bleisch) D.G. Mann ex J. Rarick, S. Wu, S.S. Lee & Edlund | Expected misidentification in “traditional” method [30] |
| Luticola mutica (Kützing) D.G. Mann | ||
| Luticola saprophila Levkov, Metzeltin & A. Pavlov | ||
| Mayamaea atomus (Kützing) Lange-Bertalot | Mayamaea atomus (Kützing) Lange-Bertalot | Possible species splitting in “traditional” method [28] |
| Mayamaea atomus var. alcimonica (E.Reichardt) E.Reichardt | ||
| Mayamaea atomus var. permitis (Hustedt) Lange-Bertalot | ||
| Nitzschia palea (Kützing) W. Smith | Nitzschia palea var. debilis (Kützing) Grunow | Possible species splitting in “traditional” method [54] |
| Nitzschia palea (Kützing) W. Smith | ||
| Psammothidium grischunum Bukhtiyarova & Round | Psammothidium bioretii (H.Germain) Buhtiyarova & Round | Possible species splitting in “traditional” method [28] |
| Psammothidium daonense (Lange-Bertalot) Lange-Bertalot | ||
| Psammothidium grischunum Bukhtiyarova & Round | ||
| Ulnaria ulna (Nitzsch) Compère | Ulnaria acus (Kützing) Aboal | Possible species splitting in “traditional” method [55] |
| Ulnaria danica (Kützing) Compère & Bukhtiyarova | ||
| Ulnaria (Kützing) Compère | ||
| Ulnaria ulna (Nitzsch) Compère |
| F-Value Richness Complete Dataset | F-Value Richness Reduced Dataset | F-Value Community Structure Reduced Dataset | |
|---|---|---|---|
| M1 per method | 14.333 *,1 | −3.333 n.s. | 3.890 n.s. |
| M2 per method | 6.667 n.s. | 0.667 n.s. | 5.004 n.s. |
| M3 per method | 8.333 n.s. | −9.000 n.s. | 3.927 n.s. |
| M4 per method | 10.000 n.s. | −9.667 n.s. | 3.528 n.s. |
| M5 per method | 4.000 n.s. | 0.333 n.s. | 1.834 n.s. |
| M6 per method | 6.000 n.s. | −2.333 n.s. | 1.695 n.s. |
| Type of Indices | Variables (-Test) | F-Value Method |
|---|---|---|
| Species richness and diversity functions | Species richness | 27.66 ***,1 |
| Shannon entropy + | 0.604 n.s. | |
| Shannon–Kruskal Wallis | 0.025 n.s. | |
| Simpson entropy + | 2.855 n.s. | |
| Simpson–Kruskal Wallis | 0.169 n.s. | |
| Trophic diatom indices | IPS | 0.478 n.s. |
| IDG | 0.387 n.s. | |
| IBD | 0.004 n.s. | |
| TDI | 0.095 n.s. | |
| Rott TI | 1.479 n.s. | |
| Saprobic diatom indices | Rott SI | 0.066 n.s. |
| Sládeček | 0.016 n.s. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Burfeid-Castellanos, A.M.; Kloster, M.; Beszteri, S.; Postel, U.; Spyra, M.; Zurowietz, M.; Nattkemper, T.W.; Beszteri, B. A Digital Light Microscopic Method for Diatom Surveys Using Embedded Acid-Cleaned Samples. Water 2022, 14, 3332. https://doi.org/10.3390/w14203332
Burfeid-Castellanos AM, Kloster M, Beszteri S, Postel U, Spyra M, Zurowietz M, Nattkemper TW, Beszteri B. A Digital Light Microscopic Method for Diatom Surveys Using Embedded Acid-Cleaned Samples. Water. 2022; 14(20):3332. https://doi.org/10.3390/w14203332
Chicago/Turabian StyleBurfeid-Castellanos, Andrea M., Michael Kloster, Sára Beszteri, Ute Postel, Marzena Spyra, Martin Zurowietz, Tim W. Nattkemper, and Bánk Beszteri. 2022. "A Digital Light Microscopic Method for Diatom Surveys Using Embedded Acid-Cleaned Samples" Water 14, no. 20: 3332. https://doi.org/10.3390/w14203332
APA StyleBurfeid-Castellanos, A. M., Kloster, M., Beszteri, S., Postel, U., Spyra, M., Zurowietz, M., Nattkemper, T. W., & Beszteri, B. (2022). A Digital Light Microscopic Method for Diatom Surveys Using Embedded Acid-Cleaned Samples. Water, 14(20), 3332. https://doi.org/10.3390/w14203332






