Removal of P-Nitrophenol by Nano Zero Valent Iron-Cobalt and Activated Persulfate Supported onto Activated Carbon
Abstract
:1. Introduction
2. Material and Methods
2.1. Reagents and Materials
2.2. Pre Experiment and Batch Experiments
2.3. Analytical Method
3. Results and Discussion
3.1. Characterization of Co-nZVI/GAC Composite
3.2. Effect of PS Concentration
3.3. Effect of Co-nZVI/GAC Dosage
3.4. Effect of Different Material System
3.5. Effects of Initial PNP Concentration
3.6. Effect of Initial pH
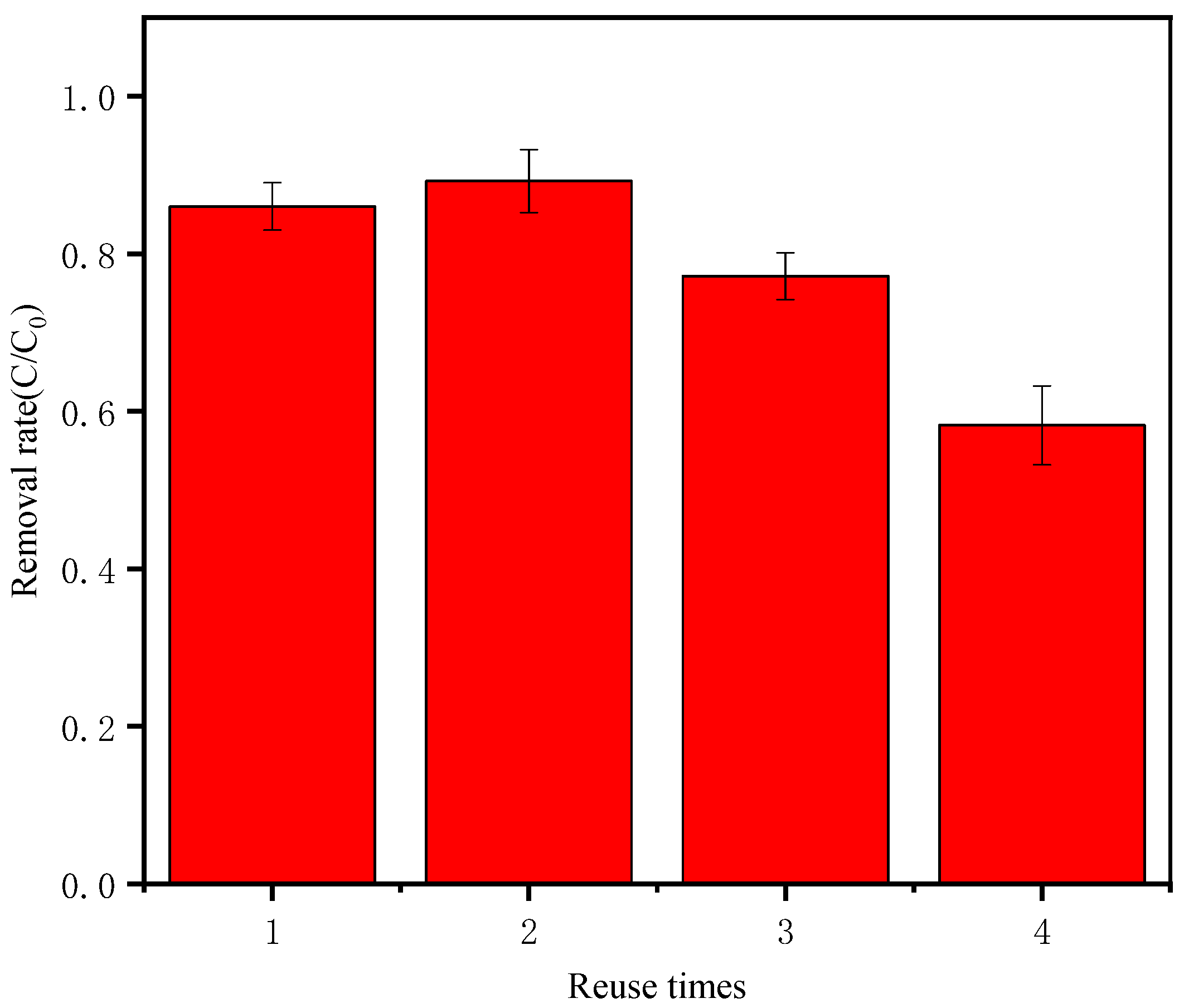
3.7. Material Reuse
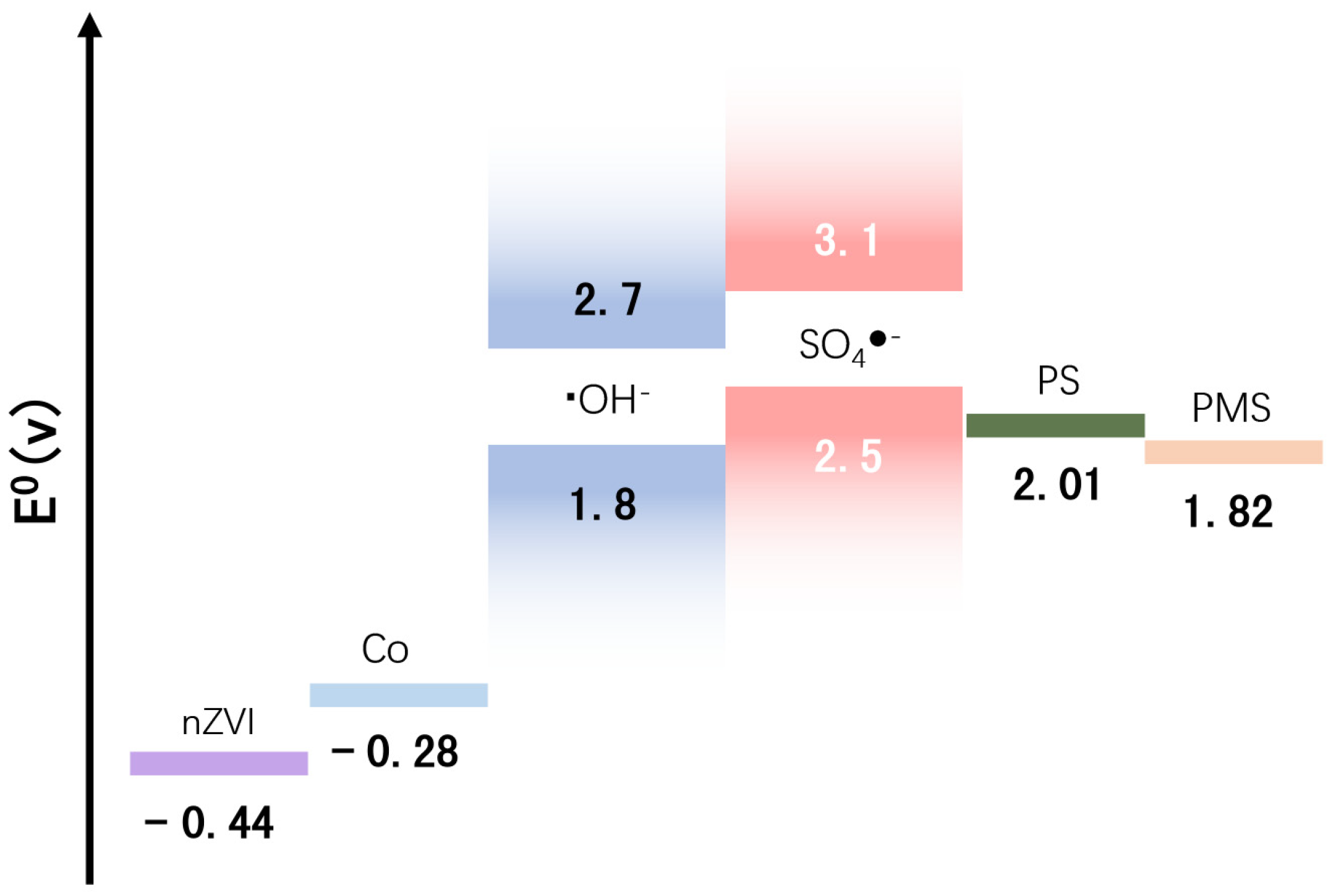
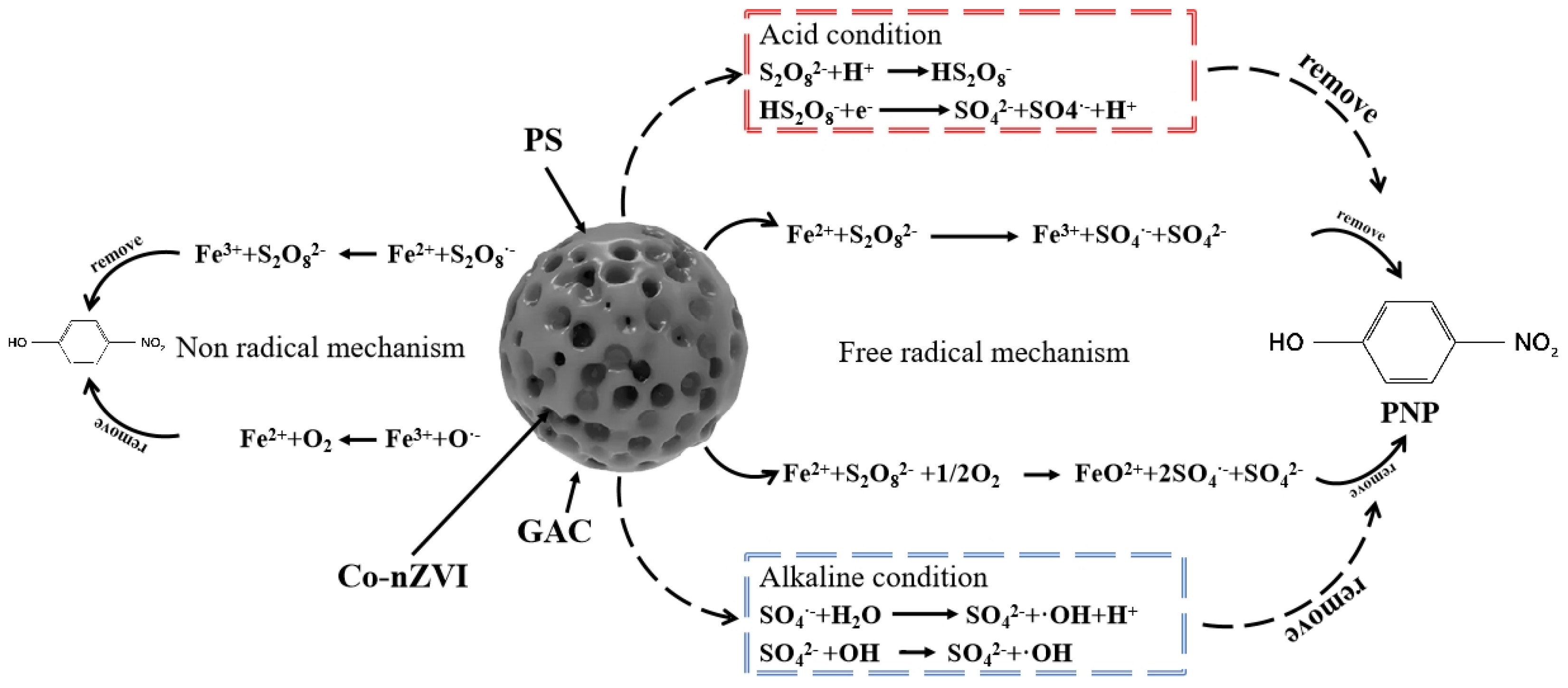
4. Degradation Mechanism
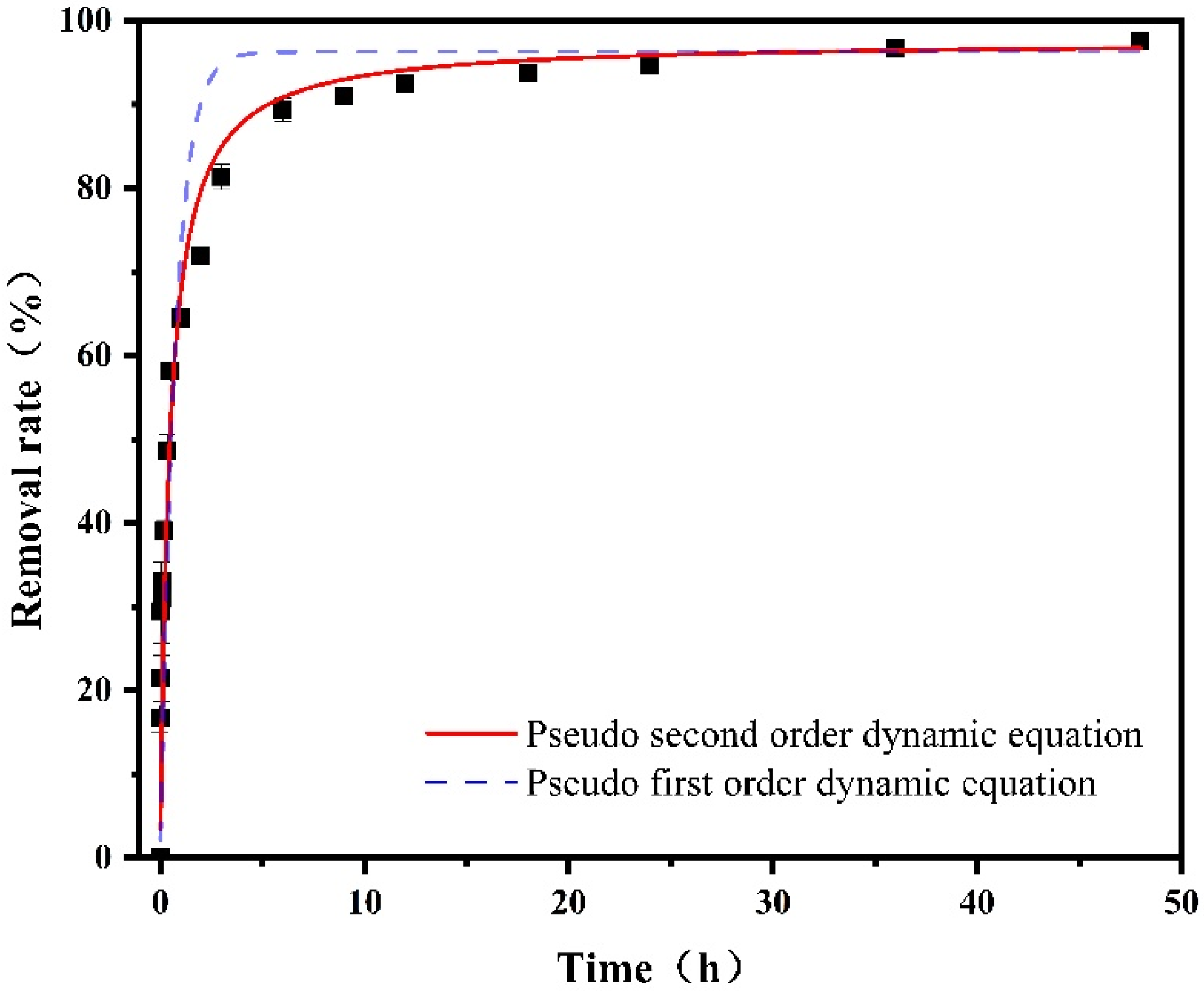
4.1. Dynamic Analysis
4.2. Fourier Infrared Spectroscopic Analysis
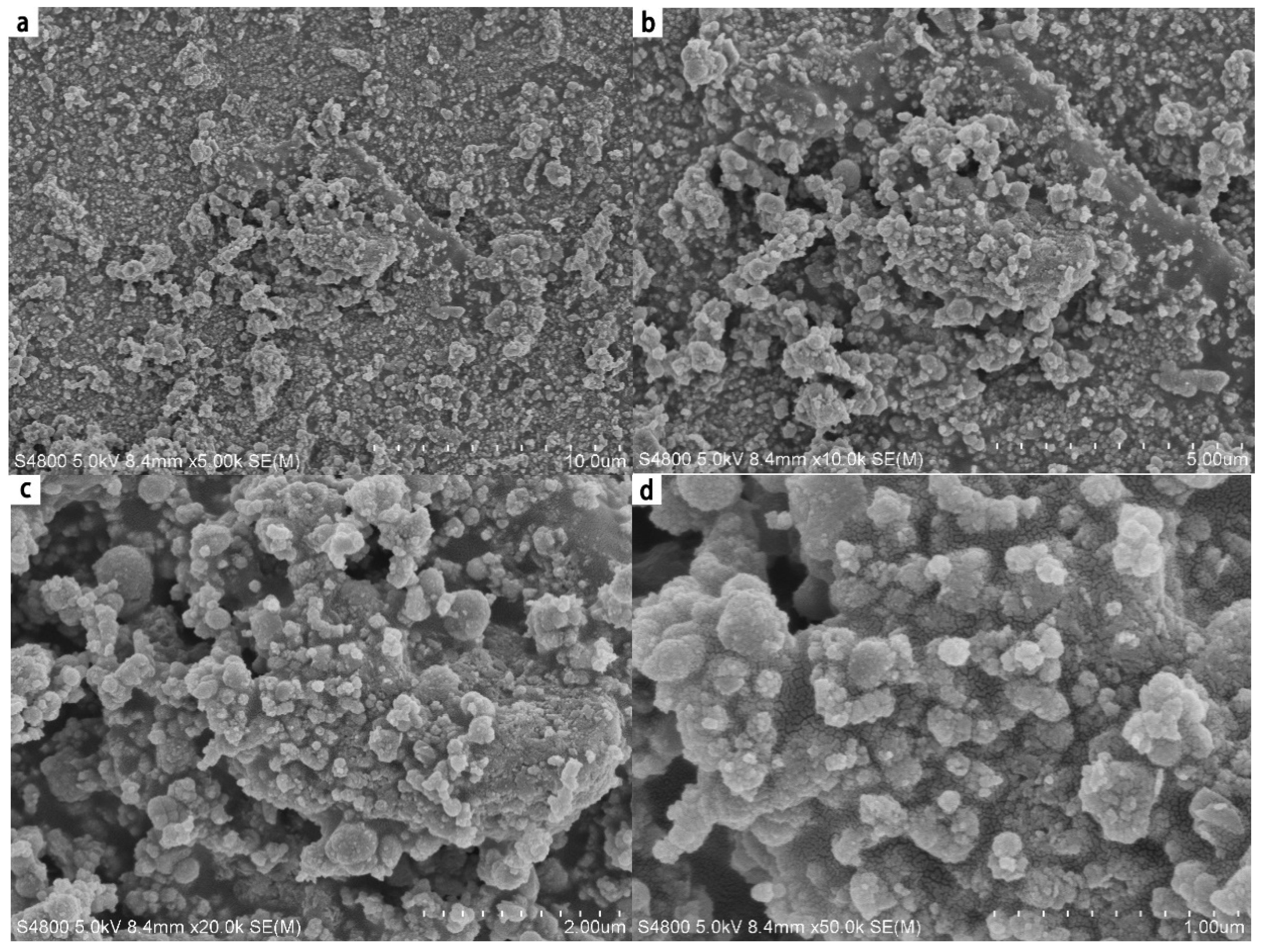
4.3. SEM Analysis
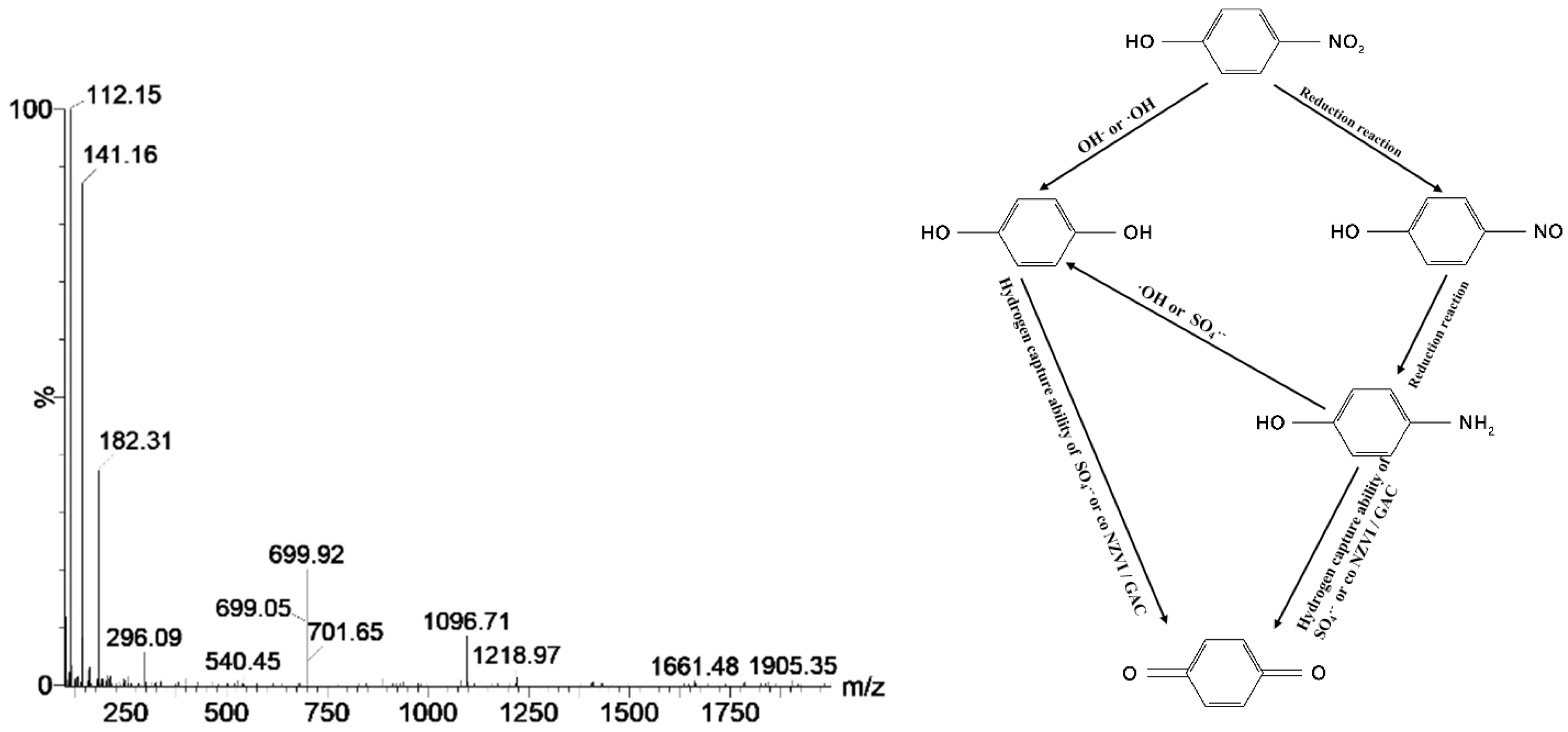
4.4. Degradation Routine and Mechanism Analysis
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Kitagawa, W.; Kimura, N.; Kamagata, Y. A Novel p-Nitrophenol Degradation Gene Cluster from a Gram-Positive Bacterium, Rhodococcus opacus SAO101. J. Bacteriol. 2004, 186, 4894–4902. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Meng, F.; Duan, W.; Lin, Y.; Sierra, J. Biodegradation of phenol in saline or hypersaline environments by bacteria: A review. Ecotoxicol. Environ. Saf. 2019, 184, 109658. [Google Scholar] [CrossRef] [PubMed]
- Hai, F.I.; Yamamoto, K.; Fukushi, K. Hybrid Treatment Systems for Dye Wastewater. Crit. Rev. Environ. Sci. Technol. 2007, 37, 315–377. [Google Scholar] [CrossRef]
- Shen, J.; Xu, X.; Jiang, X.; Hua, C.; Zhang, L.; Sun, X.; Li, J.; Mu, Y.; Wang, L. Coupling of a bioelectrochemical system for p-nitrophenol removal in an upflow anaerobic sludge blanket reactor. Water Res. 2014, 67, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.M.; Zheng, C.R.; Tu, C.; Zhu, Y.G. Heavy Metal Pollution in Soils in China: Status and Countermeasures. Ambio 1999, 28, 130–134. [Google Scholar]
- Tummala, S.; Lee, C.H.; Ho, Y.P. Boron, and nitrogen co-doped carbon dots as a multiplexing probe for sensing of p-nitrophenol, Fe (III), and temperature. Nanotechnology 2021, 32, 265502. [Google Scholar] [CrossRef]
- Jin, Z.; Hu, R.; HWang Hu, J.; Ren, T. One-step impregnation method to prepare direct Z-scheme LaCoO3/g-C3N4 heterojunction photocatalysts for phenol degradation under visible light. Appl. Surf. Sci. 2019, 491, 432–442. [Google Scholar] [CrossRef]
- Scott, T.; Zhao, H.; Deng, W.; Feng, X.; Li, Y. Photocatalytic degradation of phenol in water under simulated sunlight by an ultrathin MgO coated Ag/TiO2 nanocomposite. Chemosphere 2019, 216, 1–8. [Google Scholar] [CrossRef]
- Wai-Kay, S.; Lai, C.L.; Ip, P.; James, F.; Ka-Ho, W.D.; Chi-Hang, Y.J. A Large Population Histology Study Showing the Lack of Association between ALT Elevation and Significant Fibrosis in Chronic Hepatitis B. PLoS ONE 2012, 7, e32622. [Google Scholar]
- Lam, S.H.; Ung, C.Y.; Hlaing, M.M.; Hu, J.; Li, Z.H.; Mathavan, S.; Gong, Z. Molecular insights into 4-nitrophenol-induced hepatotoxicity in zebrafish: Transcriptomic, histological and targeted gene expression analyses. Biochim. Biophys. Acta (BBA)-Gen. Subj. 2013, 1830, 4778–4789. [Google Scholar] [CrossRef]
- Brillas, E.; Garcia-Segura, S. Benchmarking recent advances and innovative technology approaches of Fenton, photo-Fenton, electro-Fenton, and related processes: A review on the relevance of phenol as model molecule. Sep. Purif. Technol. 2020, 237, 116337. [Google Scholar] [CrossRef]
- West, L.E. Water Quality Criteria. Am. J. Public Health Nations Health 2011, 46, 1346. [Google Scholar]
- Wang, T.; Qu, G.; Sun, Q.; Liang, D.; Hu, S. Evaluation of the potential of p-nitrophenol degradation in dredged sediment by pulsed discharge plasma. Water Res. 2015, 84, 18–24. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, S. Activation of persulfate (PS) and peroxymonosulfate (PMS) and application for the degradation of emerging contaminants. Chem. Eng. J. 2018, 334, 1502–1517. [Google Scholar] [CrossRef]
- Li, Y.; Baghi, R.; Filip, J.; Islam, S.; Hope-Weeks, L.J.; Yan, W. Activation of Peroxydisulfate by Ferrite Materials for Phenol Degradation. ACS Sustain. Chem. Eng. 2019, 7, 8099–8108. [Google Scholar] [CrossRef]
- Vansek, P. Electrochemical Series. In Handbook of Chemistry and Physics; CRC Press: Boca Raton, FL, USA, 2012. [Google Scholar]
- Yi, D.A.; Zl, A.; Wei, J.; By, A.; Th, A.; Lw, B. Degradation of phenol using a peroxidase mimetic catalyst through conjugating deuterohemin-peptide onto metal-organic framework with enhanced catalytic activity. Catal. Commun. 2019, 134, 105859. [Google Scholar]
- Matzek, L.W.; Carter, K.E. Activated persulfate for organic chemical degradation: A review. Chemosphere 2016, 151, 178–188. [Google Scholar] [CrossRef] [PubMed]
- Al-Shamsi, M.A.; Thomson, N.R.; Research, E.C. Treatment of Organic Compounds by Activated Persulfate Using Nanoscale Zerovalent Iron. Ind. Eng. Chem. Res. 2013, 52, 13564–13571. [Google Scholar] [CrossRef]
- Tang, L.; Tang, J.; Zeng, G.; Yang, G.; Xie, X.; Zhou, Y. Rapid reductive degradation of aqueous p-nitrophenol using nanoscale zero-valent iron particles immobilized on mesoporous silica with enhanced antioxidation effect. Appl. Surf. Sci. 2015, 333, 220–228. [Google Scholar] [CrossRef]
- Elliott, D.; Li, X.; Zhang, W. Zero-valent Iron Nanoparticles for Abatement of Environmental Pollutants. Part. Syst. Nano-Biotechnol. 2008, 31, 111–122. [Google Scholar]
- Shiva, M.; Atashi, H.; Tabrizi, F.F.; Mirzaei, A.A. Kinetic modeling of Fischer–Tropsch synthesis on bimetallic Fe–Co catalyst with phenomenological based approaches. J. Ind. Eng. Chem. 2012, 18, 1112–1121. [Google Scholar] [CrossRef]
- Wang, G.; Chen, S.; Quan, X.; Yu, H.; Zhang, Y. Enhanced activation of peroxymonosulfate by nitrogen doped porous carbon for effective removal of organic pollutants. Carbon 2017, 115, 730–739. [Google Scholar] [CrossRef]
- Schrick, B.; Hydutsky, B.W.; Blough, J.L.; Mallouk, T.E. Delivery Vehicles for Zerovalent Metal Nanoparticles in Soil and Groundwater. Chem. Mater. 2004, 16, 2187–2193. [Google Scholar] [CrossRef]
- Liang, J.; Hassan, M.; Zhu, D.; Guo, L.; Bo, X. Cobalt nanoparticles/nitrogen-doped graphene with high nitrogen doping efficiency as noble metal-free electrocatalysts for oxygen reduction reaction. J. Colloid Interface Sci. 2017, 490, 576–586. [Google Scholar] [CrossRef] [PubMed]
- Kong, X.; Han, Z.; Wei, Z.; Le, S.; Hui, L. Synthesis of zeolite-supported microscale zero-valent iron for the removal of Cr6+ and Cd2+ from aqueous solution. J. Environ. Manag. 2016, 169, 84–90. [Google Scholar] [CrossRef] [PubMed]
- Cao, Z.; Liu, X.; Xu, J.; Zhang, J.; Yang, Y.; Zhou, J. Removal of Antibiotic Florfenicol by Sulfide-Modified Nanoscale Zero-Valent Iron. Environ. Sci. Technol. 2017, 51, 11269. [Google Scholar] [CrossRef]
- Long, D.A. Infrared and Raman characteristic group frequencies. Tables and charts. George Socrates. J. Raman Spectrosc. 2010, 35, 905. [Google Scholar] [CrossRef]
- Ying, W.; Wu, H.; Yu, A.; Wen, D. Biodegradable Polylactide/Chitosan Blend Membranes. Biomacromolecules 2006, 7, 1362–1372. [Google Scholar]
- Puziy, A.M.; Poddubnaya, O.I.; Martínez-Alonso, A.; Suárez-García, F.; Tascón, J. Surface chemistry of phosphorus-containing carbons of lignocellulosic origin. Carbon 2005, 43, 2857–2868. [Google Scholar] [CrossRef]
- Singh, P.; Raiza, P.; Kumari, S.; Kumar, A.; Thakur, P. Solar-Fenton removal of malachite green with novel Fe0-activated carbon nanocomposite. Appl. Catal. A Gen. 2014, 476, 9–18. [Google Scholar] [CrossRef]
- Gao, J.; Han, D.; Xu, Y.; Liu, Y.; Shang, J. Persulfate activation by sulfide-modified nanoscale iron supported by biochar (S-nZVI/BC) for degradation of ciprofloxacin. Sep. Purif. Technol. 2019, 235, 116202. [Google Scholar] [CrossRef]
- Arsalanfar, M.; Mirzaei, A.A.; Atashi, H.; Bozorgzadeh, H.R.; Vahid, S.; Zare, A. An investigation of the kinetics and mechanism of Fischer–Tropsch synthesis on Fe–Co–Mn supported catalyst. Fuel Processing Technol. 2014, 20, 2166–2173. [Google Scholar] [CrossRef]
- Wang, C.; Xu, Z.; Ding, G.; Wang, X.; Zhao, M.; Ho, S.S.H. Comprehensive study on the removal of chromate from aqueous solution by synthesized kaolin supported nanoscale zero-valent iron. Desalination Water Treat. 2016, 57, 5065–5078. [Google Scholar] [CrossRef]
- Zhang, W.; Zhou, S.; Sun, J. Impact of Chloride Ions on UV/H2O2 and UV/Persulfate Advanced Oxidation Processes. Environ. Sci. Technol. 2018, 52, 7380–7389. [Google Scholar] [CrossRef]
- Tang, J.; Tang, L.; Feng, H.; Zeng, G.; Dong, H.; Zhang, C. pH-dependent degradation of p-nitrophenol by sulfidated nanoscale zerovalent iron under aerobic or anoxic conditions. J. Hazard. Mater. 2016, 320, 581–590. [Google Scholar] [CrossRef]
- Dong, H.; Zhang, C.; Deng, J.; Jiang, Z.; Zhang, L.; Cheng, Y. Factors influencing degradation of trichloroethylene by sulfide-modified nanoscale zero-valent iron in aqueous solution. Water Res. 2018, 135, 1–10. [Google Scholar] [CrossRef]
- Wang, W.; Wang, J.; Guo, Y.; Zhu, C.; Pan, F.; Wu, R. Removal of multiple nitrosamines from aqueous solution by nanoscale zero-valent iron supported on granular activated carbon: Influencing factors and reaction mechanism. Sci. Total Environ. 2018, 639, 934. [Google Scholar] [CrossRef]
- Tang, L.; Wang, J.; Zeng, G.; Liu, Y.; De Ng, Y.; Zhou, Y. Enhanced photocatalytic degradation of norfloxacin in aqueous Bi2WO6 dispersions containing nonionic surfactant under visible light irradiation. J. Hazard. Mater. 2016, 306, 295–304. [Google Scholar] [CrossRef]
- Yuan, S.; Meng, T.; Cui, Y.; Lin, L.; Lu, X. Treatment of nitrophenols by cathode reduction and electro-Fenton methods. J. Hazard. Mater. 2006, 137, 573–580. [Google Scholar] [CrossRef]

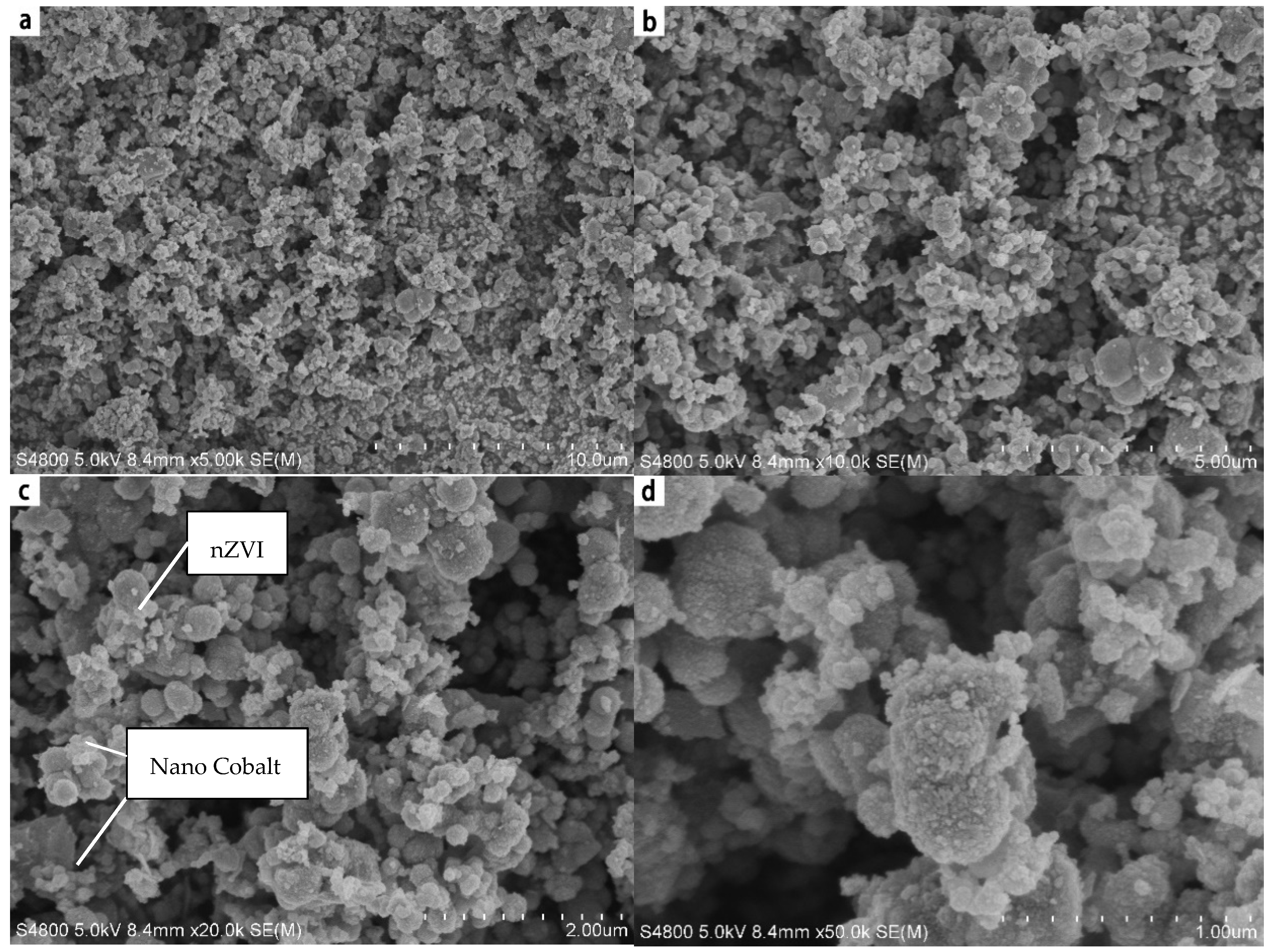

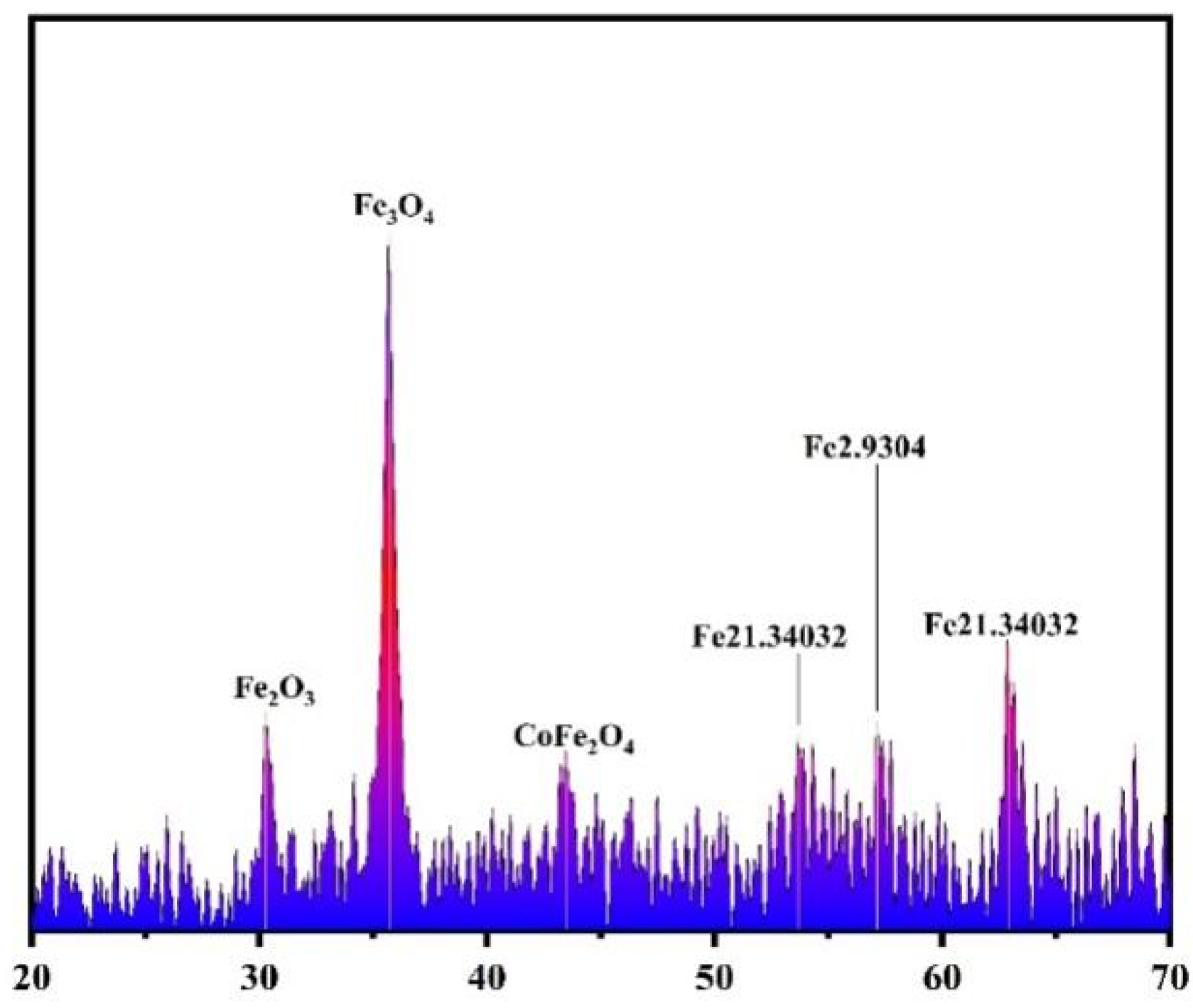


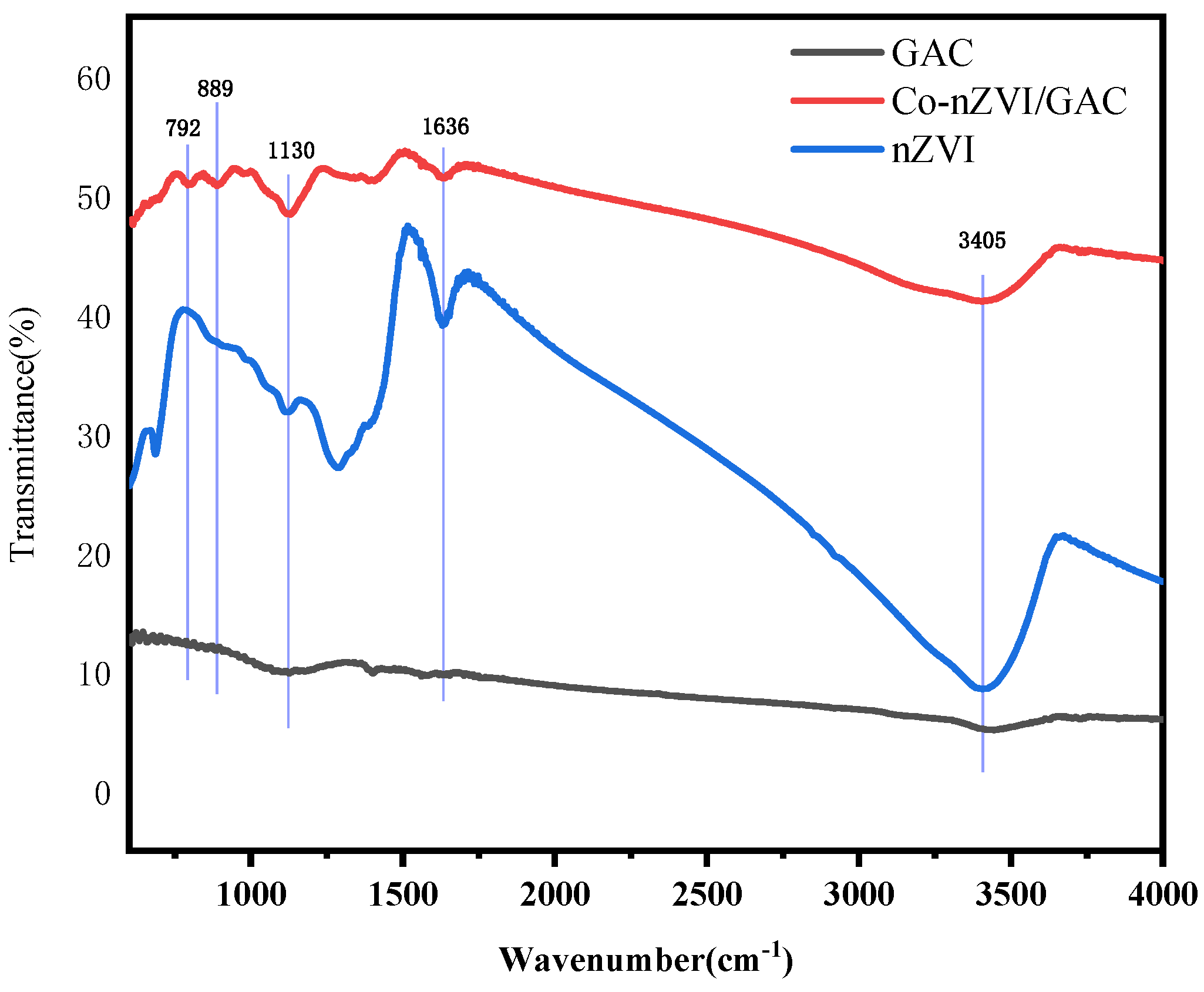
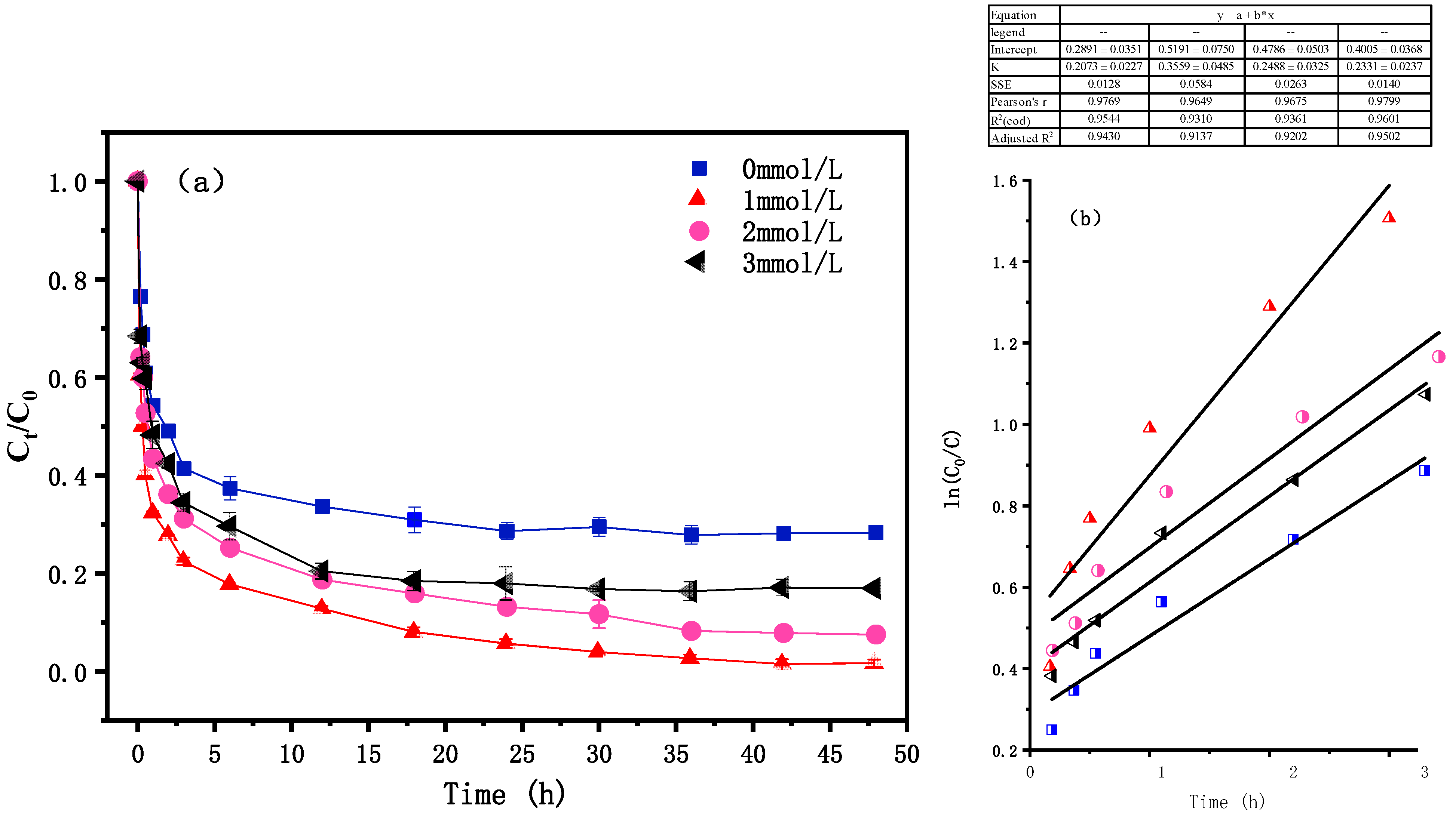

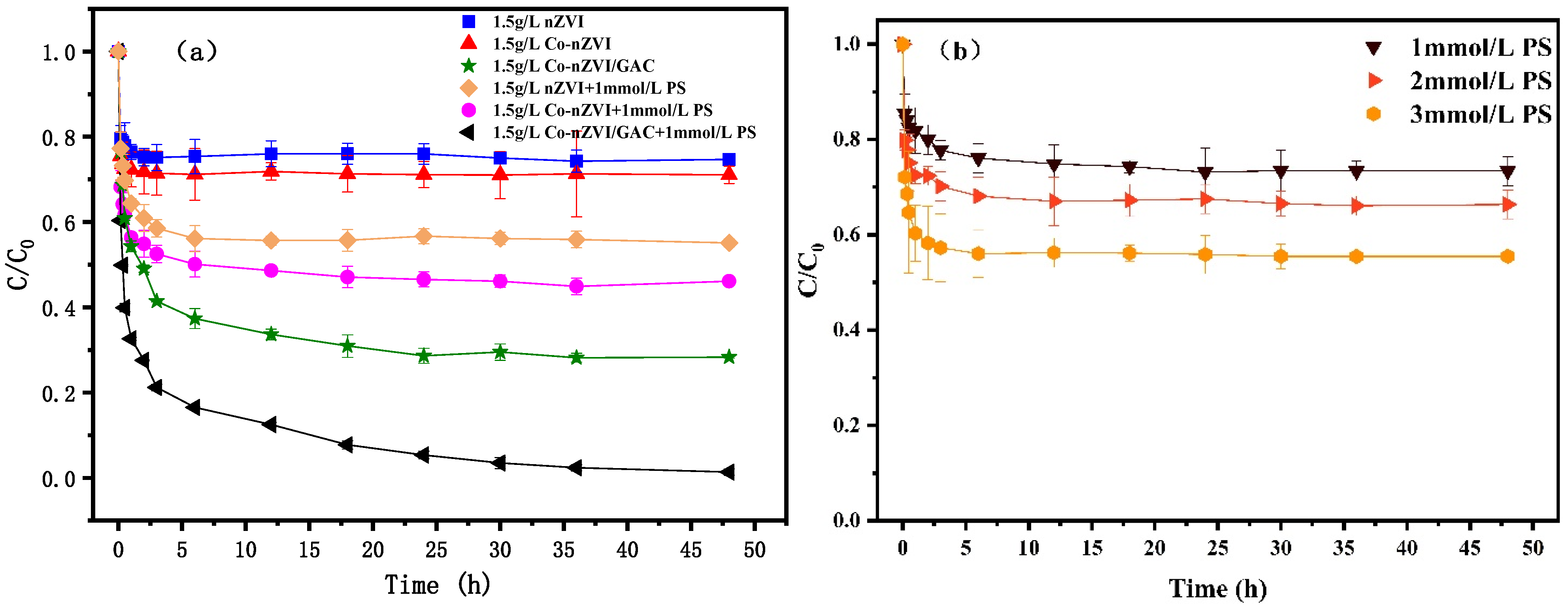

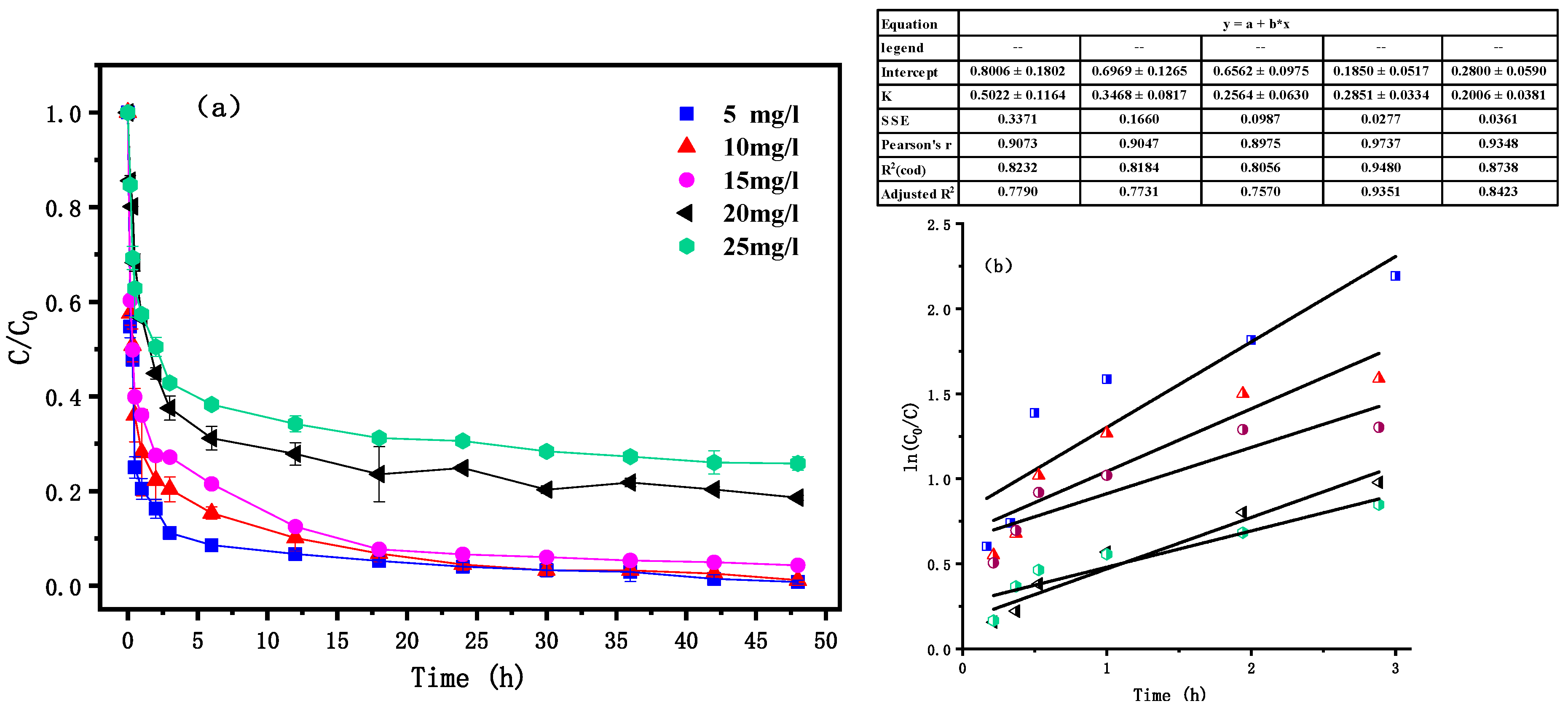
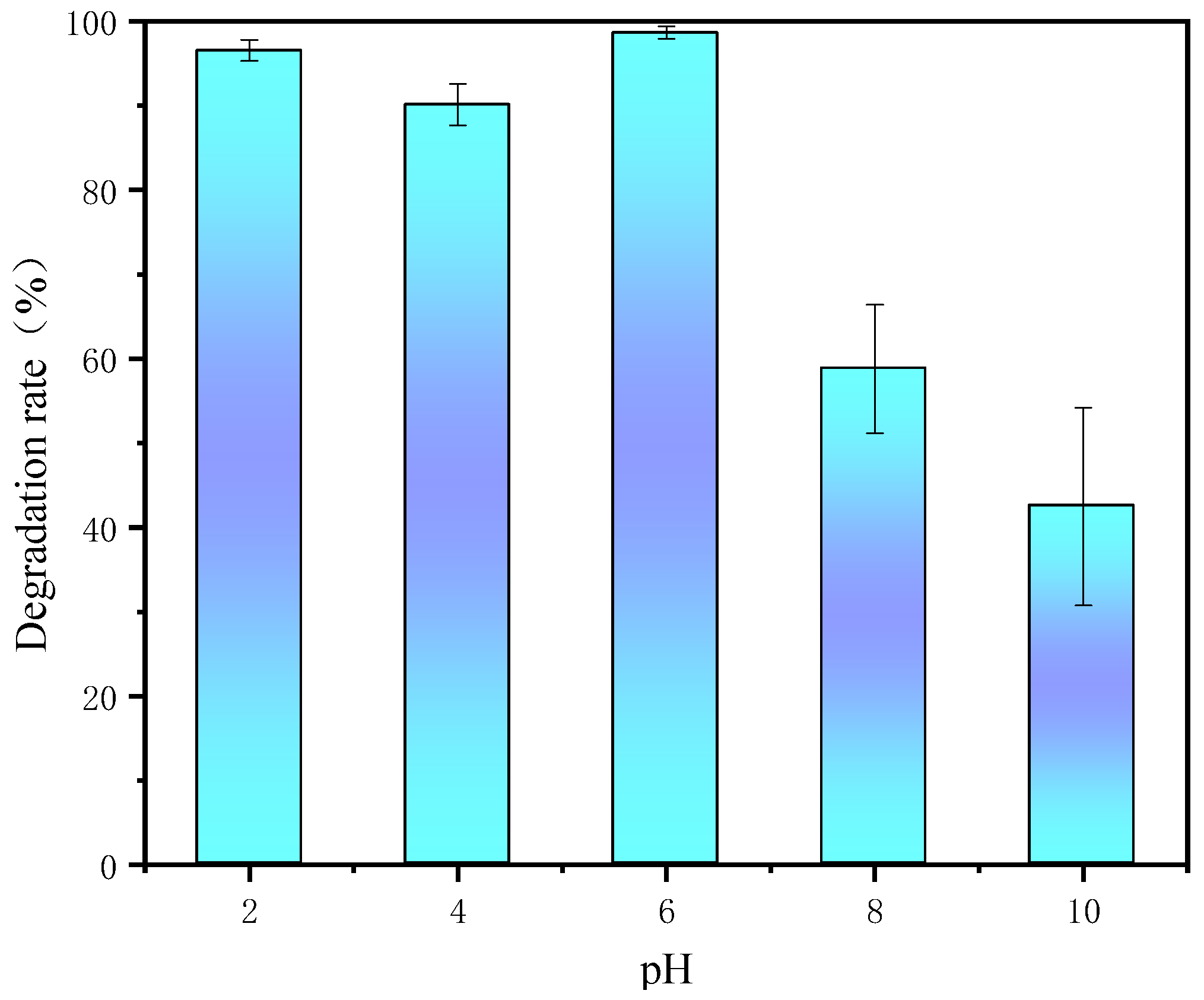

| Materials | BET/(m2·g−1) | Pore Volume/(mL·g−1) | Aperture (nm) |
|---|---|---|---|
| GAC | 557.12 | 0.28 | 1.02 |
| nZVI | 97.25 | 0.05 | 1.12 |
| Co-nZVI | 51.85 | 0.02 | 1.10 |
| Co-nZVI/GAC | 482.61 | 0.25 | 1.01 |
| Model | Pseudo First-Order Kinetics | Pseudo Second-Order Kinetics | ||||
|---|---|---|---|---|---|---|
| Equation | ||||||
| Parameter | qe/(mg·g−1) | k1/(min−1) | R2 | qe/(mg·g−1) | k2/(g·mg−1·min−1) | R2 |
| Co-nZVI/GAC | 0.9631 | 1.3856 | 0.7625 | 0.9764 | 2.2812 | 0.9022 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.; Chen, L.; Zhang, X. Removal of P-Nitrophenol by Nano Zero Valent Iron-Cobalt and Activated Persulfate Supported onto Activated Carbon. Water 2022, 14, 1387. https://doi.org/10.3390/w14091387
Zhang J, Chen L, Zhang X. Removal of P-Nitrophenol by Nano Zero Valent Iron-Cobalt and Activated Persulfate Supported onto Activated Carbon. Water. 2022; 14(9):1387. https://doi.org/10.3390/w14091387
Chicago/Turabian StyleZhang, Jiankun, Lei Chen, and Xueyang Zhang. 2022. "Removal of P-Nitrophenol by Nano Zero Valent Iron-Cobalt and Activated Persulfate Supported onto Activated Carbon" Water 14, no. 9: 1387. https://doi.org/10.3390/w14091387
APA StyleZhang, J., Chen, L., & Zhang, X. (2022). Removal of P-Nitrophenol by Nano Zero Valent Iron-Cobalt and Activated Persulfate Supported onto Activated Carbon. Water, 14(9), 1387. https://doi.org/10.3390/w14091387





