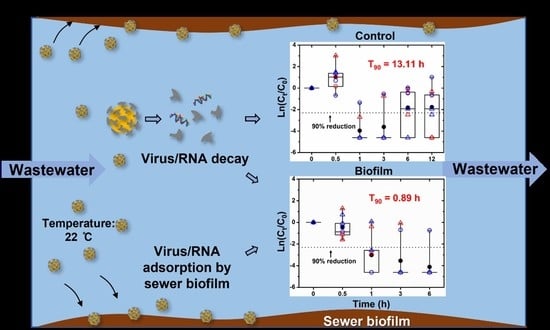
The Reduction of SARS-CoV-2 RNA Concentration in the Presence of Sewer Biofilms
Abstract
:1. Introduction
2. Materials and Methods
2.1. SARS-CoV-2 RNA Positive Wastewater
2.2. Laboratory-Scale Sewer System
2.3. Sewer Reactor Tests and Sampling Schemes
2.4. Sample Processing and RNA Extraction
2.5. RT-qPCR Assay
2.6. Data Analysis
3. Results
3.1. Reduction of SARS-CoV-2 RNA in Wastewater
3.2. In-Sewer Reduction Kinetics Analysis of SARS-CoV-2 RNA in the Wastewater Phase of Sewer Reactors
3.3. Accumulation of SARS-CoV-2 RNA in Sewer Biofilms
4. Discussion
| RT-qPCR Assay | Initial Concentration | Virus Types | Testing Method | T (°C) | k (Mean ± SD) [95% CI] (d−1) | R2 | T90 (d) | Reference |
|---|---|---|---|---|---|---|---|---|
| CDC-N1 | 107.03 ± 0.19 GC/mL | Gamma-irradiated SARS-CoV-2 | 4.99 mL of wastewater and 5 mL of gamma-irradiated SARS-CoV-2 were mixed in 15 mL sterile conical tubes. 140 μL aliquot was sampled from each tube at each sampling time point for extraction and RT-qPCR analysis. | 4 | 0.084 ± 0.013 [0.103 to 0.064] | 0.79 | 27.8 ± 4.45 [22.4 to 50.1] | [13] |
| 15 | 0.114 ± 0.012 [0.144 to 0.083] | 0.71 | 20.4 ± 2.13 [16.0 to 27.7] | |||||
| 25 | 0.183 ± 0.008 [0.219 to 0.149] | 0.87 | 12.6 ± 0.59 [10.5 to 15.5] | |||||
| 37 | 0.286 ± 0.008 [0.370 to 0.202] | 0.74 | 8.04 ± 0.23 [6.22 to 11.4] | |||||
| E-Sarbeco | 105.4 GC/mL | 1:1000 dilution of SARS-CoV-2 inoculum | Spiked wastewater influent was packed separately into 57 microcentrifuge tubes in portions of 300 μL. Triplicate tubes were stored in the dark and extracted for RT-qPCR analysis. | 4 | 0.04 ± 0.2 | 0.59 | 52 | [15] |
| CDC-N2 | 106.1 GC/mL | 0.06 ± 0.0 | 0.99 | 36 | ||||
| E-Sarbeco | High titer (105 TCID50/mL) | SARS-CoV-2 nCoV WA1-2020 (MN985325.1), isolated from a clinical patient | SARS-CoV-2 nCoV-WA1-2020 (MN985325.1) was diluted 1:10 to wastewater. 1 mL aliquots were pipetted into 2 mL screw-top vials with 3 replicates for each time point. | 20 | 0.67 [0.54 to 0.86] | 0.27 | 3.3 [2.7 to 4.3] | |
| Low titer (103 TCID50/mL) | 0.09 [0.00 * to 0.23] | −0.01 | 26 [9.8 to ∞] | |||||
| CDC-N1/N2 | 135–953 GC/mL | SARS-CoV-2 RNA positive wastewater samples without seeding | 4 | 2.16 | - | - | [39] | |
| 10 | 0.96 | - | - | |||||
| 35 | 4.31 | - | - | |||||
| CDC-N | 20.9–41.8 GC/mL | SARS-CoV-2 RNA positive wastewater samples without seeding | 50 mL of wastewater was sampled from each reactor at each time point for RNA extraction and RT-qPCR analysis. | 22 | Control: 2.1 [0.49 to 3.73] | 0.81 | 1.1 [0.00 * to 4.73] | This study |
| Biofilm k1: 94.32 [66.72 to 132.96] Biofilm k2: 1.68 [0.00 * to 7.2] | 0.66 | 0.03 [0.02 to 0.04] | ||||||
| E-Sarbeco | 36.4–230 GC/mL | Control: 6.32 [0.00 * to 15.35] | 0.53 | 0.36 [0.18 to 0.69] | ||||
| Biofilm:27.6 [17.52 to 56.4] | 0.7 | 0.08 [0.06 to 0.25] |
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Peccia, J.; Zulli, A.; Brackney, D.E.; Grubaugh, N.D.; Kaplan, E.H.; Casanovas-Massana, A.; Ko, A.I.; Malik, A.A.; Wang, D.; Wang, M.; et al. Measurement of SARS-CoV-2 RNA in wastewater tracks community infection dynamics. Nat. Biotechnol. 2020, 38, 1164–1167. [Google Scholar] [CrossRef] [PubMed]
- Galani, A.; Aalizadeh, R.; Kostakis, M.; Markou, A.; Alygizakis, N.; Lytras, T.; Adamopoulos, P.G.; Peccia, J.; Thompson, D.C.; Kontou, A.; et al. SARS-CoV-2 wastewater surveillance data can predict hospitalizations and ICU admissions. Sci. Total Environ. 2022, 804, 150151. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, A.; Lipponen, A.; Hokajärvi, A.-M.; Luomala, O.; Sarekoski, A.; Rytkönen, A.; Österlund, P.; Al-Hello, H.; Juutinen, A.; Miettinen, I.T.; et al. Detection and quantification of SARS-CoV-2 RNA in wastewater influent in relation to reported COVID-19 incidence in Finland. Water Res. 2022, 215, 118220. [Google Scholar] [CrossRef] [PubMed]
- Sherchan, S.P.; Shahin, S.; Ward, L.M.; Tandukar, S.; Aw, T.G.; Schmitz, B.; Ahmed, W.; Kitajima, M. First detection of SARS-CoV-2 RNA in wastewater in North America: A study in Louisiana, USA. Sci. Total Environ. 2020, 743, 140621. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Gao, J.; Zheng, Q.; Thai, P.K.; Duan, H.; Mueller, J.F.; Yuan, Z.; Jiang, G. Effects of pH, Temperature, Suspended Solids, and Biological Activity on Transformation of Illicit Drug and Pharmaceutical Biomarkers in Sewers. Environ. Sci. Technol. 2021, 55, 8771–8782. [Google Scholar] [CrossRef]
- Proverbio, D.; Kemp, F.; Magni, S.; Ogorzaly, L.; Cauchie, H.-M.; Gonçalves, J.; Skupin, A.; Aalto, A. Model-based assessment of COVID-19 epidemic dynamics by wastewater analysis. Sci. Total Environ. 2022, 827, 154235. [Google Scholar] [CrossRef]
- Vallejo, J.A.; Trigo-Tasende, N.; Rumbo-Feal, S.; Conde-Pérez, K.; López-Oriona, Á.; Barbeito, I.; Vaamonde, M.; Tarrío-Saavedra, J.; Reif, R.; Ladra, S.; et al. Modeling the number of people infected with SARS-COV-2 from wastewater viral load in Northwest Spain. Sci. Total Environ. 2022, 811, 152334. [Google Scholar] [CrossRef]
- Jiang, G.; Wu, J.; Weidhaas, J.; Li, X.; Chen, Y.; Mueller, J.; Li, J.; Kumar, M.; Zhou, X.; Arora, S.; et al. Artificial neural network-based estimation of COVID-19 case numbers and effective reproduction rate using wastewater-based epidemiology. Water Res. 2022, 218, 118451. [Google Scholar] [CrossRef]
- Li, X.; Zhang, S.; Shi, J.; Luby, S.P.; Jiang, G. Uncertainties in estimating SARS-CoV-2 prevalence by wastewater-based epidemiology. Chem. Eng. J. 2021, 415, 129039. [Google Scholar] [CrossRef]
- Morales Medina, W.R.; D’Elia, S.; Fahrenfeld, N.L. Accumulation of SARS-CoV-2 RNA in Sewer Biofilms. ACS EST Water 2022, 2, 1844–1851. [Google Scholar] [CrossRef]
- Fu, S.; Wang, Q.; He, F.; Zhou, C.; Zhang, J.; Xia, W. The decay of coronavirus in sewage pipes and the development of a predictive model for the estimation of SARS-CoV-2 infection cases based on wastewater surveillance. medRxiv 2022. [CrossRef]
- Bertels, X.; Demeyer, P.; Van den Bogaert, S.; Boogaerts, T.; van Nuijs, A.L.N.; Delputte, P.; Lahousse, L. Factors influencing SARS-CoV-2 RNA concentrations in wastewater up to the sampling stage: A systematic review. Sci. Total Environ. 2022, 820, 153290. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, W.; Bertsch, P.M.; Bibby, K.; Haramoto, E.; Hewitt, J.; Huygens, F.; Gyawali, P.; Korajkic, A.; Riddell, S.; Sherchan, S.P.; et al. Decay of SARS-CoV-2 and surrogate murine hepatitis virus RNA in untreated wastewater to inform application in wastewater-based epidemiology. Environ. Res. 2020, 191, 110092. [Google Scholar] [CrossRef] [PubMed]
- Bivins, A.; Greaves, J.; Fischer, R.; Yinda, K.C.; Ahmed, W.; Kitajima, M.; Munster, V.J.; Bibby, K. Persistence of SARS-CoV-2 in Water and Wastewater. Environ. Sci. Technol. Lett. 2020, 7, 937–942. [Google Scholar] [CrossRef]
- Hokajarvi, A.M.; Rytkonen, A.; Tiwari, A.; Kauppinen, A.; Oikarinen, S.; Lehto, K.M.; Kankaanpaa, A.; Gunnar, T.; Al-Hello, H.; Blomqvist, S.; et al. The detection and stability of the SARS-CoV-2 RNA biomarkers in wastewater influent in Helsinki, Finland. Sci. Total Environ. 2021, 770, 145274. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.W.; Banks, A.P.W.; Novic, A.J.; Mueller, J.F.; Jiang, G.; Ort, C.; Eaglesham, G.; Yuan, Z.; Thai, P.K. Impact of in-Sewer Degradation of Pharmaceutical and Personal Care Products (PPCPs) Population Markers on a Population Model. Environ. Sci. Technol. 2017, 51, 3816–3823. [Google Scholar] [CrossRef]
- Ort, C.; van Nuijs, A.L.N.; Berset, J.-D.; Bijlsma, L.; Castiglioni, S.; Covaci, A.; de Voogt, P.; Emke, E.; Fatta-Kassinos, D.; Griffiths, P.; et al. Spatial differences and temporal changes in illicit drug use in Europe quantified by wastewater analysis. Addiction 2014, 109, 1338–1352. [Google Scholar] [CrossRef] [Green Version]
- Hart, O.E.; Halden, R.U. Modeling wastewater temperature and attenuation of sewage-borne biomarkers globally. Water Res. 2020, 172, 115473. [Google Scholar] [CrossRef]
- McCall, C.; Fang, Z.N.; Li, D.; Czubai, A.J.; Juan, A.; LaTurner, Z.W.; Ensor, K.; Hopkins, L.; Bedient, P.B.; Stadler, L.B. Modeling SARS-CoV-2 RNA degradation in small and large sewersheds. Environ. Sci. Water Res. Technol. 2022, 8, 290–300. [Google Scholar] [CrossRef]
- Hvitved-Jacobsen, T.; Vollertsen, J.; Nielsen, A.H. Sewer Processes: Microbial and Chemical Process Engineering of Sewer Networks; CRC Press: Boca Raton, FL, USA, 2013. [Google Scholar]
- He, C.; Li, J.; Jiang, G.; Chen, S.; Niel, C.; Yuan, Z.; Mueller, J.F.; Thai, P. Transformation of phthalates and their metabolites in wastewater under different sewer conditions. Water Res. 2021, 190, 116754. [Google Scholar] [CrossRef]
- Li, J.; Gao, J.; Thai, P.K.; Mueller, J.F.; Yuan, Z.; Jiang, G. Transformation of Illicit Drugs and Pharmaceuticals in Sewer Sediments. Environ. Sci. Technol. 2020, 54, 13056–13065. [Google Scholar] [CrossRef] [PubMed]
- Thai, P.K.; O′Brien, J.; Jiang, G.; Gernjak, W.; Yuan, Z.; Eaglesham, G.; Mueller, J.F. Degradability of creatinine under sewer conditions affects its potential to be used as biomarker in sewage epidemiology. Water Res. 2014, 55, 272–279. [Google Scholar] [CrossRef] [PubMed]
- Choi, P.M.; O′Brien, J.W.; Li, J.; Jiang, G.; Thomas, K.V.; Mueller, J.F. Population histamine burden assessed using wastewater-based epidemiology: The association of 1,4-methylimidazole acetic acid and fexofenadine. Environ. Int. 2018, 120, 172–180. [Google Scholar] [CrossRef] [PubMed]
- O′Brien, J.W.; Choi, P.M.; Li, J.; Thai, P.K.; Jiang, G.; Tscharke, B.J.; Mueller, J.F.; Thomas, K.V. Evaluating the stability of three oxidative stress biomarkers under sewer conditions and potential impact for use in wastewater-based epidemiology. Water Res. 2019, 166, 115068. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Li, X.; Zhang, S.; Sharma, E.; Sivakumar, M.; Sherchan, S.P.; Jiang, G. Enhanced decay of coronaviruses in sewers with domestic wastewater. Sci. Total Environ. 2022, 813, 151919. [Google Scholar] [CrossRef]
- Zhang, S.; Li, X.; Shi, J.; Sivakumar, M.; Luby, S.; O′Brien, J.; Jiang, G. Analytical performance comparison of four SARS-CoV-2 RT-qPCR primer-probe sets for wastewater samples. Sci. Total Environ. 2022, 806, 150572. [Google Scholar] [CrossRef]
- Choi, P.M.; Li, J.; Gao, J.; O′Brien, J.W.; Thomas, K.V.; Thai, P.K.; Jiang, G.; Mueller, J.F. Considerations for assessing stability of wastewater-based epidemiology biomarkers using biofilm-free and sewer reactor tests. Sci. Total Environ. 2020, 709, 136228. [Google Scholar] [CrossRef]
- Li, J.; Gao, J.; Thai, P.K.; Sun, X.; Mueller, J.F.; Yuan, Z.; Jiang, G. Stability of Illicit Drugs as Biomarkers in Sewers: From Lab to Reality. Environ. Sci. Technol. 2018, 52, 1561–1570. [Google Scholar] [CrossRef] [Green Version]
- Kulandaivelu, J.; Gao, J.; Song, Y.; Shrestha, S.; Li, X.; Li, J.; Doederer, K.; Keller, J.; Yuan, Z.; Mueller, J.F.; et al. Removal of Pharmaceuticals and Illicit Drugs from Wastewater Due to Ferric Dosing in Sewers. Environ. Sci. Technol. 2019, 53, 6245–6254. [Google Scholar] [CrossRef]
- Jiang, G.; Sharma, K.R.; Guisasola, A.; Keller, J.; Yuan, Z. Sulfur transformation in rising main sewers receiving nitrate dosage. Water Res. 2009, 43, 4430–4440. [Google Scholar] [CrossRef]
- Li, W.; Zheng, T.; Ma, Y.; Liu, J. Current status and future prospects of sewer biofilms: Their structure, influencing factors, and substance transformations. Sci. Total Environ. 2019, 695, 133815. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, W.; Bertsch, P.M.; Bivins, A.; Bibby, K.; Farkas, K.; Gathercole, A.; Haramoto, E.; Gyawali, P.; Korajkic, A.; McMinn, B.R.; et al. Comparison of virus concentration methods for the RT-qPCR-based recovery of murine hepatitis virus, a surrogate for SARS-CoV-2 from untreated wastewater. Sci. Total Environ. 2020, 739, 139960. [Google Scholar] [CrossRef] [PubMed]
- Vogels, C.B.F.; Brito, A.F.; Wyllie, A.L.; Fauver, J.R.; Ott, I.M.; Kalinich, C.C.; Petrone, M.E.; Casanovas-Massana, A.; Catherine Muenker, M.; Moore, A.J.; et al. Analytical sensitivity and efficiency comparisons of SARS-CoV-2 RT-qPCR primer-probe sets. Nat. Microbiol. 2020, 5, 1299–1305. [Google Scholar] [CrossRef] [PubMed]
- Corman, V.M.; Landt, O.; Kaiser, M.; Molenkamp, R.; Meijer, A.; Chu, D.K.W.; Bleicker, T.; Brünink, S.; Schneider, J.; Schmidt, M.L. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance 2020, 25, 2000045. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmed, W.; Bivins, A.; Metcalfe, S.; Smith, W.J.M.; Verbyla, M.E.; Symonds, E.M.; Simpson, S.L. Evaluation of process limit of detection and quantification variation of SARS-CoV-2 RT-qPCR and RT-dPCR assays for wastewater surveillance. Water Res. 2022, 213, 118132. [Google Scholar] [CrossRef] [PubMed]
- Silverman, A.I.; Boehm, A.B. Systematic Review and Meta-Analysis of the Persistence and Disinfection of Human Coronaviruses and Their Viral Surrogates in Water and Wastewater. Environ. Sci. Technol. Lett. 2020, 7, 544–553. [Google Scholar] [CrossRef]
- Silverman, A.I.; Boehm, A.B. Systematic Review and Meta-Analysis of the Persistence of Enveloped Viruses in Environmental Waters and Wastewater in the Absence of Disinfectants. Environ. Sci. Technol. 2021, 55, 14480–14493. [Google Scholar] [CrossRef]
- Weidhaas, J.; Aanderud, Z.T.; Roper, D.K.; VanDerslice, J.; Gaddis, E.B.; Ostermiller, J.; Hoffman, K.; Jamal, R.; Heck, P.; Zhang, Y.; et al. Correlation of SARS-CoV-2 RNA in wastewater with COVID-19 disease burden in sewersheds. Sci. Total Environ. 2021, 775, 145790. [Google Scholar] [CrossRef]
- Ahmed, F.; Islam, M.A.; Kumar, M.; Hossain, M.; Bhattacharya, P.; Islam, M.T.; Hossen, F.; Hossain, M.S.; Islam, M.S.; Uddin, M.M.; et al. First detection of SARS-CoV-2 genetic material in the vicinity of COVID-19 isolation Centre in Bangladesh: Variation along the sewer network. Sci. Total Environ. 2021, 776, 145724. [Google Scholar] [CrossRef]
- Graham, K.E.; Loeb, S.K.; Wolfe, M.K.; Catoe, D.; Sinnott-Armstrong, N.; Kim, S.; Yamahara, K.M.; Sassoubre, L.M.; Mendoza Grijalva, L.M.; Roldan-Hernandez, L.; et al. SARS-CoV-2 RNA in Wastewater Settled Solids Is Associated with COVID-19 Cases in a Large Urban Sewershed. Environ. Sci. Technol. 2021, 55, 488–498. [Google Scholar] [CrossRef]
- Zhang, S.; Shi, J.; Sharma, E.; Li, X.; Gao, S.; Zhou, X.; O′Brien, J.; Coin, L.; Liu, Y.; Sivakumar, M.; et al. In-sewer decay and partitioning of Campylobacter jejuni and Campylobacter coli and implications for their wastewater surveillance. Water Res. 2023, 233, 119737. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, W.; Bertsch, P.M.; Angel, N.; Bibby, K.; Bivins, A.; Dierens, L.; Edson, J.; Ehret, J.; Gyawali, P.; Hamilton, K.A.; et al. Detection of SARS-CoV-2 RNA in commercial passenger aircraft and cruise ship wastewater: A surveillance tool for assessing the presence of COVID-19 infected travellers. J. Travel Med. 2020, 27, taaa116. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Li, S.; Deng, Y.; Xu, X.; Ding, J.; Lau, F.T.K.; In Yau, C.; Poon, L.L.M.; Tun, H.M.; Zhang, T. Quantification of SARS-CoV-2 RNA in wastewater treatment plants mirrors the pandemic trend in Hong Kong. Sci. Total Environ. 2022, 844, 157121. [Google Scholar] [CrossRef] [PubMed]


| Primer-Probe Sets | Efficiency (%) | Linearity (R2) | Slope (Mean ± SD) | Y-Intercept (Mean ± SD) |
|---|---|---|---|---|
| CCDC-N | 108.2 | 0.999 | −3.139 ± 0.12 | 40.39 ± 0.18 |
| E-Sarbeco | 96.36 | 0.999 | −3.412 ± 0.09 | 41.87 ± 0.26 |
| Targets/Reactors | k/k1 (h−1) [95% CI] | k2 (h−1) [95% CI] | t1 (h) [95% CI] | T90 (h) [95% CI] | R2 | RMSE | |
|---|---|---|---|---|---|---|---|
| Control (Monophasic) | RM | 0.23 [0.02 to 0.44] | - | - | 10.07 [4.52 to 21.63] | 0.75 | 0.89 |
| GS | 0.12 [0 * to 0.31] | - | - | 18.77 [0 * to 227.8] | 0.48 | 0.79 | |
| N gene | 0.09 [0.02 to 0.16] | - | - | 26.19 [0 * to 113.5] | 0.81 | 0.28 | |
| E gene | 0.26 [0 * to 0.64] | - | - | 8.74 [4.35 to 16.49] | 0.53 | 1.59 | |
| Total | 0.18 [0 * to 0.37] | - | - | 13.11 [7.38 to 24.14] | 0.67 | 0.8 | |
| Biofilm (Biphasic) | RM | 2.74 [1.38 to 4.69] | 0.37 [0 * to 0.78] | 1 [0 * to 2.7] | 0.84 [0.56 to 3.41] | 0.63 | 1.36 |
| GS | 2.42 [1 to 4.41] | 0.004 [0 * to 0.66] | 1.5 [0.5 to NA] | 0.95 [0.59 to 3.79] | 0.52 | 1.53 | |
| N gene | 3.92 [2.78 to 5.44] | 0.07 [0 * to 0.3] | 1 [0.72 to 1.64] | 0.59 [0.45 to 0.86] | 0.66 | 1.25 | |
| E gene | 1.15 [0.73 to 2.35] | 0.38 [NA] | NA | 2.0 [1.45 to 6] | 0.7 | 1.24 | |
| Total | 2.58 [1.63 to 3.71] | 0.19 [0 * to 0.58] | 1.25 [0.69 to 2.33] | 0.89 [0.65 to 2.67] | 0.57 | 1.45 | |
| Type | Batch Test | N Gene (GC/cm2) | E Gene (GC/cm2) | ||||
|---|---|---|---|---|---|---|---|
| Reactor-1st | Reactor-2nd | Reactor-1st | Reactor-2nd | ||||
| 0 h | 12 h | 12 h | 0 h | 12 h | 12 h | ||
| RM | Test 1 | 196.1 * | - | 285.1 * | ND | - | ND |
| Test 2 | ND | 365.5 | 187.0 | 654.5 * | 915.4 | 544.1 | |
| GS | Test 1 | ND | - | ND | 686.1 * | - | 3622.4 |
| Test 2 | ND | ND | 487.0 * | 935.9 | 1049.6 * | 2849.4 * | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, S.; Sharma, E.; Tiwari, A.; Chen, Y.; Sherchan, S.P.; Gao, S.; Zhou, X.; Shi, J.; Jiang, G. The Reduction of SARS-CoV-2 RNA Concentration in the Presence of Sewer Biofilms. Water 2023, 15, 2132. https://doi.org/10.3390/w15112132
Zhang S, Sharma E, Tiwari A, Chen Y, Sherchan SP, Gao S, Zhou X, Shi J, Jiang G. The Reduction of SARS-CoV-2 RNA Concentration in the Presence of Sewer Biofilms. Water. 2023; 15(11):2132. https://doi.org/10.3390/w15112132
Chicago/Turabian StyleZhang, Shuxin, Elipsha Sharma, Ananda Tiwari, Yan Chen, Samendra P. Sherchan, Shuhong Gao, Xu Zhou, Jiahua Shi, and Guangming Jiang. 2023. "The Reduction of SARS-CoV-2 RNA Concentration in the Presence of Sewer Biofilms" Water 15, no. 11: 2132. https://doi.org/10.3390/w15112132
APA StyleZhang, S., Sharma, E., Tiwari, A., Chen, Y., Sherchan, S. P., Gao, S., Zhou, X., Shi, J., & Jiang, G. (2023). The Reduction of SARS-CoV-2 RNA Concentration in the Presence of Sewer Biofilms. Water, 15(11), 2132. https://doi.org/10.3390/w15112132