Reviewing the Potential of Algae Species as a Green Alternative to Produce Nanoparticles: Findings from a Database Analysis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Literature Search
2.2. Search Terms and Languages
2.3. Article Screening and Study Eligibility Criteria
2.3.1. Screening Process and Eligibility Criteria
2.3.2. Eligible Organisms
2.3.3. Eligible Type of Synthesis
2.3.4. Date Range
2.3.5. Reasons for Exclusion
2.3.6. Study Validity Assessment
2.4. Post-Selection Information Retrieval and Summary of the Basic Features of the Dataset
- Bibliographic information—title, authors, year, cited sources.
- Organisms—algae—(micro, macro, eukaryote, prokaryote) scientific name, and any other relevant phylogenetic information, algae pre-process (dying, centrifuging, filtering, lyophilizing, whole living cultures, grounding, and cutting).
- Final nanoparticle—nanoparticle characteristics—the type of NPs produced using the algae, chemical compound(s) used to produce the NPs, size of the produced NPs (measured and stated in the study), if the color of the reaction, shape, and surface of the NPs were investigated in the research and clearly mentioned.
- Use of the synthesized nanoparticle—(in the specific article), categorized following the classes in Table 1.
2.5. Study Presentation, Visualization, and Descriptive Statistics
2.6. Semantic and Citation Network Building
2.7. Bibliometric and Network Analysis (SNA)
3. Results and Discussion
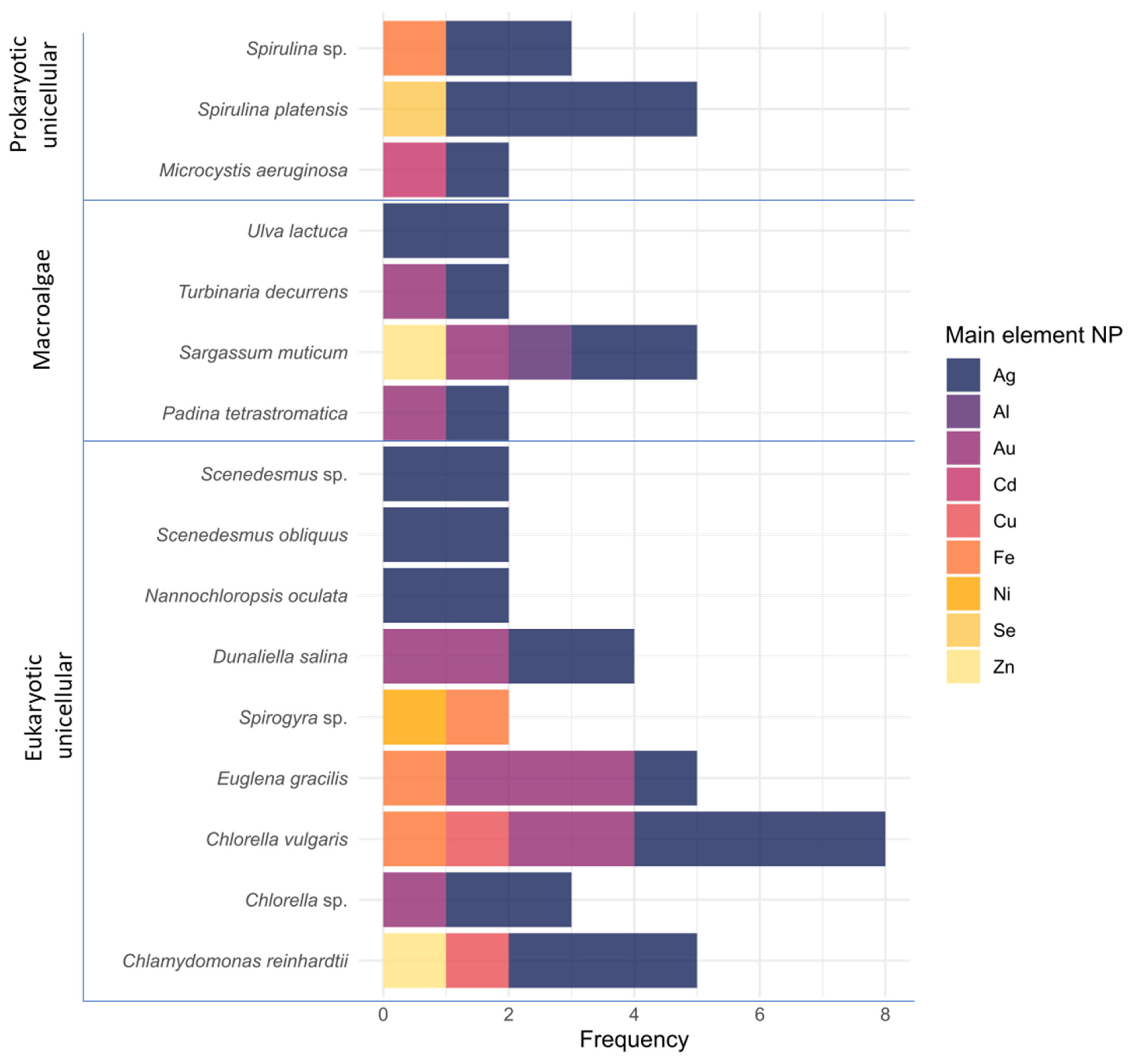
3.1. Relevant Species and the Importance of Species Characterization
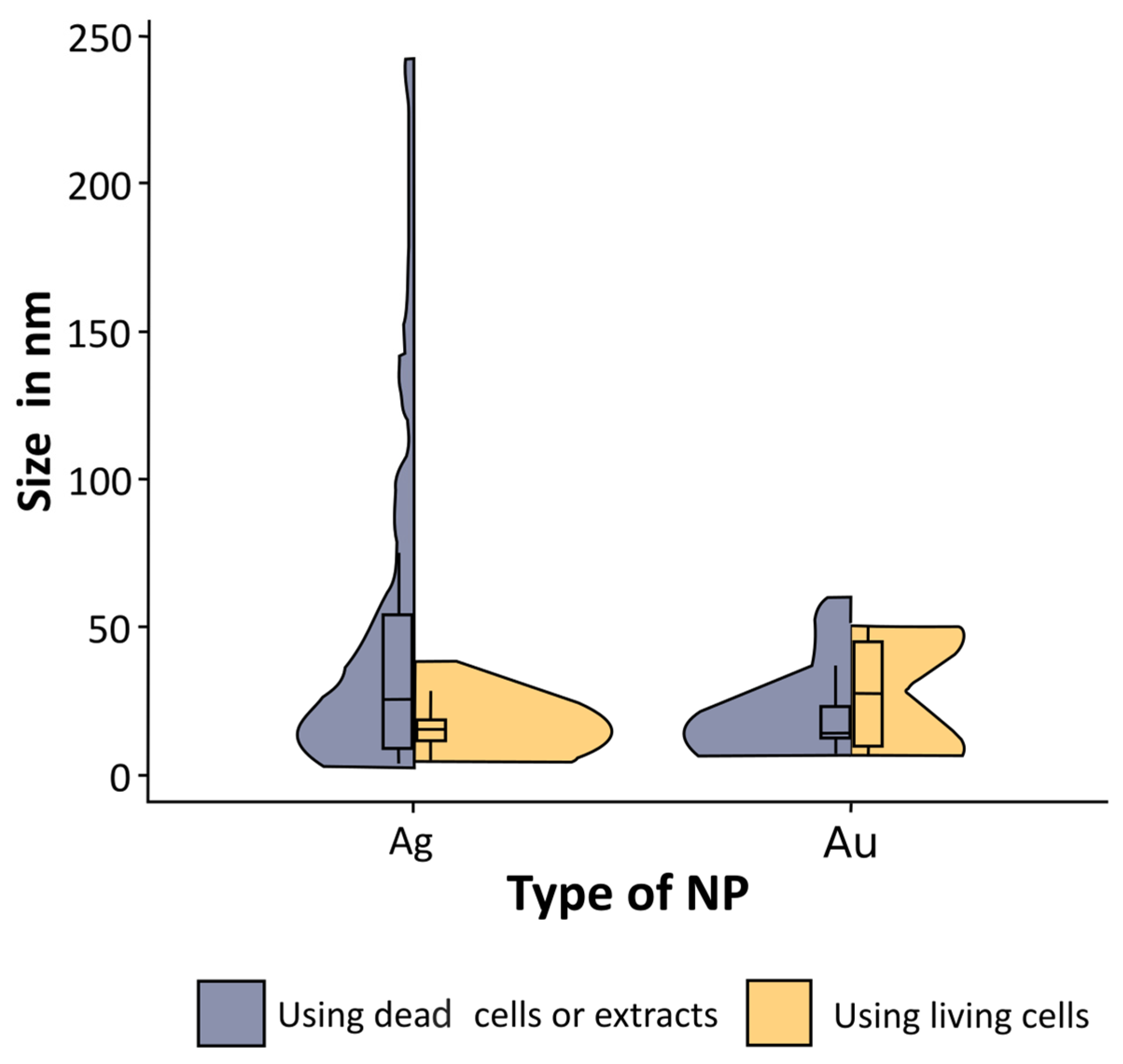
3.2. Green Synthesis and Particle Size
- (i) Factors related to the algae culture conditions:
- (ii) Changes related to the synthesis conditions:
3.3. Algae Pre-Process Dominant Methodologies and Solvents
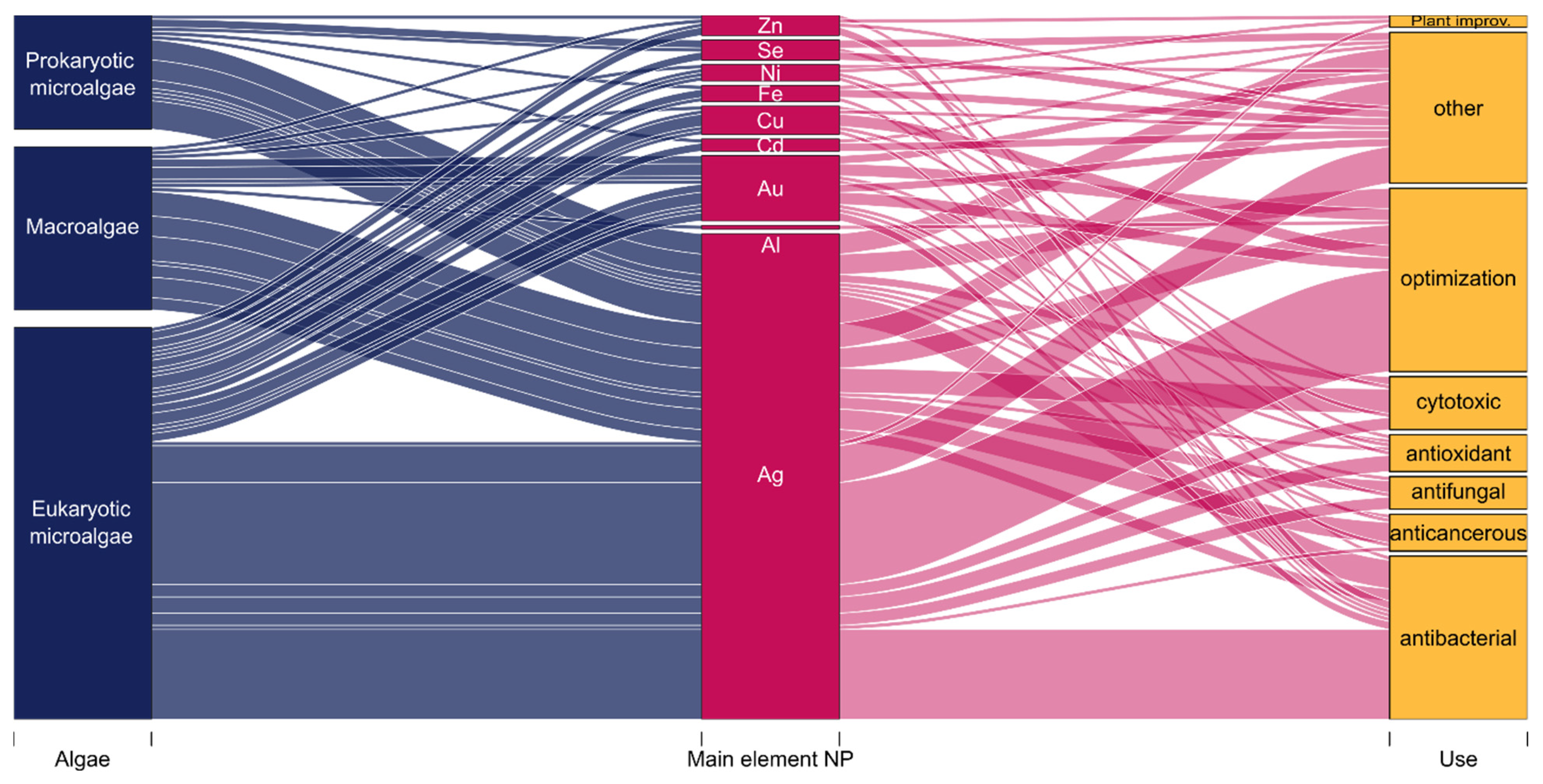
3.4. Leading Types of Nanoparticles and Possible Uses
3.5. Network Analysis
3.5.1. Qualitative Network Analysis
3.5.2. Quantitative Networks Analysis
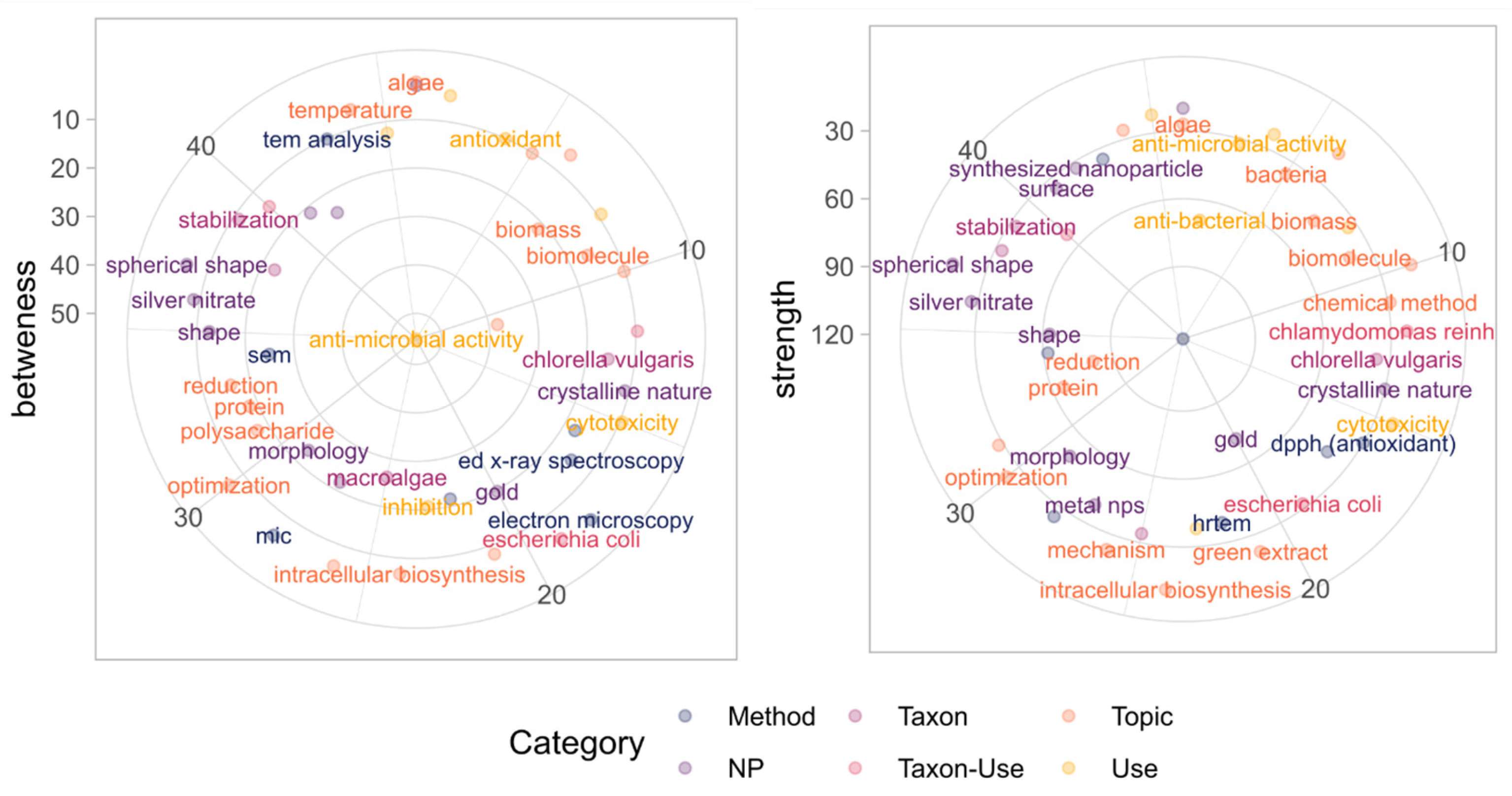
3.6. Clustering Analysis of the Semantic Network
3.7. Importance of Node Attributes
3.8. Comments on the Methodology Used to Compile the Data
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Morris, J.; Willis, J. Nanotechnology White Paper; U.S. Environmental Protection Agency: Washington, DC, USA, 2007; p. 137.
- Sanvicens, N.; Marco, M.P. Multifunctional Nanoparticles—Properties and Prospects for Their Use in Human Medicine. Trends Biotechnol. 2008, 26, 425–433. [Google Scholar] [CrossRef] [PubMed]
- Sajid, M.; Płotka-Wasylka, J. Nanoparticles: Synthesis, Characteristics, and Applications in Analytical and Other Sciences. Microchem. J. 2020, 154, 104623. [Google Scholar] [CrossRef]
- Mourdikoudis, S.; Pallares, M.R.; Thanh, K.N.T. Characterization Techniques for Nanoparticles: Comparison and Complementarity upon Studying Nanoparticle Properties. Nanoscale 2018, 10, 12871–12934. [Google Scholar] [CrossRef] [Green Version]
- Hasan, A.; Morshed, M.; Memic, A.; Hassan, S.; Webster, T.J.; Marei, H.E.-S. Nanoparticles in Tissue Engineering: Applications, Challenges and Prospects. Int. J. Nanomed. 2018, 13, 5637–5655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, I.; Saeed, K.; Khan, I. Nanoparticles: Properties, Applications and Toxicities. Arab. J. Chem. 2019, 12, 908–931. [Google Scholar] [CrossRef]
- Keat, C.L.; Aziz, A.; Eid, A.M.; Elmarzugi, N.A. Biosynthesis of Nanoparticles and Silver Nanoparticles. Bioresour. Bioprocess. 2015, 2, 47. [Google Scholar] [CrossRef] [Green Version]
- Roduner, E. Size Matters: Why Nanomaterials Are Different. Chem. Soc. Rev. 2006, 35, 583. [Google Scholar] [CrossRef]
- Jabbar, K.Q.; Barzinjy, A.A.; Hamad, S.M. Iron Oxide Nanoparticles: Preparation Methods, Functions, Adsorption and Coagulation/Flocculation in Wastewater Treatment. Environ. Nanotechnol. Monit. Manag. 2022, 17, 100661. [Google Scholar] [CrossRef]
- Schacht, V.J.; Neumann, L.V.; Sandhi, S.K.; Chen, L.; Henning, T.; Klar, P.J.; Theophel, K.; Schnell, S.; Bunge, M. Effects of Silver Nanoparticles on Microbial Growth Dynamics. J. Appl. Microbiol. 2013, 114, 25–35. [Google Scholar] [CrossRef]
- Lee, N.-Y.; Ko, W.-C.; Hsueh, P.-R. Nanoparticles in the Treatment of Infections Caused by Multidrug-Resistant Organisms. Front. Pharmacol. 2019, 10, 1153. [Google Scholar] [CrossRef] [Green Version]
- Rai, M.; Ingle, A.P.; Birla, S.; Yadav, A.; Santos, C.A.D. Strategic Role of Selected Noble Metal Nanoparticles in Medicine. Crit. Rev. Microbiol. 2016, 42, 696–719. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Gu, F.; Chan, J.; Wang, A.; Langer, R.; Farokhzad, O. Nanoparticles in Medicine: Therapeutic Applications and Developments. Clin. Pharmacol. Ther. 2008, 83, 761–769. [Google Scholar] [CrossRef] [PubMed]
- Priyadarsini, S.; Mukherjee, S.; Mishra, M. Nanoparticles Used in Dentistry: A Review. J. Oral. Biol. Craniofac. Res. 2018, 8, 58–67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khosravi-Katuli, K.; Prato, E.; Lofrano, G.; Guida, M.; Vale, G.; Libralato, G. Effects of Nanoparticles in Species of Aquaculture Interest. Environ. Sci. Pollut. Res. 2017, 24, 17326–17346. [Google Scholar] [CrossRef]
- Swain, P.; Nayak, S.K.; Sasmal, A.; Behera, T.; Barik, S.K.; Swain, S.K.; Mishra, S.S.; Sen, A.K.; Das, J.K.; Jayasankar, P. Antimicrobial Activity of Metal Based Nanoparticles against Microbes Associated with Diseases in Aquaculture. World J. Microbiol. Biotechnol. 2014, 30, 2491–2502. [Google Scholar] [CrossRef]
- Singh, R.P.; Handa, R.; Manchanda, G. Nanoparticles in Sustainable Agriculture: An Emerging Opportunity. J. Control. Release 2021, 329, 1234–1248. [Google Scholar] [CrossRef]
- Vargas-Estrada, L.; Torres-Arellano, S.; Longoria, A.; Arias, D.M.; Okoye, P.U.; Sebastian, P.J. Role of Nanoparticles on Microalgal Cultivation: A Review. Fuel 2020, 280, 118598. [Google Scholar] [CrossRef]
- Dev Sarkar, R.; Singh, H.B.; Chandra Kalita, M. Enhanced Lipid Accumulation in Microalgae through Nanoparticle-Mediated Approach, for Biodiesel Production: A Mini-Review. Heliyon 2021, 7, e08057. [Google Scholar] [CrossRef]
- Sarma, S.J.; Das, R.K.; Brar, S.K.; Le Bihan, Y.; Buelna, G.; Verma, M.; Soccol, C.R. Application of Magnesium Sulfate and Its Nanoparticles for Enhanced Lipid Production by Mixotrophic Cultivation of Algae Using Biodiesel Waste. Energy 2014, 78, 16–22. [Google Scholar] [CrossRef]
- Comotto, M.; Casazza, A.A.; Aliakbarian, B.; Caratto, V.; Ferretti, M.; Perego, P. Influence of TiO2 Nanoparticles on Growth and Phenolic Compounds Production in Photosynthetic Microorganisms. Sci. World J. 2014, 2014, e961437. [Google Scholar] [CrossRef] [Green Version]
- Huang, J.; Cheng, J.; Yi, J. Impact of Silver Nanoparticles on Marine Diatom Skeletonema Costatum. J. Appl. Toxicol. 2016, 36, 1343–1354. [Google Scholar] [CrossRef]
- Perreault, F.; Bogdan, N.; Morin, M.; Claverie, J.; Popovic, R. Interaction of Gold Nanoglycodendrimers with Algal Cells (Chlamydomonas Reinhardtii) and Their Effect on Physiological Processes. Nanotoxicology 2012, 6, 109–120. [Google Scholar] [CrossRef]
- Abid, N.; Khan, A.M.; Shujait, S.; Chaudhary, K.; Ikram, M.; Imran, M.; Haider, J.; Khan, M.; Khan, Q.; Maqbool, M. Synthesis of Nanomaterials Using Various Top-down and Bottom-up Approaches, Influencing Factors, Advantages, and Disadvantages: A Review. Adv. Colloid Interface Sci. 2022, 300, 102597. [Google Scholar] [CrossRef]
- Moraes, L.C.; Figueiredo, R.C.; Ribeiro-Andrade, R.; Pontes-Silva, A.V.; Arantes, M.L.; Giani, A.; Figueredo, C.C. High Diversity of Microalgae as a Tool for the Synthesis of Different Silver Nanoparticles: A Species-Specific Green Synthesis. Colloid Interface Sci. Commun. 2021, 42, 100420. [Google Scholar] [CrossRef]
- Pieper, D.; Puljak, L. Language Restrictions in Systematic Reviews Should Not Be Imposed in the Search Strategy but in the Eligibility Criteria If Necessary. J. Clin. Epidemiol. 2021, 132, 146–147. [Google Scholar] [CrossRef] [PubMed]
- van Eck, N.J.; Waltman, L. Visualizing Bibliometric Networks. In Measuring Scholarly Impact: Methods and Practice; Ding, Y., Rousseau, R., Wolfram, D., Eds.; Springer International Publishing: Cham, Switzerland, 2014; pp. 285–320. ISBN 978-3-319-10377-8. [Google Scholar]
- van Eck, N.J.; Waltman, L. Text Mining and Visualization Using VOSviewer. ISSI Newsl. 2011, 7, 50–54. [Google Scholar] [CrossRef]
- Waltman, L.; van Eck, N.J.; Noyons, E.C.M. A Unified Approach to Mapping and Clustering of Bibliometric Networks. J. Informetr. 2010, 4, 629–635. [Google Scholar] [CrossRef] [Green Version]
- Luke, D.A. Subgroups. In A User’s Guide to Network Analysis in R; Springer International Publishing: Cham, Switzerland, 2015; pp. 105–123. ISBN 978-3-319-23883-8. [Google Scholar]
- Callon, M.; Courtial, J.P.; Laville, F. Co-Word Analysis as a Tool for Describing the Network of Interactions between Basic and Technological Research: The Case of Polymer Chemsitry. Scientometrics 1991, 22, 155–205. [Google Scholar] [CrossRef]
- Handcock, M.S.; Hunter, D.R.; Butts, C.T.; Goodreau, S.M.; Krivitsky, P.N.; Morris, M. Ergm: Fit, Simulate and Diagnose Exponential-Family Models for Networks, R package version 4.4.0; 2023. Available online: https://github.com/statnet/ergm (accessed on 27 February 2023).
- Hunter, D.R.; Handcock, M.S.; Butts, C.T.; Goodreau, S.M.; Morris, M. Ergm: A Package to Fit, Simulate and Diagnose Exponential-Family Models for Networks. J. Stat. Softw. 2008, 24, nihpa54860. [Google Scholar] [CrossRef] [Green Version]
- Krivitsky, P.N.; Hunter, D.R.; Morris, M.; Klumb, C. Ergm 4: New Features for Analyzing Exponential-Family Random Graph Models. J. Stat. Softw. 2023, 105, 1–44. [Google Scholar] [CrossRef]
- Luke, D.A. Statistical Network Models. In A User’s Guide to Network Analysis in R; Luke, D., Ed.; Use R! Springer International Publishing: Cham, Switzerland, 2015; pp. 163–187. ISBN 978-3-319-23883-8. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019. [Google Scholar]
- Csardi, G.; Nepusz, T. The Igraph Software Package for Complex Network Research. InterJournal 2006, 1695, 1–9. [Google Scholar]
- Bojanowski, M. Intergraph: Coercion Routines for Network Data Objects. R package version 2.0-2; 2016. Available online: https://mbojan.github.io/intergraph (accessed on 27 February 2023).
- Mourão, M.M.; Xavier, L.P.; Urbatzka, R.; Figueiroa, L.B.; da Costa, C.E.F.; Dias, C.G.B.T.; Schneider, M.P.C.; Vasconcelos, V.; Santos, A.V. Characterization and Biotechnological Potential of Intracellular Polyhydroxybutyrate by Stigeoclonium Sp. B23 Using Cassava Peel as Carbon Source. Polymers 2021, 13, 687. [Google Scholar] [CrossRef] [PubMed]
- Moavi, J.; Buazar, F.; Sayahi, M.H. Algal Magnetic Nickel Oxide Nanocatalyst in Accelerated Synthesis of Pyridopyrimidine Derivatives. Sci. Rep. 2021, 11, 6296. [Google Scholar] [CrossRef] [PubMed]
- Marella, T.K.; Saxena, A.; Tiwari, A.; Datta, A.; Dixit, S. Treating Agricultural Non-Point Source Pollutants Using Periphyton Biofilms and Biomass Volarization. J. Environ. Manag. 2022, 301, 113869. [Google Scholar] [CrossRef] [PubMed]
- Jamil, Z.; Naqvi, S.T.Q.; Rasul, S.; Hussain, S.B.; Fatima, N.; Qadir, M.I.; Muhammad, S.A. Antibacterial Activity and Characterization of Zinc Oxide Nanoparticles Synthesized by Microalgae. Pak. J. Pharm. Sci. 2020, 33, 2497–2504. [Google Scholar] [CrossRef] [PubMed]
- Koçer, A.T.; Özçimen, D. Eco-Friendly Synthesis of Silver Nanoparticles from Macroalgae: Optimization, Characterization and Antimicrobial Activity. Biomass Conv. Bioref. 2022. [Google Scholar] [CrossRef]
- El-Sheekh, M.M.; Alwaleed, E.A.; Kassem, W.M.A.; Saber, H. Antialgal and Antiproliferative Activities of the Algal Silver Nanoparticles against the Toxic Cyanobacterium Microcystis Aeruginosa and Human Tumor Colon Cell Line. Environ. Nanotechnol. Monit. Manag. 2020, 14, 100352. [Google Scholar] [CrossRef]
- Kashyap, M.; Samadhiya, K.; Ghosh, A.; Anand, V.; Shirage, P.M.; Bala, K. Screening of Microalgae for Biosynthesis and Optimization of Ag/AgCl Nano Hybrids Having Antibacterial Effect. RSC Adv. 2019, 9, 25583–25591. [Google Scholar] [CrossRef] [Green Version]
- El-Kassas, H.Y.; Ghobrial, M.G. Biosynthesis of Metal Nanoparticles Using Three Marine Plant Species: Anti-Algal Efficiencies against “Oscillatoria Simplicissima”. Environ. Sci. Pollut. Res. 2017, 24, 7837–7849. [Google Scholar] [CrossRef]
- Hoshyar, N.; Gray, S.; Han, H.; Bao, G. The Effect of Nanoparticle Size on in Vivo Pharmacokinetics and Cellular Interaction. Nanomedicine 2016, 11, 673–692. [Google Scholar] [CrossRef] [Green Version]
- De Matteis, V.; Rizzello, L.; Cascione, M.; Liatsi-Douvitsa, E.; Apriceno, A.; Rinaldi, R. Green Plasmonic Nanoparticles and Bio-Inspired Stimuli-Responsive Vesicles in Cancer Therapy Application. Nanomaterials 2020, 10, 1083. [Google Scholar] [CrossRef] [PubMed]
- Osonga, F.J.; Akgul, A.; Yazgan, I.; Akgul, A.; Eshun, G.B.; Sakhaee, L.; Sadik, O.A. Size and Shape-Dependent Antimicrobial Activities of Silver and Gold Nanoparticles: A Model Study as Potential Fungicides. Molecules 2020, 25, 2682. [Google Scholar] [CrossRef] [PubMed]
- Al-Amri, N.; Tombuloglu, H.; Slimani, Y.; Akhtar, S.; Barghouthi, M.; Almessiere, M.; Alshammari, T.; Baykal, A.; Sabit, H.; Ercan, I.; et al. Size Effect of Iron (III) Oxide Nanomaterials on the Growth, and Their Uptake and Translocation in Common Wheat (Triticum aestivum L.). Ecotoxicol. Environ. Saf. 2020, 194, 110377. [Google Scholar] [CrossRef] [PubMed]
- Paul, M.J.; Foyer, C.H. Sink Regulation of Photosynthesis. J. Exp. Bot. 2001, 52, 1383–1400. [Google Scholar] [CrossRef] [PubMed]
- Long, S.P.; Marshall-Colon, A.; Zhu, X.-G. Meeting the Global Food Demand of the Future by Engineering Crop Photosynthesis and Yield Potential. Cell 2015, 161, 56–66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lavens, P.; Sorgeloos, P. Manual on the Production and Use of Live Food for Aquaculture; FAO Fisheries Technical Paper; Food and Agriculture Organization of the United Nations: Rome, Italy, 1996; pp. 10–14. ISBN 92-5-103934-8. [Google Scholar]
- Salas-Herrera, G.; González-Morales, S.; Benavides-Mendoza, A.; Castañeda-Facio, A.O.; Fernández-Luqueño, F.; Robledo-Olivo, A. Impact of Microalgae Culture Conditions over the Capacity of Copper Nanoparticle Biosynthesis. J. Appl. Phycol. 2019, 31, 2437–2447. [Google Scholar] [CrossRef]
- Darwesh, O.M.; Matter, I.A.; Eida, M.F.; Moawad, H.; Oh, Y.-K. Influence of Nitrogen Source and Growth Phase on Extracellular Biosynthesis of Silver Nanoparticles Using Cultural Filtrates of Scenedesmus Obliquus. Appl. Sci. 2019, 9, 1465. [Google Scholar] [CrossRef] [Green Version]
- Alipour, S.; Kalari, S.; Morowvat, M.H.; Sabahi, Z.; Dehshahri, A. Green Synthesis of Selenium Nanoparticles by Cyanobacterium Spirulina platensis (Abdf2224): Cultivation Condition Quality Controls. BioMed Res. Int. 2021, 2021, e6635297. [Google Scholar] [CrossRef]
- Chand Mali, S.; Dhaka, A.; Sharma, S.; Trivedi, R. Review on Biogenic Synthesis of Copper Nanoparticles and Its Potential Applications. Inorg. Chem. Commun. 2023, 149, 110448. [Google Scholar] [CrossRef]
- de Jaeger, L.; Carreres, B.M.; Springer, J.; Schaap, P.J.; Eggink, G.; Martins Dos Santos, V.A.; Wijffels, R.H.; Martens, D.E. Neochloris Oleoabundans Is Worth Its Salt: Transcriptomic Analysis under Salt and Nitrogen Stress. PLoS ONE 2018, 13, e0194834. [Google Scholar] [CrossRef] [Green Version]
- Gualtieri, P.; Barsanti, L. Algae: Anatomy, Biochemistry, and Biotechnology; Taylor & Francis: Boca Raton, FL, USA, 2006; ISBN 978-0-8493-1467-4. [Google Scholar]
- Yaakob, M.A.; Mohamed, R.M.S.R.; Al-Gheethi, A.; Aswathnarayana Gokare, R.; Ambati, R.R. Influence of Nitrogen and Phosphorus on Microalgal Growth, Biomass, Lipid, and Fatty Acid Production: An Overview. Cells 2021, 10, 393. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.Y.; Durbin, E.G. Effects of PH on the Growth and Carbon Uptake of Marine Phytoplankton. Mar. Ecol. Prog. Ser. 1994, 109, 83–94. [Google Scholar] [CrossRef]
- Rahman, A.; Kumar, S.; Bafana, A.; Dahoumane, S.A.; Jeffryes, C. Individual and Combined Effects of Extracellular Polymeric Substances and Whole Cell Components of Chlamydomonas Reinhardtii on Silver Nanoparticle Synthesis and Stability. Molecules 2019, 24, 956. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jena, J.; Pradhan, N.; Nayak, R.R.; Dash, B.P.; Sukla, L.B.; Panda, P.K.; Mishra, B.K. Microalga Scenedesmus Sp.: A Potential Low-Cost Green Machine for Silver Nanoparticle Synthesis. J. Microbiol. Biotechnol. 2014, 24, 522–533. [Google Scholar] [CrossRef]
- Li, Y.; Tang, X.; Song, W.; Zhu, L.; Liu, X.; Yan, X.; Jin, C.; Ren, Q. Biosynthesis of Silver Nanoparticles Using Euglena Gracilis, Euglena Intermedia and Their Extract. IET Nanobiotechnol. 2015, 9, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Siddiqi, K.S.; Husen, A.; Rao, R.A.K. A Review on Biosynthesis of Silver Nanoparticles and Their Biocidal Properties. J. Nanobiotechnol. 2018, 16, 14. [Google Scholar] [CrossRef]
- Singh, M.; Kalaivani, R.; Manikandan, S.; Sangeetha, N.; Kumaraguru, A.K. Facile Green Synthesis of Variable Metallic Gold Nanoparticle Using Padina Gymnospora, a Brown Marine Macroalga. Appl. Nanosci. 2013, 3, 145–151. [Google Scholar] [CrossRef] [Green Version]
- Monika, M.; Prema Krishna, K. Phytochemical Analysis of Commercially Available Spirulina, Their Activities and Biosynthesis of Nanoparticles. Res. J. Pharm. Technol. 2021, 14, 6189–6193. [Google Scholar] [CrossRef]
- Cele, T. Preparation of Nanoparticles; IntechOpen: London, UK, 2020; ISBN 978-1-83880-412-1. [Google Scholar]
- Merin, D.D.; Prakash, S.; Bhimba, B.V. Antibacterial Screening of Silver Nanoparticles Synthesized by Marine Micro Algae. Asian Pac. J. Trop. Med. 2010, 3, 797–799. [Google Scholar] [CrossRef] [Green Version]
- Abou El-Nour, K.M.M.; Eftaiha, A.; Al-Warthan, A.; Ammar, R.A.A. Synthesis and Applications of Silver Nanoparticles. Arab. J. Chem. 2010, 3, 135–140. [Google Scholar] [CrossRef] [Green Version]
- Gupta, R.; Xie, H. Nanoparticles in Daily Life: Applications, Toxicity and Regulations. J. Environ. Pathol. Toxicol. Oncol. 2018, 37, 209–230. [Google Scholar] [CrossRef] [PubMed]
- Hano, C.; Abbasi, B.H. Plant-Based Green Synthesis of Nanoparticles: Production, Characterization and Applications. Biomolecules 2021, 12, 31. [Google Scholar] [CrossRef]
- Tolaymat, T.M.; El Badawy, A.M.; Genaidy, A.; Scheckel, K.G.; Luxton, T.P.; Suidan, M. An Evidence-Based Environmental Perspective of Manufactured Silver Nanoparticle in Syntheses and Applications: A Systematic Review and Critical Appraisal of Peer-Reviewed Scientific Papers. Sci. Total Environ. 2010, 408, 999–1006. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muñoz-Écija, T.; Vargas-Quesada, B.; Chinchilla-Rodríguez, Z. Identification and Visualization of the Intellectual Structure and the Main Research Lines in Nanoscience and Nanotechnology at the Worldwide Level. J. Nanopart. Res. 2017, 19, 62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Darvish, H.; Tonta, Y. Diffusion of Nanotechnology Knowledge in Turkey and Its Network Structure. Scientometrics 2016, 107, 569–592. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Guo, X.; Jovanovic, A. Bibliometrics Analysis of Nanosafety Research. Collnet J. Scientometr. Inf. Manag. 2014, 8, 437–455. [Google Scholar] [CrossRef]
- Resch, S.; Farina, M.C. Mapa do conhecimento em nanotecnologia no setor agroalimentar. RAM Rev. Adm. Mackenzie 2015, 16, 51–75. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.-F.; Liu, Z.-G.; Shen, W.; Gurunathan, S. Silver Nanoparticles: Synthesis, Characterization, Properties, Applications, and Therapeutic Approaches. Int. J. Mol. Sci. 2016, 17, 1534. [Google Scholar] [CrossRef]
- Joudeh, N.; Linke, D. Nanoparticle Classification, Physicochemical Properties, Characterization, and Applications: A Comprehensive Review for Biologists. J. Nanobiotechnol. 2022, 20, 262. [Google Scholar] [CrossRef]
- Dahoumane, S.A.; Yéprémian, C.; Djédiat, C.; Couté, A.; Fiévet, F.; Coradin, T.; Brayner, R. Improvement of Kinetics, Yield, and Colloidal Stability of Biogenic Gold Nanoparticles Using Living Cells of Euglena Gracilis Microalga. J. Nanopart Res. 2016, 18, 79. [Google Scholar] [CrossRef] [Green Version]
- Mohseniazar, M.; Barin, M.; Zarredar, H.; Alizadeh, S.; Shanehbandi, D. Potential of Microalgae and Lactobacilli in Biosynthesis of Silver Nanoparticles. Bioimpacts 2011, 1, 149–152. [Google Scholar] [CrossRef]
- Duygu, D.Y.; Erkaya, I.A.; Erdem, B.; Yalcin, B.M. Characterization of Silver Nanoparticle Produced by Pseudopediastrum Boryanum (Turpin) E. Hegewald and Its Antimicrobial Effects on Some Pathogens. Int. J. Environ. Sci. Technol. 2019, 16, 7093–7102. [Google Scholar] [CrossRef]
- Dziergowska, K.; Wełna, M.; Szymczycha-Madeja, A.; Chęcmanowski, J.; Michalak, I. Valorization of Cladophora Glomerata Biomass and Obtained Bioproducts into Biostimulants of Plant Growth and as Sorbents (Biosorbents) of Metal Ions. Molecules 2021, 26, 6917. [Google Scholar] [CrossRef] [PubMed]
- Dahoumane, S.A.; Yéprémian, C.; Djédiat, C.; Couté, A.; Fiévet, F.; Coradin, T.; Brayner, R. A Global Approach of the Mechanism Involved in the Biosynthesis of Gold Colloids Using Micro-Algae. J. Nanopart Res. 2014, 16, 2607. [Google Scholar] [CrossRef]
- Brayner, R.; Coradin, T.; Beaunier, P.; Grenèche, J.-M.; Djediat, C.; Yéprémian, C.; Couté, A.; Fiévet, F. Intracellular Biosynthesis of Superparamagnetic 2-Lines Ferri-Hydrite Nanoparticles Using Euglena Gracilis Microalgae. Colloids Surf. B Biointerfaces 2012, 93, 20–23. [Google Scholar] [CrossRef] [PubMed]
- Singh, Y.; Sodhi, R.S.; Singh, P.P.; Kaushal, S. Biosynthesis of NiO Nanoparticles Using Spirogyra Sp. Cell-Free Extract and Their Potential Biological Applications. Mater. Adv. 2022, 3, 4991–5000. [Google Scholar] [CrossRef]
- El-Naggar, N.E.-A.; Hussein, M.H.; Shaaban-Dessuuki, S.A.; Dalal, S.R. Production, Extraction and Characterization of Chlorella Vulgaris Soluble Polysaccharides and Their Applications in AgNPs Biosynthesis and Biostimulation of Plant Growth. Sci. Rep. 2020, 10, 3011. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, G.; Jasuja, N.D.; Kumar, M.; Ali, M.I. Biological Synthesis of Silver Nanoparticles by Cell-Free Extract of Spirulina platensis. J. Nanotechnol. 2015, 2015, e132675. [Google Scholar] [CrossRef] [Green Version]
- Ebrahiminezhad, A.; Bagheri, M.; Taghizadeh, S.-M.; Berenjian, A.; Ghasemi, Y. Biomimetic Synthesis of Silver Nanoparticles Using Microalgal Secretory Carbohydrates as a Novel Anticancer and Antimicrobial. Adv. Nat. Sci. Nanosci. Nanotechnol. 2016, 7, 015018. [Google Scholar] [CrossRef]
- Cepoi, L.; Zinicovscaia, I.; Rudi, L.; Chiriac, T.; Turchenko, V. Changes in the Dunaliella Salina Biomass Composition during Silver Nanoparticles Formation. Nanotechnol. Environ. Eng. 2022, 7, 235–243. [Google Scholar] [CrossRef]
- Thangaswamy, S.J.K.; Mir, M.A.; Muthu, A. Green Synthesis of Mono and Bimetallic Alloy Nanoparticles of Gold and Silver Using Aqueous Extract of Chlorella Acidophile for Potential Applications in Sensors. Prep. Biochem. Biotechnol. 2021, 51, 1026–1035. [Google Scholar] [CrossRef] [PubMed]
- Azizi, S.; Namvar, F.; Mahdavi, M.; Ahmad, M.B.; Mohamad, R. Biosynthesis of Silver Nanoparticles Using Brown Marine Macroalga, Sargassum Muticum Aqueous Extract. Materials 2013, 6, 5942–5950. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koopi, H.; Buazar, F. A Novel One-Pot Biosynthesis of Pure Alpha Aluminum Oxide Nanoparticles Using the Macroalgae Sargassum Ilicifolium: A Green Marine Approach. Ceram. Int. 2018, 44, 8940–8945. [Google Scholar] [CrossRef]
- Zhang, Z.; Yan, K.; Zhang, L.; Wang, Q.; Guo, R.; Yan, Z.; Chen, J. A Novel Cadmium-Containing Wastewater Treatment Method: Bio-Immobilization by Microalgae Cell and Their Mechanism. J. Hazard. Mater. 2019, 374, 420–427. [Google Scholar] [CrossRef] [PubMed]
- Feurtet-Mazel, A.; Mornet, S.; Charron, L.; Mesmer-Dudons, N.; Maury-Brachet, R.; Baudrimont, M. Biosynthesis of Gold Nanoparticles by the Living Freshwater Diatom Eolimna Minima, a Species Developed in River Biofilms. Environ. Sci. Pollut. Res. 2016, 23, 4334–4339. [Google Scholar] [CrossRef] [PubMed]
- Bao, Z.; Cao, J.; Kang, G.; Lan, C.Q. Effects of Reaction Conditions on Light-Dependent Silver Nanoparticle Biosynthesis Mediated by Cell Extract of Green Alga Neochloris Oleoabundans. Environ. Sci. Pollut Res. 2019, 26, 2873–2881. [Google Scholar] [CrossRef] [PubMed]
- Chokshi, K.; Pancha, I.; Ghosh, T.; Paliwal, C.; Maurya, R.; Ghosh, A.; Mishra, S. Green Synthesis, Characterization and Antioxidant Potential of Silver Nanoparticles Biosynthesized from de-Oiled Biomass of Thermotolerant Oleaginous Microalgae Acutodesmus Dimorphus. RSC Adv. 2016, 6, 72269–72274. [Google Scholar] [CrossRef]
- Rahman, A.; Kumar, S.; Bafana, A.; Dahoumane, S.A.; Jeffryes, C. Biosynthetic Conversion of Ag+ to Highly Stable Ag0 Nanoparticles by Wild Type and Cell Wall Deficient Strains of Chlamydomonas Reinhardtii. Molecules 2019, 24, 98. [Google Scholar] [CrossRef] [Green Version]
- Azizi, S.; Ahmad, M.B.; Namvar, F.; Mohamad, R. Green Biosynthesis and Characterization of Zinc Oxide Nanoparticles Using Brown Marine Macroalga Sargassum Muticum Aqueous Extract. Mater. Lett. 2014, 116, 275–277. [Google Scholar] [CrossRef]
- Arévalo-Gallegos, A.; Garcia-Perez, J.S.; Carrillo-Nieves, D.; Ramirez-Mendoza, R.; Iqbal, H.M.; Parra-Saldívar, R. Botryococcus Braunii as a Bioreactor for the Production of Nanoparticles with Antimicrobial Potentialities. Int. J. Nanomed. 2018, 13, 5591–5604. [Google Scholar] [CrossRef] [Green Version]
- Kashyap, M.; Samadhiya, K.; Ghosh, A.; Anand, V.; Lee, H.; Sawamoto, N.; Ogura, A.; Ohshita, Y.; Shirage, P.M.; Bala, K. Synthesis, Characterization and Application of Intracellular Ag/AgCl Nanohybrids Biosynthesized in Scenedesmus Sp. as Neutral Lipid Inducer and Antibacterial Agent. Environ. Res. 2021, 201, 111499. [Google Scholar] [CrossRef] [PubMed]
- Cepoi, L.; Rudi, L.; Zinicovscaia, I.; Chiriac, T.; Miscu, V.; Rudic, V. Biochemical Changes in Microalga Porphyridium Cruentum Associated with Silver Nanoparticles Biosynthesis. Arch Microbiol. 2021, 203, 1547–1554. [Google Scholar] [CrossRef] [PubMed]
- Fathy, W.; Elsayed, K.; Essawy, E.; Tawfik, E.; Zaki, A.; Abdelhameed, M.S.; Hammouda, O. Biosynthesis of Silver Nanoparticles from Synechocystis Sp to Be Used as a Flocculant Agent with Different Microalgae Strains. CNM 2020, 5, 175–187. [Google Scholar] [CrossRef]
- Salari, Z.; Danafar, F.; Dabaghi, S.; Ataei, S.A. Sustainable Synthesis of Silver Nanoparticles Using Macroalgae Spirogyra Varians and Analysis of Their Antibacterial Activity. J. Saudi Chem. Soc. 2016, 20, 459–464. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Hu, C.; Shao, L. The Antimicrobial Activity of Nanoparticles: Present Situation and Prospects for the Future. Int. J. Nanomed. 2017, 12, 1227–1249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ozdal, M.; Gurkok, S. Recent Advances in Nanoparticles as Antibacterial Agent. Admet. Dmpk. 2022, 10, 115–129. [Google Scholar] [CrossRef]
- Kostoff, R.N.; Stump, J.A.; Johnson, D.; Murday, J.S.; Lau, C.G.Y.; Tolles, W.M. The Structure and Infrastructure of the Global Nanotechnology Literature. J. Nanopart. Res. 2006, 8, 301–321. [Google Scholar] [CrossRef]
- Bejan, A.; Lorente, S. The Physics of Spreading Ideas. Int. J. Heat Mass Transf. 2012, 55, 802–807. [Google Scholar] [CrossRef]
- Khan, M.I.; Shin, J.H.; Kim, J.D. The Promising Future of Microalgae: Current Status, Challenges, and Optimization of a Sustainable and Renewable Industry for Biofuels, Feed, and Other Products. Microb. Cell Factories 2018, 17, 36. [Google Scholar] [CrossRef]
- Lane, T.W. Barriers to Microalgal Mass Cultivation. Curr. Opin. Biotechnol. 2022, 73, 323–328. [Google Scholar] [CrossRef]
- Rai, P.K.; Kumar, V.; Lee, S.; Raza, N.; Kim, K.-H.; Ok, Y.S.; Tsang, D.C.W. Nanoparticle-Plant Interaction: Implications in Energy, Environment, and Agriculture. Environ. Int. 2018, 119, 1–19. [Google Scholar] [CrossRef] [PubMed]
- El-Sheekh, M.M.; Morsi, H.H.; Hassan, L.H.S.; Ali, S.S. The Efficient Role of Algae as Green Factories for Nanotechnology and Their Vital Applications. Microbiol. Res. 2022, 263, 127111. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, A.; Sarkar, D.; Sasmal, S. A Review of Green Synthesis of Metal Nanoparticles Using Algae. Front. Microbiol. 2021, 12, 693899. [Google Scholar] [CrossRef] [PubMed]

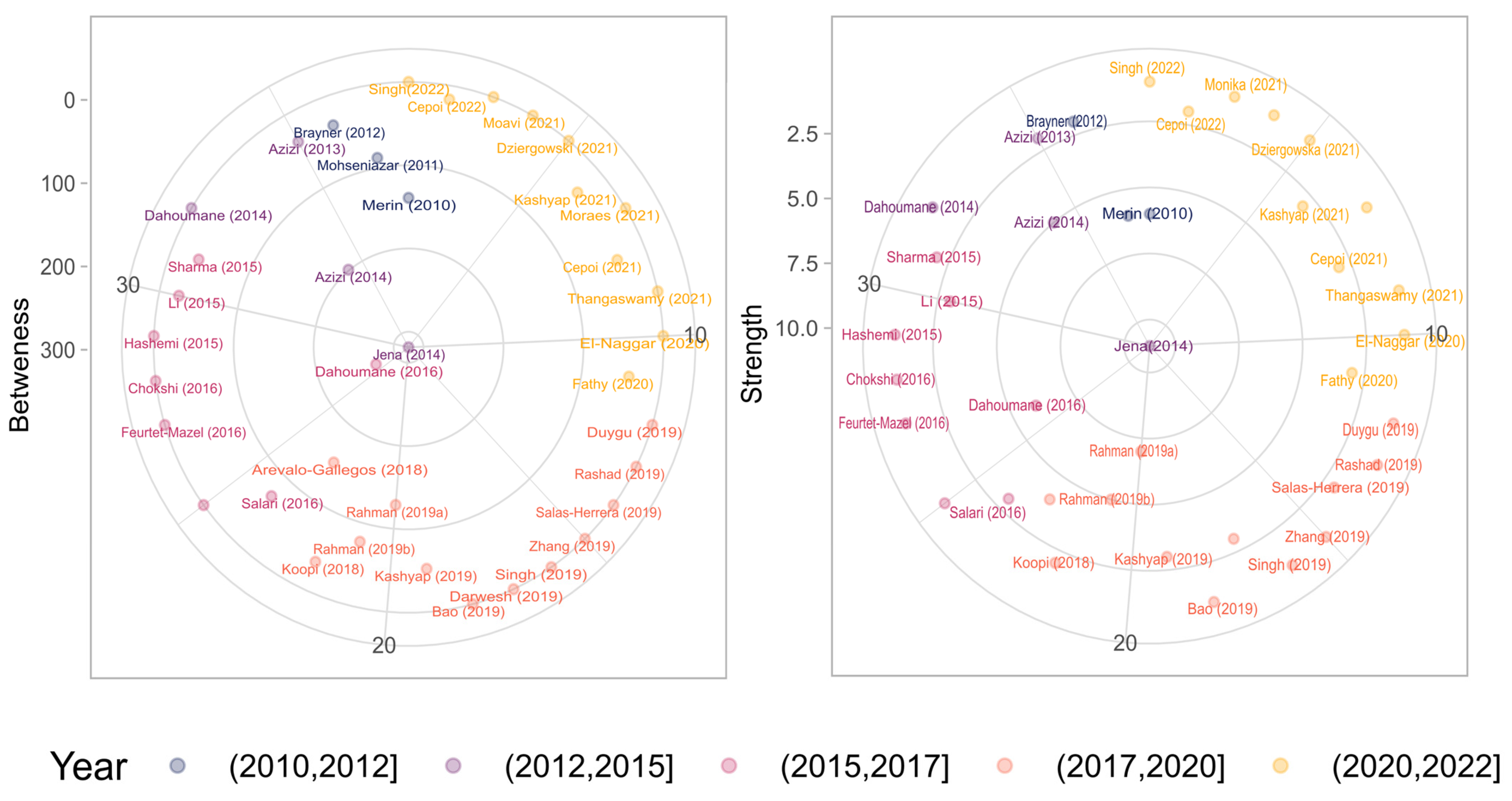
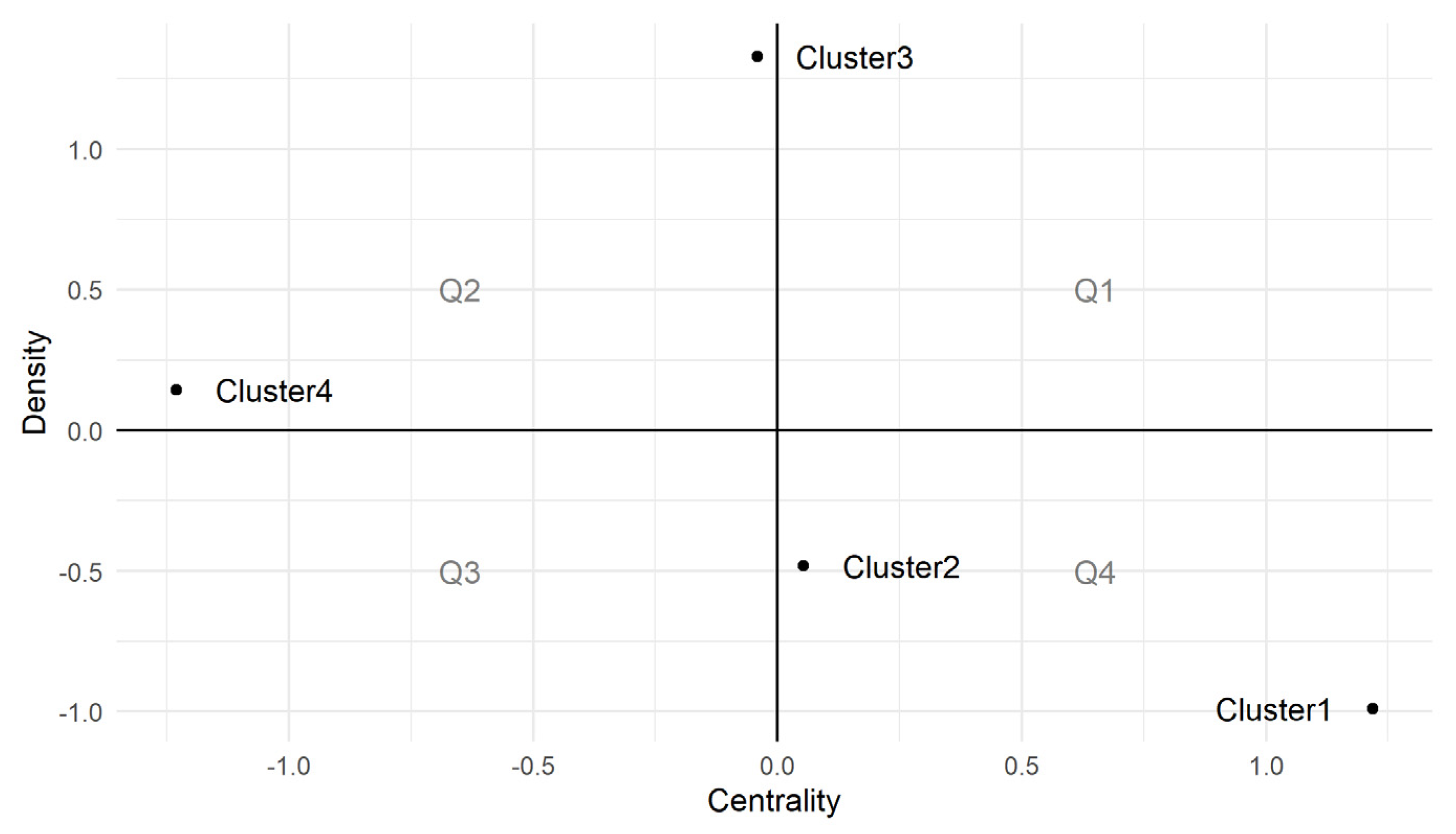
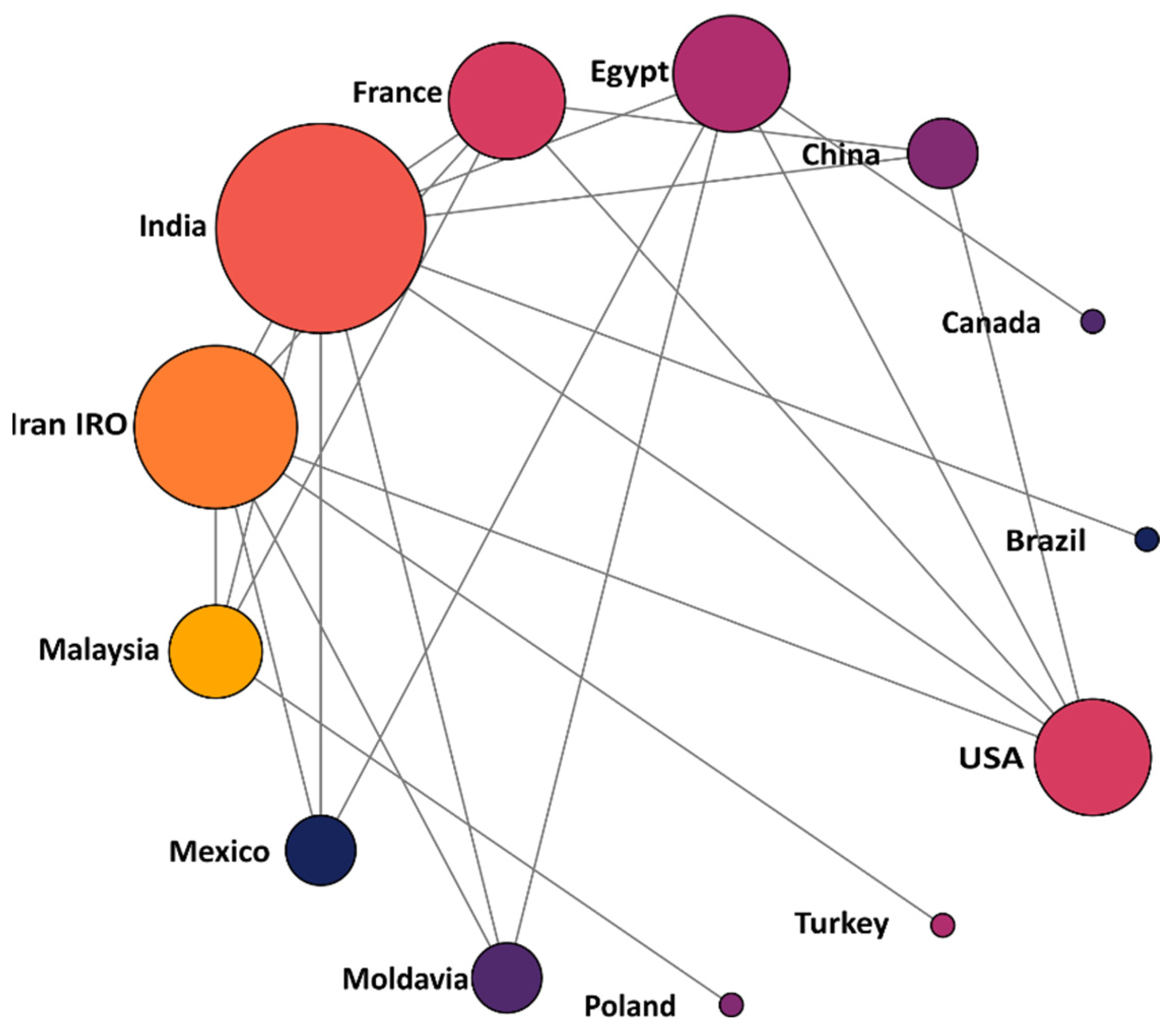

| Uses | Criteria |
|---|---|
| Antibacterial | Used to destroy bacteria or suppresses their growth or their ability to reproduce |
| Anticancerous | Used against or tending to arrest or prevent cancer |
| Antifungal | (also called antimycotic): Kill or stop the growth of fungi that cause infections |
| Antioxidant | Lessen or prevent the effects of free radicals |
| Cytotoxic | Potency to bring variations in cellular functions and results in cell death |
| Optimization | Improvement of the conditions to produce NPs using algae |
| Plant improvement | (and protection): Increased agricultural productivity is the primary concern |
| Other | Different industrial or technical applications from the stated before |
| Categories | Criteria |
|---|---|
| Topic | A general word that refers to a comprehensive concept or group, e.g., temperature, biosynthesis, algae |
| Use | Specific uses of the NPs in the paper or methods related to the study of the benefits, e.g., antibacterial, inhibition zone |
| Taxon-Use | Name of a taxon (at any taxonomic level) that was used for testing the uses of the NPs, e.g., Escherichia coli, Pseudomonas sp. |
| Taxon | Name of a taxon of algae (at any taxonomic level) that was used for the synthesis of the NPs |
| NP | Characteristics related to the structure or class of NP, e.g., gold NPs, particle size, shape, color |
| Method | Analytical approaches used for the characterization of the NPs |
| NP Type | Minimum Size Obtained (nm) | Maximum Size Obtained (nm) | |
|---|---|---|---|
| Using death cells or extracts | Ag | 1.8 | 100 |
| Au–Ag | 5 | 45 | |
| Al2O3 | |||
| Au | 2 | 67 | |
| Cu | 35 | 45 | |
| CuFe2O4@Ag | |||
| CuO/Perlite | 13 | 24 | |
| FeO | 4.8 | 9.07 | |
| NiO * | 30.2 | ||
| Zn (nanoflowers) * | 4000 | ||
| ZnO | 30 | 57 | |
| Using living cells | Ag | 3 | 60 |
| Au | 5 | 80 | |
| Cd, Se * | 7.0 | ||
| Cu | 15 | 65 | |
| FeO * | 45.0 | ||
| ferrihydrite | 0.6 | 1 | |
| Se | 36 | 190 | |
| Type of NPs | Precursor | Number of Experiments |
|---|---|---|
| Ag | AgNO3 | 42 |
| Au | HAuCl4 | 13 |
| Gold (III) chloride solution | 1 | |
| HAuCl5 | 1 | |
| KAuCl4 | 1 | |
| Au–Ag | AgNO3 + Gold (III) chloride solution | 1 |
| Al2O3 | Al2(SO4)3 | 1 |
| Cd | CdCl2 | 1 |
| Se | Na2SeO3 | 2 |
| Cu | CuCl2 | 1 |
| CuSO4 | 1 | |
| CuFe2O4@Ag | CuCl2, FeCl3, AgNO3 | 1 |
| CuO/Perlite | CuCl2 + Perlite powder | 1 |
| FeO | FeCl2 + FeCl3 | 2 |
| Ferrihydrite | equimolar Fe(II)/Fe(III) ions | 1 |
| NiO | Ni(NO3)2 | 1 |
| NiCl2 | 1 | |
| Zn (nanoflowers) | ZnC₄H₆O₄ | 1 |
| ZnO | Zinc acetate | 1 |
| Zn(NO3)2 | 1 | |
| ZnSO4 | 1 |
| Cluster 1 | Cluster 2 | Cluster 3 | Cluster 4 |
|---|---|---|---|
| algae | biomass | antibacterial | bioactive compound |
| antimicrobial activity | chemical method | bacteria | carbohydrates |
| antioxidant | Chlamydomonas reinhardtii | biomedical application | cytotoxicity |
| Chlorella vulgaris | ed x-ray spectroscopy | biomolecule | DPPH (antioxidant) |
| crystalline nature | electron microscopy | Escherichia coli | HRTEM |
| gold | green extract | inhibition | macroalgae |
| mechanism | intracellular biosynthesis | morphology | metal nps |
| polysaccharide | optimization | Spirulina | MIC |
| protein | SEM | Staphylococcus aureus | |
| reduction | synthesized nanoparticle | TEM analysis | |
| shape | temperature | ||
| silver nitrate | |||
| spherical shape | |||
| stabilization | |||
| surface | |||
| toxicity | |||
| XRD pattern |
| Semantic Networks Nodes | Degree | Citation Networks Nodes | Degree |
|---|---|---|---|
| electron microscopy | 40 | Jena J. (2014) [63] | 11 |
| gold | 39 | Rahman A. (2019a) [62] | 7 |
| reduction | 37 | Dahoumane S.A. (2016) [80] | 6 |
| protein | 36 | Mohseniazar M. (2011) [81] | 6 |
| SEM | 33 | Merin D.D. (2010) [69] | 6 |
| antibacterial | 31 | Rahman A. (2019b) [98] | 5 |
| morphology | 31 | Azizi S. (2014) [99] | 5 |
| shape | 31 | Arévalo-Gallegos A. (2018) [100] | 4 |
| biomass | 30 | Kashyap M. (2021) [101] | 3 |
| antimicrobial activity | 29 | Cepoi L. (2021) [102] | 3 |
| Staphylococcus aureus | 29 | Fathy W. (2020) [103] | 3 |
| biomolecule | 28 | Darwesh O.M. (2019) [55] | 3 |
| HRTEM | 28 | Kashyap M. (2019) [45] | 3 |
| chemical method | 27 | Salari Z. (2016) [104] | 3 |
| inhibition | 27 | Li Y. (2015) [64] | 3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lamilla-Tamayo, L.; Escobar-Calderón, F.; Skalický, M. Reviewing the Potential of Algae Species as a Green Alternative to Produce Nanoparticles: Findings from a Database Analysis. Water 2023, 15, 2208. https://doi.org/10.3390/w15122208
Lamilla-Tamayo L, Escobar-Calderón F, Skalický M. Reviewing the Potential of Algae Species as a Green Alternative to Produce Nanoparticles: Findings from a Database Analysis. Water. 2023; 15(12):2208. https://doi.org/10.3390/w15122208
Chicago/Turabian StyleLamilla-Tamayo, Laura, Felipe Escobar-Calderón, and Milan Skalický. 2023. "Reviewing the Potential of Algae Species as a Green Alternative to Produce Nanoparticles: Findings from a Database Analysis" Water 15, no. 12: 2208. https://doi.org/10.3390/w15122208






