Characteristics of Nitrogen Removal and Functional Gene Transcription of Heterotrophic Nitrification-Aerobic Denitrification Strain, Acinetobacter sp. JQ1004
Abstract
:1. Introduction
2. Materials and Methods
2.1. Media and Strains
2.2. Nitrogen Balance Analysis
2.3. Kinetics of Nitrogen Degradation and Cell Growth
2.4. The Effect of Environmental Conditions on Nitrogen Removal
2.5. Functional Genes Analysis
2.6. Whole Genome Sequencing and Analysis
2.7. Analytical Methods
3. Results and Discussion
3.1. Nitrogen Balance Analysis
3.2. Kinetic Analysis of Nitrogen Degradation and Cell Growth
3.3. Amplification and Expression of Functional Genes
3.3.1. PCR Amplification of Functional Genes
3.3.2. The Patterns of Functional Genes Expression
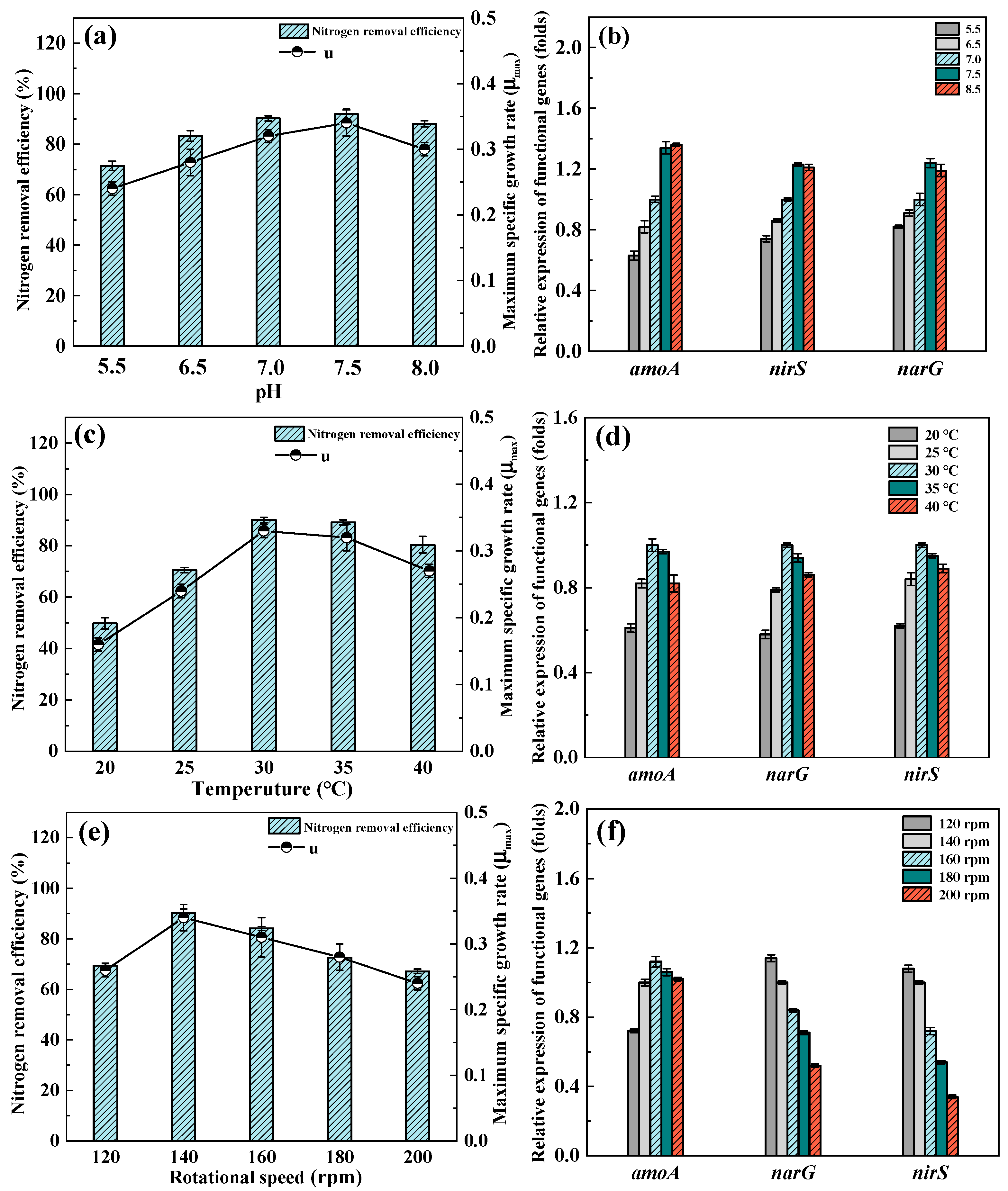
3.4. Effect of Environmental Conditions
3.4.1. Different pH
3.4.2. Different Temperatures
3.4.3. Different Rotational Speeds
3.5. Analysis of the Whole Genome
3.5.1. Molecular Characterization of the Genome
3.5.2. Functional Genes Annotation
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Shu, H.; Sun, H.; Huang, W.; Zhao, Y.; Ma, Y.; Chen, W.; Sun, Y.; Chen, X.; Zhong, P.; Yang, H.; et al. Nitrogen removal characteristics and potential application of the heterotrophic nitrifying-aerobic denitrifying bacteria Pseudomonas mendocina S16 and Enterobacter cloacae DS’5 isolated from aquaculture wastewater ponds. Bioresour. Technol. 2022, 345, 126541. [Google Scholar] [CrossRef] [PubMed]
- Joo, H.; Mitsuyo, H.; Makoto, S. Characteristics of ammonium removal by heterotrophic nitrification-aerobic denitrification by Alcaligenes faecalis No. 4. J. Biosci. Bioeng. 2005, 100, 184–191. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Hou, H.; Liu, P.; Hou, L.; Yang, T.; Dai, H.; Li, J. Acceleration of nitrogen removal performance in a biofilm reactor augmented with Pseudomonas sp. using polycaprolactone as carbon source for treating low carbon to nitrogen wastewater. Bioresour. Technol. 2023, 386, 129507. [Google Scholar] [CrossRef]
- Hou, L.; Li, J.; Liu, Y. Microbial communities variation analysis of denitrifying bacteria immobilized particles. Process Biochem. 2019, 87, 151–156. [Google Scholar] [CrossRef]
- Gu, X.; Leng, J.; Zhu, J.; Zhang, K.; Zhao, J.; Wu, P.; Xing, Q.; Tang, K.; Li, X.; Hu, B. Influence mechanism of C/N ratio on heterotrophic nitrification-aerobic denitrification process. Bioresour. Technol. 2022, 343, 126116. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Tian, M.; An, Q.; Ye, J.; Guo, J. Characteristics of a heterotrophic nitrogen removal bacterium and its potential application on treatment of ammonium-rich wastewater. Bioresour. Technol. 2017, 226, 46–54. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Di, F. Sensitivity analysis strategy to assess the salting-out problem during CO2 geological storage process. Shock Vib. 2021, 2021, 2809467. [Google Scholar] [CrossRef]
- Fu, G.; Zhao, L.; Huangshen, L.; Wu, J. Isolation and identification of a salt-tolerant aerobic denitrifying bacterial strain and its application to saline wastewater treatment in constructed wetlands. Bioresour. Technol. 2019, 290, 121725. [Google Scholar] [CrossRef] [PubMed]
- Xie, F.; Myat, T.; Wang, H. Simultaneous heterotrophic nitrification and aerobic denitrification by a novel isolated Pseudomonas mendocina X49. Bioresour. Technol. 2021, 319, 124198. [Google Scholar] [CrossRef]
- Yao, S.; Ni, J.; Ma, T.; Li, C. Heterotrophic nitrification and aerobic denitrification at low temperature by a newly isolated bacterium, Acinetobacter sp. HA2. Bioresour. Technol. 2013, 139, 80–86. [Google Scholar] [CrossRef]
- Xi, H.; Zhou, X.; Arslan, M.; Luo, Z.; Wei, J.; Wu, Z.; El-Din, M. Heterotrophic nitrification and aerobic denitrification process: Promising but a long way to go in the wastewater treatment. Sci. Total Environ. 2022, 805, 150212. [Google Scholar] [CrossRef] [PubMed]
- Song, T.; Zhang, X.; Li, J.; Wu, X.; Feng, H.; Dong, W. A review of research progress of heterotrophic nitrification and aerobic denitrification microorganisms (HNADMs). Sci. Total Environ. 2021, 801, 149319. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Liu, H.; Liang, S.; Chen, W.; Tan, S.; Yang, C.; Qin, S.; Long, K. Comparison of moving bed biofilm reactor and bio-contact oxidation reactor start-up with heterotrophic nitrification-aerobic denitrification bacteria and activated sludge inoculation under high ammonia nitrogen conditions. Bioresour. Technol. 2024, 395, 130408. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, W.; Zhang, Y.; Zhang, J.; Li, J.; Wang, S.; Chen, G. Isolation and characterization of Acinetobacter sp. JQ1004 and evaluation of its inhibitory kinetics by free ammonia. Desalin. Water Treat. 2019, 147, 316–325. [Google Scholar] [CrossRef]
- Wang, X.; Wang, W.; Zhang, Y.; Sun, Z.; Zhang, J.; Chen, G.; Li, J. Simultaneous nitrification and denitrification by a novel isolated Pseudomonas sp. JQ-H3 using polycaprolactone as carbon source. Bioresour. Technol. 2019, 288, 121506. [Google Scholar] [CrossRef]
- Li, J.; Gu, J.; Pan, L. Transformation of dimethyl phthalate, dimethyl isophthalate and dimethyl terephthalate by Rhodococcus rubber Sa and modeling the processes using the modified Gompertz model. Int. Biodeter. Biodegr. 2005, 55, 223–232. [Google Scholar] [CrossRef]
- Liu, H.; Zeng, W.; Fan, Z.; Li, J.; Zhan, M.; Peng, Y. Effect of iron on enhanced nitrogen removal from wastewater by sulfur autotrophic denitrification coupled to heterotrophic denitrification under different substrate ratios. Chem. Eng. J. 2021, 421, 129828. [Google Scholar] [CrossRef]
- Chai, Y.; Huang, C.; Sui, M.; Yin, Y.; Sun, N.; Chen, Y.; Liao, Z.; Sun, X.; Shen, W.; Tang, S. Fe-loaded alginate hydrogel beads activating peroxymonosulfate for enhancing anaerobic fermentation of waste activated sludge: Performance and potential mechanism. J. Environ. Manag. 2023, 341, 118079. [Google Scholar] [CrossRef]
- Zhou, X.; Wang, Y.; Tan, X.; Sheng, Y.; Li, Y.; Zhang, Q.; Xu, J.; Shi, Z. Genomics and nitrogen metabolic characteristics of a novel heterotrophic nitrifying-aerobic denitrifying bacterium Acinetobacter oleivorans AHP123. Bioresour. Technol. 2023, 375, 128822. [Google Scholar] [CrossRef]
- APHA; AWWA; WEF. Standards Methods for the Examination of Water and Wastewater; American Public Health Association: Washington, DC, USA, 2005. [Google Scholar]
- Xu, M.; Cui, Y.; Huang, M.; Sui, Y. Simultaneous inorganic nitrogen and phosphate removal by aerobic-heterotrophic fungus Fusarium keratoplasticum FSP1: Performance, pathway and application. Bioresour. Technol. 2024, 393, 130141. [Google Scholar] [CrossRef]
- Chen, M.; He, T.; Wu, Q.; Zhang, M.; He, K. Enhanced heterotrophic nitrification and aerobic denitrification performance of Glutamicibacter arilaitensis EM-H8 with different carbon sources. Chemosphere 2023, 323, 138266. [Google Scholar] [CrossRef]
- Chen, X.; Li, S.; Zhang, W.; Li, S.; Gu, Y.; Ouyang, L. A newly isolated Rhodococcus sp. S2 from landfill leachate capable of heterotrophic nitrification and aerobic denitrification. Water 2024, 16, 431. [Google Scholar] [CrossRef]
- Ren, Y.; Yang, L.; Liang, X. The characteristics of a novel heterotrophic nitrifying and aerobic denitrifying bacterium, Acinetobacter junii YB. Bioresour. Technol. 2014, 171, 1–9. [Google Scholar] [CrossRef]
- Ke, X.; Liu, C.; Tang, S.; Guo, T.; Pan, L.; Xue, Y.; Zheng, Y. Characterization of Acinetobacter indicus ZJB20129 for heterotrophic nitrification and aerobic denitrification isolated from an urban sewage treatment plant. Bioresour. Technol. 2022, 347, 126423. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Ansari, F.; Nasr, M.; Rawat, I.; Nayunigari, K.; Bux, F. Cultivation of Chlorella sorokiniana and Scenedesmus obliquus in wastewater: Fuzzy intelligence for evaluation of growth parameters and metabolites extraction. J. Clean. Prod. 2017, 147, 419–430. [Google Scholar] [CrossRef]
- Huang, X.; Yi, X.; He, T.; Jia, H.; Feng, M.; Xiang, S.; Wang, S.; Ni, J.; Xie, D.; Li, Z. Ammonium transformed into nitrous oxide via nitric oxide by Pseudomonas putida Y-9 under aerobic conditions without hydroxylamine as intermediate. Bioresour. Technol. 2019, 277, 87–93. [Google Scholar] [CrossRef]
- Chen, P.; Wang, J.; Lv, J.; Wang, Q.; Zhang, C.; Zhao, W.; Li, S. Nitrogen removal by Rhodococcus sp. SY24 under linear alkylbenzene sulphonate stress: Carbon source metabolism activity, kinetics, and optimum culture conditions. Bioresour. Technol. 2023, 368, 128348. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Yang, C.; Pan, T.; An, Q.; Guo, J. Characterization of an aerobic denitrifier Pseudomonas stutzeri strain XL-2 to achieve efficient nitrate removal. Bioresour. Technol. 2018, 250, 564–573. [Google Scholar] [CrossRef] [PubMed]
- Braker, G.; Zhou, J.; Wu, L.; Devol, A.; Tiedje, J. Nitrite reductase genes (nirK and nirS) as functional markers to investigate diversity of denitrifying bacteria in pacific northwest marine sediment communities. Appl. Environ. Microb. 2000, 66, 2096–2104. [Google Scholar] [CrossRef]
- Härtig, E.; Walter, Z. Kinetics of nirS expression (cytochromecd1 nitrite reductase) in Pseudomonas stutzeri during the transition from aerobic respiration to denitrification: Evidence for a denitrification specific nitrate and nitrite responsive regulatory system. J. Bacteriol. 1999, 181, 161–166. [Google Scholar] [CrossRef]
- Loh, X. The effect of pH on the hydrolytic degradation of poly(ε-caprolactone)-block-poly(ethylene glycol) copolymers. J. Appl. Polym. Sci. 2013, 127, 2046–2056. [Google Scholar] [CrossRef]
- Chen, Q.; Ni, J. Ammonium removal by Agrobacterium sp. LAD9 capable of heterotrophic nitrification-aerobic denitrification. J. Biosci. Bioeng. 2012, 113, 619–623. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Ying, L.; Ai, G.; Miao, L.; Zheng, H.; Liu, Z. The characteristics of a novel heterotrophic nitrification-aerobic denitrification bacterium, Bacillus methylotrophicus strain L7. Bioresour. Technol. 2012, 108, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Wehrfritz, J.; Reilly, A.; Spiro, S.; Richardson, D. Purification of hydroxylamine oxidase from Thiosphaera pantotropha. Identification of electron acceptors that couple heterotrophic nitrification to aerobic denitrification. FEBS Lett. 1993, 335, 246–250. [Google Scholar] [CrossRef] [PubMed]
- Ka, J.; Urbance, J.; Ye, W.; Ahn, T.; Tiedje, J. Diversity of oxygen and N-oxide regulation of nitrite reductases in denitrifying bacteria. FEMS Microbiol. Lett. 1997, 156, 55–60. [Google Scholar] [CrossRef] [PubMed]
- Shoda, M.; Yoichi, I. Heterotrophic nitrification and aerobic denitrification of high-strength ammonium in anaerobically digested sludge by Alcaligenes faecalis strain No. 4. J. Biosci. Bioeng. 2014, 117, 737–741. [Google Scholar] [CrossRef] [PubMed]
- Pous, N.; Bañeras, L.; Corvini, P.; Liu, S.; Puig, S. Direct ammonium oxidation to nitrogen gas (Dirammox) in Alcaligenes strain HO-1: The electrode role. Environ. Sci. Ecotechnol. 2023, 15, 100253. [Google Scholar] [CrossRef]
- Pan, Y.; Liu, D. Tapping the potential of wastewater treatment with Direct ammonia oxidation (Dirammox). Environ. Sci. Technol. 2023, 57, 7106–7108. [Google Scholar] [CrossRef]

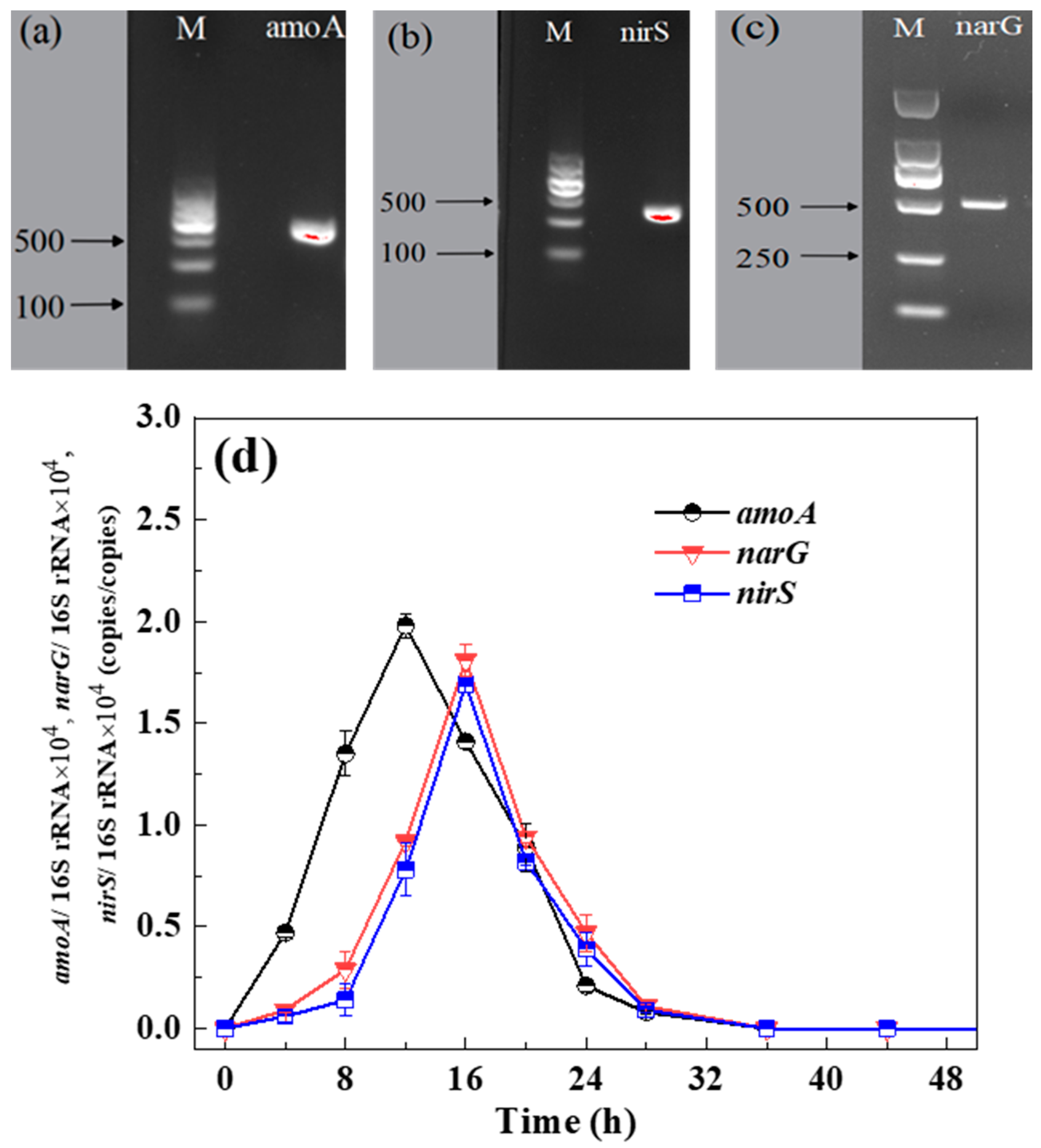


| Functional Genes | Primer Sequence (5′-3′) | Product Length (bp) |
|---|---|---|
| amoA | AAGGATTGGCCATTGCTCTG | 606 |
| GTGGACCTAAAATCCAAGCATT | ||
| nirS | TAAAAGTTCACACACAAAAAGCAACGC | 517 |
| ACAAGTACTGCACCCAGTAATTTGG | ||
| narG | AATCGCAGATCAATTCCAAGCG | 537 |
| TCAGCTTCAGTCTGACTAGATTCTAGT |
| Initial TN(mg/L) | Nitrogen Content (mg/L) | Error | |||||
|---|---|---|---|---|---|---|---|
| NH4+ | NO3− | NO2− | Intracellular N | N2O | N2 | ||
| 50.58 ± 0.74 | 4.37 ± 0.08 | 0.04 ± 0.01 | 0.06 ± 0.02 | 27.62 ± 0.57 | 6.21 ± 0.14 | 11.26 ± 0.94 | 2.02% |
| Kinetic Parameters | Ammonia | Nitrate |
|---|---|---|
| S0 (mg/L) | 101.19 | 114.86 |
| t0 (h) | 5.91 | 12.74 |
| Rm [mg/(L·h)] | 7.93 | 4.08 |
| R2 | 0.997 | 0.985 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hou, L.; Huang, F.; Pan, Z.; Chen, W.; Wang, X. Characteristics of Nitrogen Removal and Functional Gene Transcription of Heterotrophic Nitrification-Aerobic Denitrification Strain, Acinetobacter sp. JQ1004. Water 2024, 16, 1799. https://doi.org/10.3390/w16131799
Hou L, Huang F, Pan Z, Chen W, Wang X. Characteristics of Nitrogen Removal and Functional Gene Transcription of Heterotrophic Nitrification-Aerobic Denitrification Strain, Acinetobacter sp. JQ1004. Water. 2024; 16(13):1799. https://doi.org/10.3390/w16131799
Chicago/Turabian StyleHou, Liangang, Feng Huang, Zhengwei Pan, Wei Chen, and Xiujie Wang. 2024. "Characteristics of Nitrogen Removal and Functional Gene Transcription of Heterotrophic Nitrification-Aerobic Denitrification Strain, Acinetobacter sp. JQ1004" Water 16, no. 13: 1799. https://doi.org/10.3390/w16131799





