Contrasting Distribution of Microbial Communities, Functional Genes, and Antibiotic Resistance Genes in Produced Water Treatment Plants with Different Treatment Technologies
Abstract
:1. Introduction
2. Materials and Methods
2.1. Treatment Plants and Sample Collection
2.2. Analytical Methods
2.3. DNA Extraction and PCR Amplification
2.4. Metagenomic Sequencing
2.5. Statistical Analysis and Data Visualization
3. Results and Discussion
3.1. The Water Quality Indexes of Different PWTPs
3.2. Analysis of Microbial Community Structure in Different PWTPs
3.2.1. Overview of Microbial Diversity
3.2.2. Microbial Community Analysis at the Phylum Level
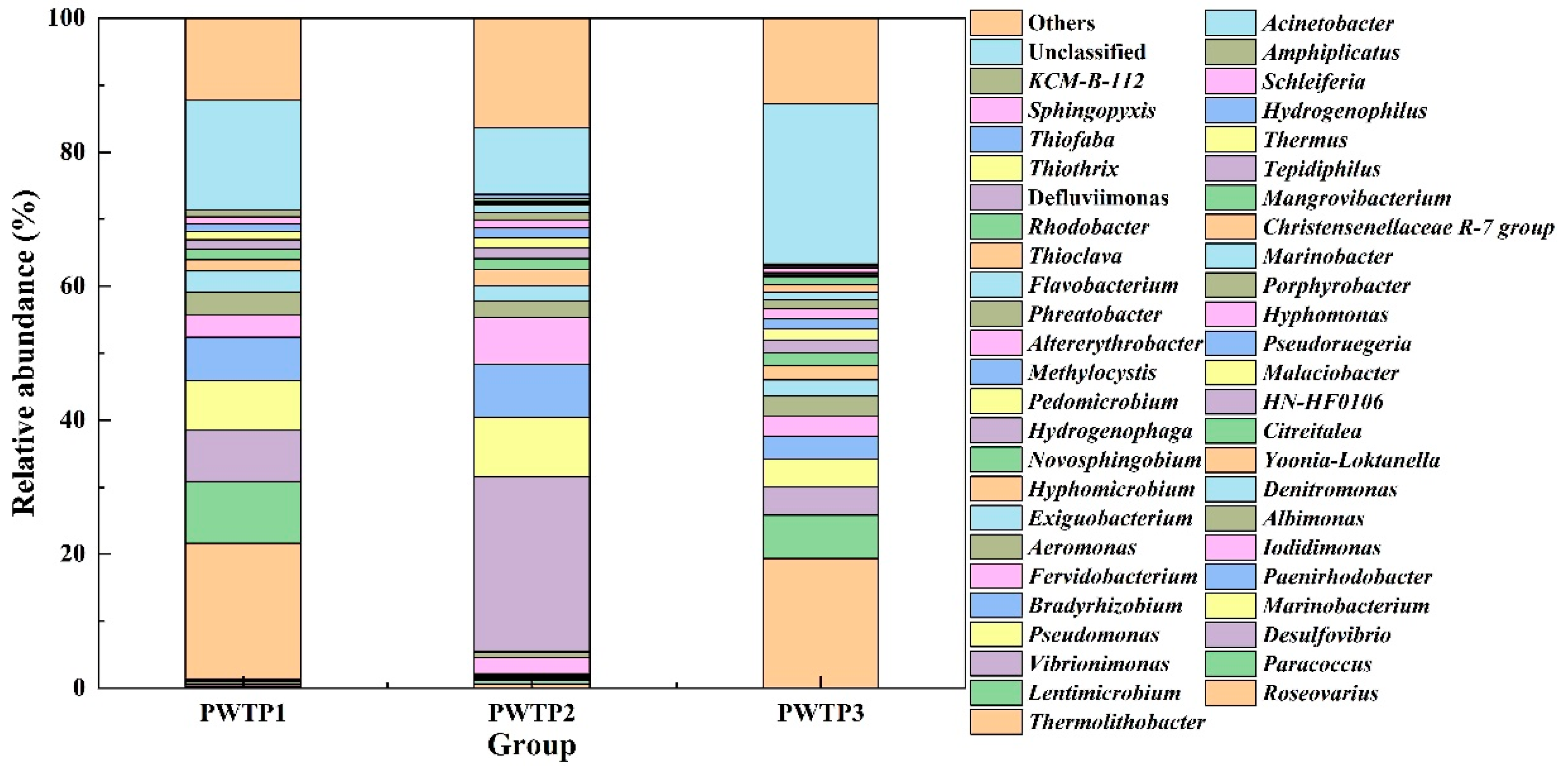
3.2.3. Microbial Community Analysis at the Genus Level
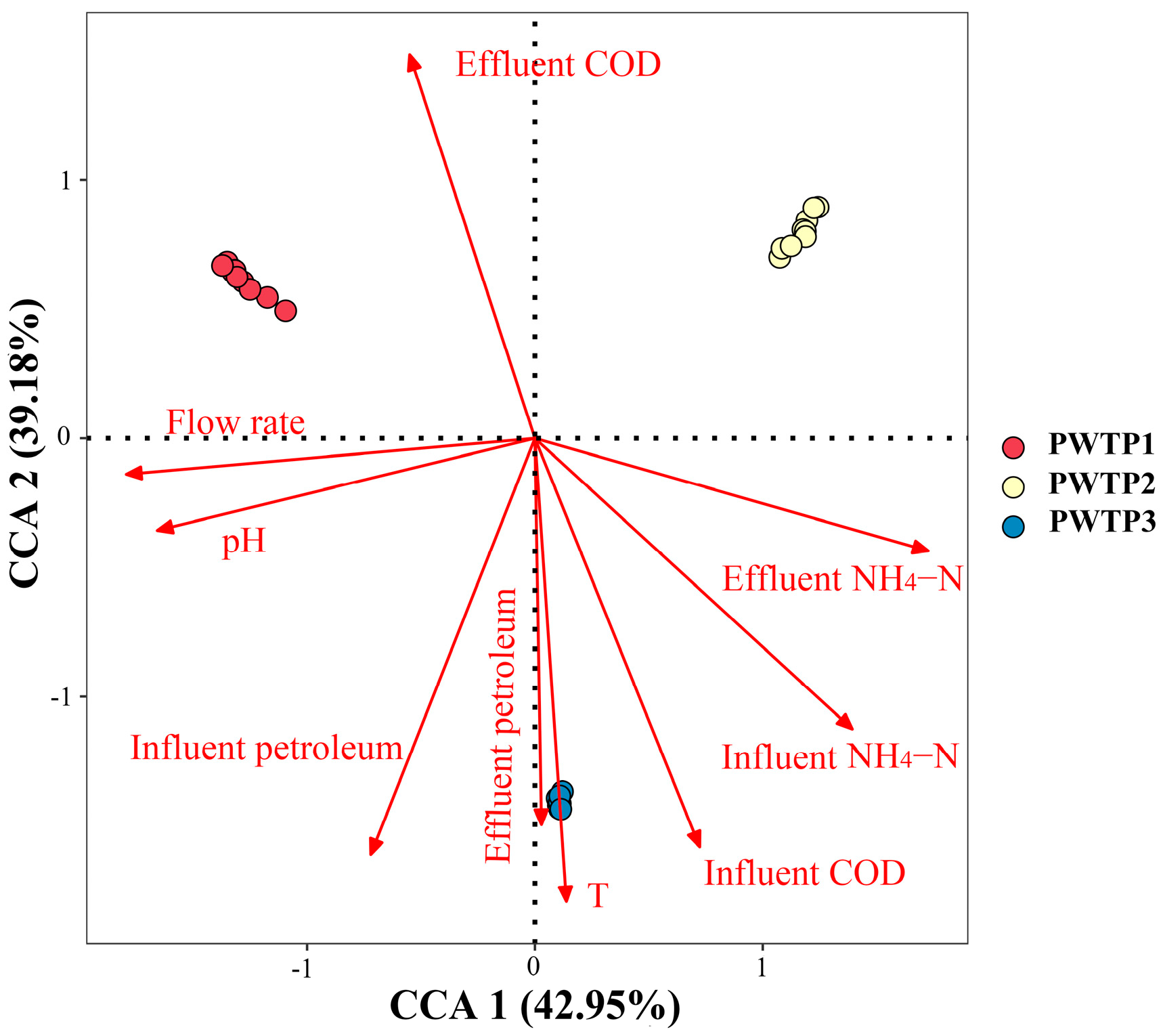
3.2.4. Correlation Analysis of Environmental Factors and Microbial Community Structure
3.2.5. Microbial Community Assembly in in Different PWTPs
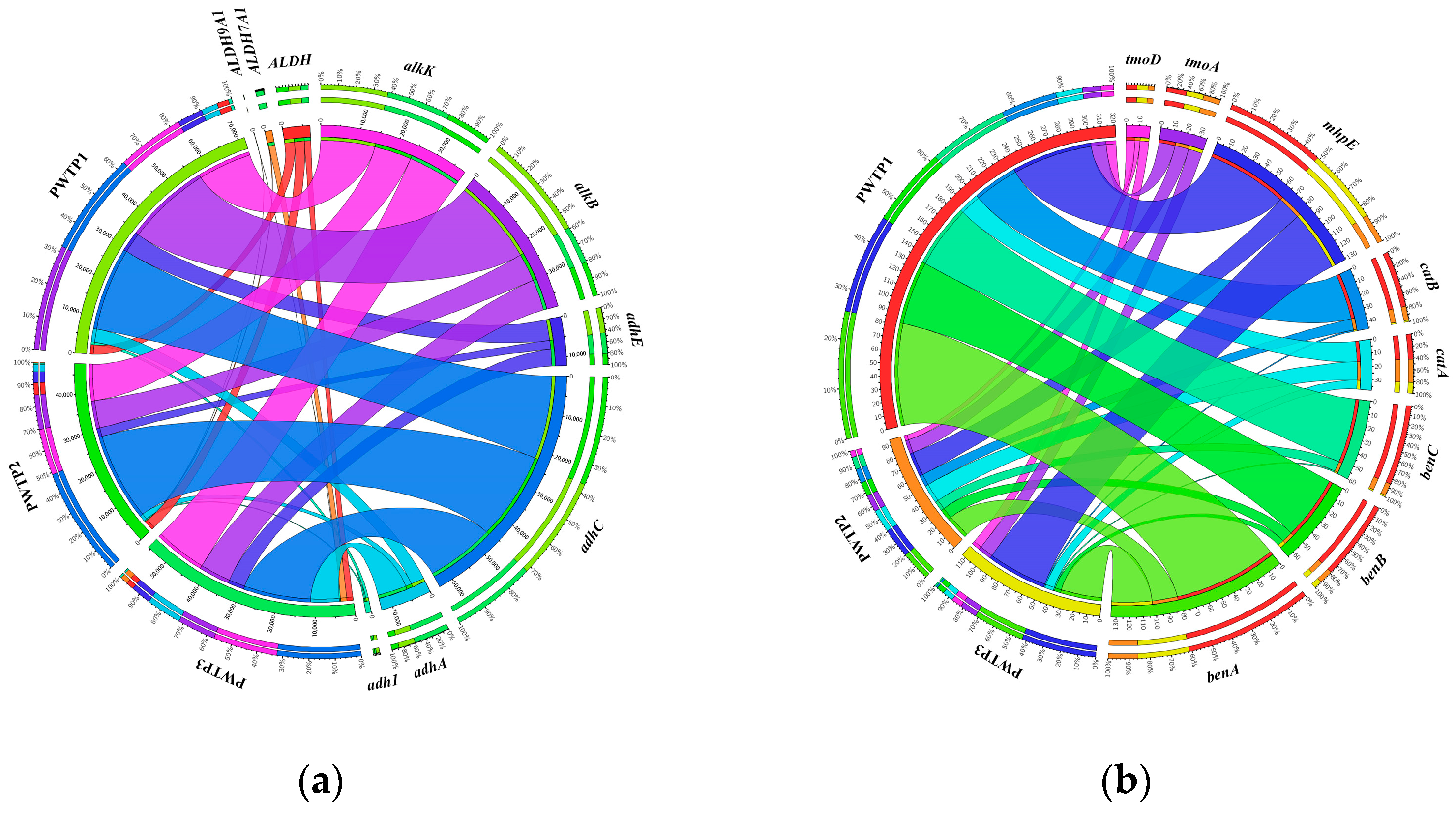
3.3. Analysis of Petroleum Hydrocarbon Degradation-Related Genes in Different PWTPs
3.4. Abundance and Diversity of ARGs in Different PWTPs
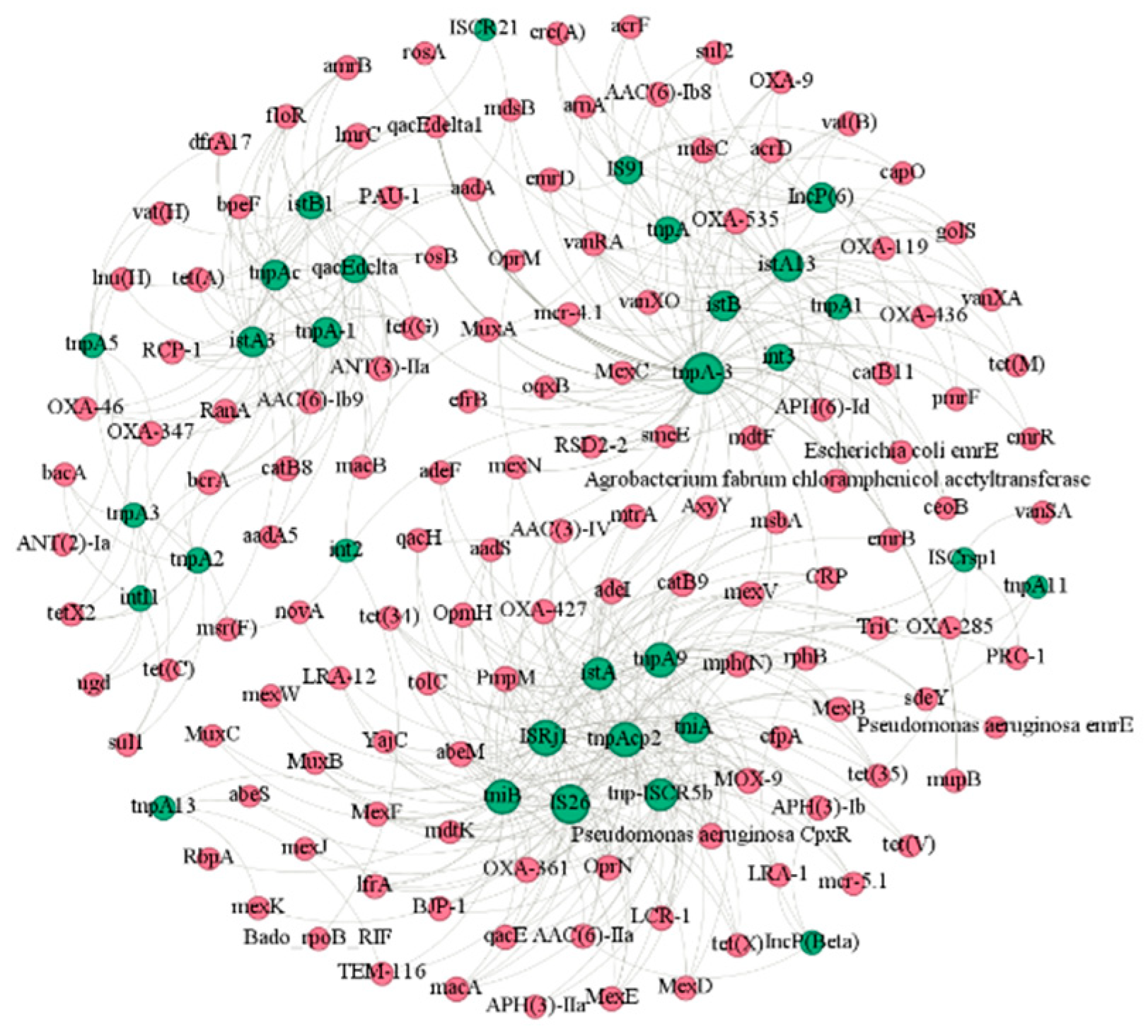
3.5. Co-Occurrence between ARGs and MGEs
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Olajire, A.A. Recent advances on the treatment technology of oil and gas produced water for sustainable energy industry-mechanistic aspects and process chemistry perspectives. Chem. Eng. J. Adv. 2020, 4, 100049. [Google Scholar] [CrossRef]
- Samuel, O.; Othman, M.H.D.; Kamaludin, R.; Sinsamphanh, O.; Abdullah, H.; Puteh, M.H.; Kurniawan, T.A.; Li, T.; Ismail, A.F.; Rahman, M.A.; et al. Oilfield-produced water treatment using conventional and membrane-based technologies for beneficial reuse: A critical review. J. Environ. Manag. 2022, 308, 114556. [Google Scholar] [CrossRef] [PubMed]
- Chittick, E.A.; Srebotnjak, T. An analysis of chemicals and other constituents found in produced water from hydraulically fractured wells in California and the challenges for wastewater management. J. Environ. Manag. 2017, 204, 502–509. [Google Scholar] [CrossRef] [PubMed]
- Al-Ghouti, M.A.; Al-Kaabi, M.A.; Ashfaq, M.Y.; Da’na, D.A. Produced water characteristics, treatment and reuse: A review. J. Water Process. 2019, 28, 222–239. [Google Scholar] [CrossRef]
- Husain, A.; Al-Harthi, M.A. Chemical treatment of oilfield wastewater and the effect of temperature on treatment efficiency: A review. J. Pet. Sci. Eng. 2023, 220, 111089. [Google Scholar] [CrossRef]
- Yang, Y.; Jie, B.; Zhai, Y.; Zeng, Y.; Kang, J.; Cheng, G.; Zhang, X. Performance and mechanism of efficient activation of peroxymonosulfate by Co-Mn-ZIF derived layered double hydroxide for the degradation of enrofloxacin. J. Mol. Liq. 2024, 394, 123723. [Google Scholar] [CrossRef]
- Wang, Y.; Lin, N.; Xu, J.; Jiang, H.; Chen, R.; Zhang, X.; Liu, N. Construction of microwave/PMS combined dual responsive perovskite-MXene system for antibiotic degradation: Synergistic effects of thermal and non-thermal. Appl. Surf. Sci. 2023, 639, 158263. [Google Scholar] [CrossRef]
- Ghafoori, S.; Omar, M.; Koutahzadeh, N.; Zendehboudi, S.; Malhas, R.N.; Mohamed, M.; Al-Zubaidi, S.; Redha, K.; Baraki, F.; Mehrvar, M. New advancements, challenges, and future needs on treatment of oilfield produced water: A state-of-the-art review. Sep. Purif. Technol. 2022, 289, 120652. [Google Scholar] [CrossRef]
- Patni, H.; Ragunathan, B. Recycling and re-usage of oilfield produced water—A review. Mater. Today Proc. 2023, 77, 307–313. [Google Scholar] [CrossRef]
- Akmirza, I.; Pascual, C.; Carvajal, A.; Pérez, R.; Muñoz, R.; Lebrero, R. Anoxic biodegradation of BTEX in a biotrickling filter. Sci. Total Environ. 2017, 587–588, 457–465. [Google Scholar] [CrossRef]
- Forero, J.E.; Ortíz, O.P.; Nariño, F.A.; Díaz, J.; Peña, H. Design and development of a high efficiency tank for crude oil dehydration (I). CTF-Cienc. Tecnol. Futuro 2008, 3, 185–199. [Google Scholar] [CrossRef]
- Wang, C.; Wang, Z.; Wei, X.; Li, X. A numerical study and flotation experiments of bicyclone column flotation for treating of produced water from ASP flooding. J. Water Process Eng. 2019, 32, 100972. [Google Scholar] [CrossRef]
- Kogiso, M.; Aoyagi, M. Produced water treatment using self-assembled organic nanotubes as adsorbents. J. Jpn. Pet. Inst. 2018, 61, 44–49. [Google Scholar] [CrossRef]
- Hassan, A.A.; Al-zobai, K.M.M. Chemical oxidation for oil separation from oilfield produced water under UV irradiation using Titanium dioxide as a nano-photocatalyst by batch and continuous techniques. Int. J. Chem. Eng. 2019, 2019, 9810728. [Google Scholar] [CrossRef]
- Mokhbi, Y.; Korichi, M.; Akchiche, Z. Combined photocatalytic and Fenton oxidation for oily wastewater treatment. Appl. Water Sci. 2019, 9, 35. [Google Scholar] [CrossRef]
- Duan, M.; He, Z.; Wang, X.; Jing, B.; Xu, Z.; Xiong, Y.; Fang, S. A novel interface-active cationic flocculant for the oil-water separation of oily wastewater produced from polymer flooding. J. Mol. Liq. 2019, 286, 110868. [Google Scholar] [CrossRef]
- Lusinier, N.; Seyssiecq, I.; Sambusiti, C.; Jacob, M.; Lesage, N.; Roche, N. Biological treatments of oilfield produced water: A comprehensive review. SPE J. 2019, 24, 2135–2147. [Google Scholar] [CrossRef]
- Guo, C.; Chen, Y.; Chen, J.; Wang, X.; Zhang, G.; Wang, J.; Cui, W.; Zhang, Z. Combined hydrolysis acidification and bio-contact oxidation system with air-lift tubes and activated carbon bioreactor for oilfield wastewater treatment. Bioresour. Technol. 2014, 169, 630–636. [Google Scholar] [CrossRef]
- Sharghi, E.A.; Bonakdarpour, B. The study of organic removal efficiency and halophilic bacterial mixed liquor characteristics in a membrane bioreactor treating hypersaline produced water at varying organic loading rates. Bioresour. Technol. 2013, 149, 486–495. [Google Scholar] [CrossRef]
- Liu, H.; Lam, J.C.W.; Li, W.; Yu, H.; Lam, P.K.S. Spatial distribution and removal performance of pharmaceuticals in municipal wastewater treatment plants in China. Sci. Total Environ. 2017, 586, 1162–1169. [Google Scholar] [CrossRef]
- Cao, J.; Zhang, T.; Wu, Y.; Sun, Y.; Zhang, Y.; Huang, B.; Fu, B.; Yang, E.; Zhang, Q.; Luo, J. Correlations of nitrogen removal and core functional genera in full-scale wastewater treatment plants: Influences of different treatment processes and influent characteristics. Bioresour. Technol. 2020, 297, 122455. [Google Scholar] [CrossRef] [PubMed]
- Wagner, M.; Loy, A. Bacterial community composition and function in sewage treatment systems. Curr. Opin. Biotechnol. 2002, 13, 218–227. [Google Scholar] [CrossRef] [PubMed]
- Jiao, S.; Liu, Z.S.; Lin, Y.B.; Yang, J.; Chen, W.M.; Wei, G.H. Bacterial communities in oil contaminated soils: Biogeography and co-occurrence patterns. Soil Biol. Biochem. 2016, 98, 64–73. [Google Scholar] [CrossRef]
- Wang, X.; Jiang, L.; Gai, Z.; Tao, F.; Tang, H.; Xu, P. The plasticity of indigenous microbial community in a full-scale heavy oil-produced water treatment plant. J. Hazard. Mater. 2018, 358, 155–164. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Zhou, J.; Zheng, P.; Wan, X.; Zhu, L.; Huang, J.; Jiang, J.; Chen, Y.; Song, Z. Seasonal variation significantly influenced the stochasticity of community assembly of amphibian symbiotic bacteria. Environ. Microbiol. 2022, 24, 5734–5748. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.Z.; Ning, D.L. Stochastic Community Assembly: Does It Matter in Microbial Ecology? Microbiol. Mol. Biol. Rev. 2017, 81, 10–1128. [Google Scholar] [CrossRef]
- Chave, J. Neutral theory and community ecology. Ecol. Lett. 2004, 7, 241–253. [Google Scholar] [CrossRef]
- Chesson, P. Mechanisms of maintenance of species diversity. Annu. Rev. Ecol. Syst. 2000, 31, 343–366. [Google Scholar] [CrossRef]
- Hubbell, S.P. The Unified Neutral Theory of Biodiversity and Biogeography; Princeton University Press: Princeton, NJ, USA, 2001. [Google Scholar]
- Dini-Andreote, F.; Stegen, J.C.; van Elsas, J.D.; Salles, J.F. Disentangling mechanisms that mediate the balance between stochastic and deterministic processes in microbial succession. Proc. Natl. Acad. Sci. USA 2015, 112, E1326–E1332. [Google Scholar] [CrossRef]
- Stegen, J.C.; Lin, X.; Fredrickson, J.K.; Chen, X.; Kennedy, D.W.; Murray, C.J.; Rockhold, M.L. Quantifying community assembly processes and identifying features that impose them. ISME J. 2013, 7, 2069. [Google Scholar] [CrossRef]
- Stegen, J.C.; Lin, X.; Fredrickson, J.K.; Konopka, A.E. Estimating and mapping ecological processes influencing microbial community assembly. Front. Microbiol. 2015, 6, 370. [Google Scholar] [CrossRef] [PubMed]
- Vellend, M. Conceptual synthesis in community ecology. Q. Rev. Biol. 2010, 85, 183–206. [Google Scholar] [CrossRef] [PubMed]
- Vellend, M.; Srivastava, D.S.; Anderson, K.M.; Brown, C.D.; Jankowski, J.E.; Kleynhans, E.J.; Kraft, N.J.B.; Letaw, A.D.; Macdonald, A.A.M.; Maclean, J.E.; et al. Assessing the relative importance of neutral stochasticity in ecological communities. Oikos 2014, 123, 1420–1430. [Google Scholar] [CrossRef]
- Ning, D.; Yuan, M.; Wu, L.; Zhang, Y.; Guo, X.; Zhou, X.; Yang, Y.; Arkin, A.P.; Firestone, M.K. A quantitative framework reveals ecological drivers of grassland microbial community assembly in response to warming. Nat. Commun. 2020, 11, 4717. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.; Xu, R.; Wang, D.; Lin, Q.; Zhou, S.; Lin, J.; Xia, L.; Fu, Y.; Gan, Z.; Meng, F. Ecological linkages between a biofilm ecosystem and reactor performance: The specificity of biofilm development phases. Environ. Sci. Technol. 2021, 55, 11948–11960. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Deng, Y.; Feng, K.; Cai, W.; Li, S.; Yin, H.; Xu, M.; Ning, D.; Qu, Y. Deterministic assembly and diversity gradient altered the biofilm community performances of bioreactors. Environ. Sci. Technol. 2019, 53, 1315–1324. [Google Scholar] [CrossRef]
- Zhou, J.; Liu, W.; Deng, Y.; Jiang, Y.H.; Xue, K.; He, Z.; Van Nostrand, J.D.; Wu, L.; Yang, Y.; Wang, A. Stochastic assembly leads to alternative communities with distinct functions in a bioreactor microbial community. Mbio 2013, 4, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Zielinski, W.; Korzeniewska, E.; Harnisz, M.; Drzymala, J.; Felis, E.; Bajkacz, S. Wastewater treatment plants as a reservoir of integrase and antibiotic resistance genes—An epidemiological threat to workers and environment. Environ. Int. 2021, 156, 106641. [Google Scholar] [CrossRef]
- Pruden, A. Balancing water sustainability and public health goals in the face of growing concerns about antibiotic resistance. Environ. Sci. Technol. 2014, 48, 5–14. [Google Scholar] [CrossRef]
- Berendonk, T.U.; Manaia, C.M.; Merlin, C.; Fatta-Kassinos, D.; Cytryn, E.; Walsh, F.; Bürgmann, H.; Sørum, H.; Norström, M.; Pons, M.N.; et al. Tackling antibiotic resistance: The environmental framework. Nat. Rev. Microbiol. 2015, 13, 310–317. [Google Scholar] [CrossRef]
- Jäger, T.; Hembach, N.; Elpers, C.; Wieland, A.; Alexander, J.; Hiller, C.; Krauter, G.; Schwartz, T. Reduction of antibiotic resistant bacteria during conventional and advanced wastewater treatment, and the disseminated loads released to the environment. Front. Microbiol. 2018, 9, 2599. [Google Scholar] [CrossRef] [PubMed]
- Osińska, A.; Korzeniewska, E.; Harnisz, M.; Felis, E.; Bajkacz, S.; Jachimowicz, P.; Niestępski, S.; Konopka, I. Small-scale wastewater treatment plants as a source of the dissemination of antibiotic resistance genes in the aquatic environment. J. Hazard. Mater. 2020, 381, 121221. [Google Scholar] [CrossRef] [PubMed]
- Orem, W.; Tatu, C.; Varonka, M.; Lerch, H.; Bates, A.; Engle, M.; Crosby, L.; McIntosh, J. Organic substances in produced and formation water from unconventional natural gas extraction in coal and shale. Int. J. Coal Geol. 2014, 126, 20–31. [Google Scholar] [CrossRef]
- Acharya, S.M.; Chakraborty, R.; Tringe, S.G. Emerging trends in biological treatment of wastewater from unconventional oil and gas extraction. Front. Microbiol. 2020, 11, 569019. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Xiang, J.; Zeng, Q.; Liu, Q.; Li, Y.; Sun, Y.; Xie, K.; Shi, S.; Gong, Z. Mitigation of the negative impacts of 84 disinfectant exposure for phenol treatment by an electro-enhanced SBR: Performance, oxidative stress, iron homeostasis and antibiotic resistance genes. J. Water Process. 2023, 55, 104161. [Google Scholar] [CrossRef]
- Zhou, S.; Peng, S.; Li, Z.; Zhang, D.; Zhu, Y.; Li, X.; Hong, M.; Li, W.; Lu, P. Characterization of microbial communities and functions in shale gas wastewaters and sludge: Implications for pretreatment. J. Hazard. Mater. 2022, 424, 127649. [Google Scholar] [CrossRef]
- Liu, Q.; Li, Y.; Sun, Y.; Xie, K.; Zeng, Q.; Hao, Y.; Yang, Q.; Pu, Y.; Shi, S.; Gong, Z. Deterioration of sludge characteristics and promotion of antibiotic resistance genes spread with the co-existing of polyvinylchloride microplastics and tetracycline in the sequencing batch reactor. Sci. Total Environ. 2024, 906, 167544. [Google Scholar] [CrossRef]
- Revelle, W.; Revelle, M.W. Package ‘psych’. Compr. R Arch. Netw. 2015, 337. Available online: https://personality-project.org/r/psych/ (accessed on 1 January 2024).
- Ma, F.; Guo, J.B.; Zhao, L.J.; Chang, C.C.; Cui, D. Application of bioaugmentation to improve the activated sludge system into the contact oxidation system treating petrochemical wastewater. Bioresour. Technol. 2009, 100, 597–602. [Google Scholar] [CrossRef]
- Ma, W.; Han, Y.; Xu, C.; Han, H.; Ma, W.; Zhu, H.; Li, K.; Wang, D. Enhanced degradation of phenolic compounds in coal gasification wastewater by a novel integration of micro-electrolysis with biological reactor (MEBR) under the micro-oxygen condition. Bioresour. Technol. 2018, 251, 303–310. [Google Scholar] [CrossRef]
- Huang, C.; Shi, Y.; Sheng, Z.; El-Din, M.G.; Liu, Y. Characterization of microbial communities during start-up of integrated fixed-film activated sludge (IFAS) systems for the treatment of oil sands process-affected water (OSPW). Biochem. Eng. J. 2017, 122, 123–132. [Google Scholar] [CrossRef]
- Kim, E.; Yulisa, A.; Kim, S.; Hwang, S. Monitoring microbial community structure and variations in a full-scale petroleum refinery wastewater treatment plant. Bioresour. Technol. 2020, 306, 123178. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Kong, J.; Liang, J.; El-Din, M.G.; Zhao, P.; Xie, W.; Chen, C. Nitrogen removal intensification of aerobic granular sludge through bioaugmentation with “heterotrophic nitrification-aerobic denitrification” consortium during petroleum wastewater treatment. Bioresour. Technol. 2022, 361, 127719. [Google Scholar] [CrossRef] [PubMed]
- Chu, L.; Wang, J. Treatment of oil-field produced wastewater by electron beam technology: Demulsification, disinfection and oil removal. J. Clean. Prod. 2022, 378, 134532. [Google Scholar] [CrossRef]
- Graham, E.D.; Tully, B.J. Marine Dadabacteria exhibit genome streamlining and phototrophy-driven niche partitioning. ISME J. 2021, 15, 1248–1256. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Sun, H.; Wang, X.; Li, F.; Cao, L.; Li, Y.; Dong, R.; Sun, Y.; Sun, P.; Bao, M. Temporal and spatial variation of petroleum hydrocarbons and microbial communities during static release of oil pollution sediments. Front. Mar. Sci. 2022, 9, 1025612. [Google Scholar] [CrossRef]
- Gao, Y.; Yuan, L.; Du, J.; Wang, H.; Yang, X.; Duan, L.; Zheng, L.; Bahar, M.M.; Zhao, Q.; Zhang, W.; et al. Bacterial community profile of the crude oil-contaminated saline soil in the Yellow River Delta Natural Reserve, China. Chemosphere 2022, 289, 133207. [Google Scholar] [CrossRef] [PubMed]
- Hu, P.; Tom, L.; Singh, A.; Thomas, B.C.; Baker, B.J.; Piceno, Y.M.; Andersen, G.L.; Banfield, J.F. Genome-resolved metagenomic analysis reveals roles for Candidate phyla and other microbial community members in biogeochemical transformations in oil reservoirs. mBio 2016, 7, 10–1128. [Google Scholar] [CrossRef]
- Nesbø, C.L.; Farrell, A.A.; Zhaxybayeva, O.; L’Haridon, S. Thermotoga, Bergey’s Manual of Systematics of Archaea and Bacteria; Wiley: Hoboken, NJ, USA, 2019; pp. 1–13. [Google Scholar] [CrossRef]
- Shahi, A.; Aydin, S.; Ince, B.; Ince, O. Reconstruction of bacterial community structure and variation for enhanced petroleum hydrocarbons degradation through biostimulation of oil contaminated soil. Chem. Eng. J. 2016, 306, 60–66. [Google Scholar] [CrossRef]
- He, J.; Luo, T.; Shi, Z.; Angelidaki, I.; Zhang, S.; Luo, G. Microbial shifts in anaerobic digestion towards phenol inhibition with and without hydrochar as revealed by metagenomic binning. J. Hazard. Mater. 2022, 440, 129718. [Google Scholar] [CrossRef]
- Yoshikawa, M.; Zhang, M.; Toyota, K. Integrated anaerobic-aerobic biodegradation of multiple contaminants including chlorinated ethylenes, benzene, toluene, and dichloromethane. Water Air Soil Pollut. 2017, 228, 25. [Google Scholar] [CrossRef] [PubMed]
- Segura, A.; Hernández-Sánchez, V.; Marqués, S.; Molina, L. Insights in the regulation of the degradation of PAHs in Novosphingobium sp. HR1a and utilization of this regulatory system as a tool for the detection of PAHs. Sci. Total Environ. 2017, 590, 381–393. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.Y.; Liu, W.X.; Wang, B.B.; Wang, Q.L.; Luo, Y.M.; Franks, A.E. PGPR enhanced phytoremediation of petroleum contaminated soil and rhizosphere microbial community response. Chemosphere 2015, 138, 592–598. [Google Scholar] [CrossRef] [PubMed]
- de la Cueva, S.C.; Rodríguez, C.H.; Cruz, N.O.S.; Contreras, J.A.R.; Miranda, J.L. Changes in bacterial populations during bioremediation of soil contaminated with petroleum hydrocarbons. Water Air Soil Pollut. 2016, 227, 91. [Google Scholar] [CrossRef]
- Magdy, M.M.; Gaber, Y.; Sebak, M.; Azmy, A.F.; AbdelGhani, S. Different metabolic pathways involved in anthracene biodegradation by Brevibacillus, Pseudomonas and Methylocystis Species. Beni-Suef Univ. J. Basic Appl. Sci. 2022, 11, 4. [Google Scholar] [CrossRef]
- Liang, J.; Chen, C.; Yoza, B.A.; Liang, Y.; Li, J.; Ke, M. Hydrolysis and acidification of activated sludge from a petroleum refinery. Pet. Sci. 2019, 16, 428–438. [Google Scholar] [CrossRef]
- Feitkenhauer, H.; Müller, R. Degradation of polycyclic aromatic hydrocarbons and long chain alkanes at 6070 °C by Thermus and Bacillus spp. Biodegradation 2003, 14, 367–372. [Google Scholar] [CrossRef]
- Verde, L.C.L.; Silva, T.R.E.; Dellagnezze, B.M.; Neto, E.V.D.S.; de Oliveira, V.M. Diversity of Hydrocarbon-Related Catabolic Genes in Oil Samples from Potiguar Basin (Rn, Brazil). Biotechnology 2013, 2013, 1–7. [Google Scholar]
- Albuquerque, L.; Rainey, F.A.; Nobre, M.F.; da Costa, M.S. Schleiferia thermophila gen. nov., sp. nov., a slightly thermophilic bacterium of the phylum ‘Bacteroidetes’ and the proposal of Schleiferiaceae fam. Nov. Int. J. Syst. Evol. Microbiol. 2011, 61, 2450–2455. [Google Scholar] [CrossRef]
- McKew, B.A.; Coulon, F.; Osborn, A.M.; Timmis, K.N.; McGenity, T.J. Determining the identity and roles of oil-metabolizing marine bacteria from the Thames estuary, UK. Environ. Microbiol. 2007, 9, 165–176. [Google Scholar] [CrossRef]
- Wang, B.; Lai, Q.; Cui, Z.; Tan, T.; Shao, Z. A pyrene-degrading consortium from deep-sea sediment of the West Pacific and its key member Cycloclasticus sp. P1. Environ. Microbiol. 2008, 10, 1948–1963. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Wu, X.; Liu, S.; Zhang, Y.; Cai, Z. Screening of three ammonia-oxidizing bacteria and construction of compounding agent CCZU C6 in high-efficiency ammonia-oxidizing. J. Water Process Eng. 2021, 40, 101862. [Google Scholar] [CrossRef]
- Qian, Y.; Xu, M.; Deng, T.; Hu, W.; He, Z.; Yang, X.; Wang, B.; Song, D.; Chen, L.; Huang, Y.; et al. Synergistic interactions of Desulfovibrio and Petrimonas for sulfate-reduction coupling polycyclic aromatic hydrocarbon degradation. J. Hazard. Mater. 2021, 407, 124385. [Google Scholar] [CrossRef] [PubMed]
- Dworkin, M.; Falkow, S.; Rosenberg, E.; Schleifer, K.H.; Stackebrandt, E. The Prokaryotes: A Handbook on the Biology of Bacteria; Archaea. Bacteria: Firmicutes, Actinomycetes; Springer: New York, NY, USA, 2006; Volume 1. [Google Scholar] [CrossRef]
- Van Houghton, B.D.; Acharya, S.M.; Rosenblum, J.S.; Chakraborty, R.; Tringe, S.G.; Cath, T.Y. Membrane bioreactor pretreatment of high-salinity O&G produced water. ACS ES&T Water 2022, 2, 484–494. [Google Scholar] [CrossRef]
- Valentín-Vargas, A.; Toro-Labrador, G.; Massol-Deyá, A.A. Bacterial community dynamics in full-scale activated sludge bioreactors: Operational and ecological factors driving community assembly and performance. PLoS ONE 2012, 7, e42524. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Zhang, B.; Ning, D.; Zhang, Y.; Dai, T.; Wu, L.; Li, T.; Liu, W.; Zhou, J.; Wen, X. Seasonal dynamics of the microbial community in two full-scale wastewater treatment plants: Diversity, composition, phylogenetic group based assembly and co-occurrence pattern. Water Res. 2021, 200, 117295. [Google Scholar] [CrossRef] [PubMed]
- Sierra-Garcia, I.N.; Belgini, D.R.B.; Torres-Ballesteros, A.; Paez-Espino, D.; Capilla, R.; Neto, E.V.S.; Gray, N.; de Oliveira, V.M. In depth metagenomic analysis in contrasting oil wells reveals syntrophic bacterial and archaeal associations for oil biodegradation in petroleum reservoirs. Sci. Total Environ. 2020, 715, 136646. [Google Scholar] [CrossRef]
- Das, N.; Chandran, P. Microbial degradation of petroleum hydrocarbon contaminants: An overview. Biotechnol. Res. Int. 2011, 2011, 941810. [Google Scholar] [CrossRef]
- Li, B.B.; Zhi, L.L.; Peng, Z.Y.; Ma, X.X.; Li, J. Contrasting distribution of antibiotic resistance genes and microbial communities in suspended activated sludge versus attached biofilms in an integrated fixed film activated sludge (IFAS) system. Sci. Total Environ. 2020, 742, 140481. [Google Scholar] [CrossRef]
- Geissen, V.; Gomez-Rivera, P.; Lwanga, E.H.; Mendoza, R.B.; Narcías, A.T.; Mar-cías, E.B. Using earthworms to test the efficiency of remediation of oil-polluted soil in tropical Mexico. Ecotox. Environ. Saf. 2008, 71, 638–642. [Google Scholar] [CrossRef]
- Du, J.; Ren, H.Q.; Geng, J.J.; Zhang, Y.; Xu, K.; Ding, L.L. Occurrence and abundance of tetracycline, sulfonamide resistance genes, and class 1 integron in five wastewater treatment plants. Environ. Sci. Pollut. Res. 2014, 21, 7276–7284. [Google Scholar] [CrossRef]
- Sun, H.; Zhang, H.; Wu, D.; Ding, J.; Niu, Y.; Jiang, T.; Yang, X.; Liu, Y. Deciphering the antibiotic resistome and microbial community in municipal wastewater treatment plants at different elevations in eastern and western China. Water Res. 2023, 229, 119461. [Google Scholar] [CrossRef]
- Xu, M.; Gao, P.; Chen, H.Q.; Shen, X.X.; Xu, R.Z.; Cao, J.S. Metagenomic insight into the prevalence and driving forces of antibiotic resistance genes in the whole process of three full-scale wastewater treatment plants. J. Environ. Manag. 2023, 344, 118369. [Google Scholar] [CrossRef]
- Petrovich, M.; Chu, B.; Wright, D.; Griffin, J.; Elfeki, M.; Murphy, B.T.; Poretsky, R.; Wells, G. Antibiotic resistance genes show enhanced mobilization through suspended growth and biofilm-based wastewater treatment processes. FEMS Microbiol. Ecol. 2018, 94, fiy041. [Google Scholar] [CrossRef]
- Li, B.; Qiu, Y.; Zhang, J.; Huang, X.; Shi, H.C.; Yin, H.B. Real-time study of rapid spread of antibiotic resistance plasmid in biofilm using microfluidics. Environ. Sci. Technol. 2018, 52, 11132–11141. [Google Scholar] [CrossRef]
- Qiu, Y.; Zhang, J.; Li, B.; Wen, X.H.; Liang, P.; Huang, X. A novel microfluidic system enables visualization and analysis of antibiotic resistance gene transfer to activated sludge bacteria in biofilm. Sci. Total Environ. 2018, 642, 582–590. [Google Scholar] [CrossRef]
- Król, J.E.; Nguyen, H.D.; Rogers, L.M.; Beyenal, H.; Krone, S.M.; Top, E.M. Increased transfer of a multidrug resistance plasmid in Escherichia coli biofilms at the air-liquid interface. Appl. Environ. Microbiol. 2011, 77, 5079–5088. [Google Scholar] [CrossRef] [PubMed]
- Stalder, T.; Top, E. Plasmid transfer in biofilms: A perspective on limitations and opportunities. NPJ Biofilms Microbiomes 2016, 2, 16022. [Google Scholar] [CrossRef]
- Wu, Y.; Qi, D.; Yao, H.; Ren, J.; Hu, J.; Lyu, Y.; Yang, S.; Sun, W. Antibiotic resistome and its driving factors in an urban river in northern China. Sci. Total Environ. 2022, 838, 156536. [Google Scholar] [CrossRef] [PubMed]
- Zeng, J.Y.; Li, Y.Y.; Jin, G.P.; Su, J.Q.; Yao, H.Y. Short-term benzalkonium chloride (C12) exposure induced the occurrence of wide-spectrum antibiotic resistance in agricultural soils. Environ. Sci. Technol. 2022, 56, 15054–15063. [Google Scholar] [CrossRef] [PubMed]
- Ochman, H.; Lawrence, J.G.; Groisman, E.A. Lateral gene transfer and the nature of bacterial innovation. Nature 2000, 405, 299–304. [Google Scholar] [CrossRef] [PubMed]


| PWTP1 | PWTP2 | PWTP3 | ||
|---|---|---|---|---|
| Location | Karamay, Xinjiang | Karamay, Xinjiang | Karamay, Xinjiang | |
| Name | Heavy oil sewage standard discharge station | Heavy oil saline sewage standard discharge station | Fracturing flowback fluid standard discharge station | |
| Bioprocess | Activated sludge process | Biological aerated filter | Biological contact oxidation process | |
| COD (mg/L) | Influent | 300–350 | 350–400 | 400–500 |
| Effluent | 180–200 | 150–200 | 100–150 | |
| NH4-N (mg/L) | Influent | 7–8 | 13–14 | 15–16 |
| Effluent | 1–2 | 6–7 | 5–6 | |
| Petroleum (mg/L) | Influent | 7–8 | 5–6 | 9–10 |
| Effluent | 1–2 | 1–2 | 2–3 | |
| Flow rate (m3/h) | 80 | 60 | 70 | |
| T (°C) | 30–32 | 25–27 | 28–30 | |
| pH | 7.0–7.5 | 7.0–7.5 | 7.5–8.0 | |
| DO (mg/L) | 2–4 | 2–3 | 3–4 | |
| Volatile Phenol (mg/L) | 0.074 | 0.135 | 0.035 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xie, K.; Zeng, Q.; Yu, S.; Luo, H.; Zhang, Y.; Ma, C.; Hu, H.; Shi, S.; Gong, Z. Contrasting Distribution of Microbial Communities, Functional Genes, and Antibiotic Resistance Genes in Produced Water Treatment Plants with Different Treatment Technologies. Water 2024, 16, 195. https://doi.org/10.3390/w16020195
Xie K, Zeng Q, Yu S, Luo H, Zhang Y, Ma C, Hu H, Shi S, Gong Z. Contrasting Distribution of Microbial Communities, Functional Genes, and Antibiotic Resistance Genes in Produced Water Treatment Plants with Different Treatment Technologies. Water. 2024; 16(2):195. https://doi.org/10.3390/w16020195
Chicago/Turabian StyleXie, Kunpeng, Qianzhi Zeng, Sihui Yu, Hongjing Luo, Yongsheng Zhang, Changwei Ma, Haoyu Hu, Shengnan Shi, and Zheng Gong. 2024. "Contrasting Distribution of Microbial Communities, Functional Genes, and Antibiotic Resistance Genes in Produced Water Treatment Plants with Different Treatment Technologies" Water 16, no. 2: 195. https://doi.org/10.3390/w16020195
APA StyleXie, K., Zeng, Q., Yu, S., Luo, H., Zhang, Y., Ma, C., Hu, H., Shi, S., & Gong, Z. (2024). Contrasting Distribution of Microbial Communities, Functional Genes, and Antibiotic Resistance Genes in Produced Water Treatment Plants with Different Treatment Technologies. Water, 16(2), 195. https://doi.org/10.3390/w16020195





