Could Land Abandonment with Human Intervention Benefit Cropland Restoration? From the Perspective of Soil Microbiota
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Site Description
2.2. Experimental Design
- LA, spontaneous vegetation, no manure input, spontaneous vegetation, as agricultural land abandonment;
- HIS, no manure input, spontaneous vegetation sown with alfalfa (Medicago sativa L.) and Dahurian wild rye (Elymus dahuricus Turcz.) in LA, as slight human intervention in LA;
- HIM, spontaneous vegetation, LA with composted cattle manure (400 g kg−1 organic C, 7.0 g kg−1 TN, 11.5 g kg−1 total P, and 9.8 g kg−1 total K), 1500 kg ha−1, as medium human intervention in LA;
- HII, HIS with composted cattle manure (same as HIM), 1500 kg ha−1, as intensive human intervention in LA.
2.3. Preparation of Soil Samples
2.4. Edaphic Properties Analysis
2.5. Bacterial and Fungal Analysis
2.6. Sequences Processing and Statistical Analysis
3. Results
3.1. The Effects of Management Scenarios on Edaphic Properties
3.2. The Effects of Management Scenarios on Microbial Biodiversity
3.3. The Effects of Management Scenarios on Microbial Communities
3.4. The Effects of Management and Edaphic Properties on Classified Microbial Communities
4. Discussion
4.1. The Risks and Benefits of LA
4.2. The Risks and Benefits of Planting Forage Grass on LA
4.3. The Risks and Benefits of Cow Manure Inputs on LA
4.4. The Risks and Benefits of Intensive Human Interference on LA
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Xu, D.; Deng, X.; Guo, S.; Liu, S. Labor migration and farmland abandonment in rural China: Empirical results and policy implications. J. Environ. Manag. 2019, 232, 738–750. [Google Scholar] [CrossRef]
- Ferrucci, G.; Jiménez-Rodríguez, R.; Onorantea, L. Food Price Pass-Through in the Euro Area: Non-Linearities and the Role of the Common Agricultural Policy. Int. J. Cent. Bank. 2012, 8, 179–218. [Google Scholar]
- Lasanta, T.; Arnáez, J.; Pascual, N.; Ruiz, P.; Errea, M.; Lana-Renault, N. Space–time process and drivers of land abandonment in Europe. CATENA 2017, 149, 810–823. [Google Scholar] [CrossRef]
- Yang, Y.; Hobbie, S.E.; Hernandez, R.R.; Fargione, J.; Grodsky, S.M.; Tilman, D.; Zhu, Y.-G.; Luo, Y.; Smith, T.M.; Jungers, J.M.; et al. Restoring Abandoned Farmland to Mitigate Climate Change on a Full Earth. One Earth 2020, 3, 176–186. [Google Scholar] [CrossRef]
- Tian, H.; Wang, H.; Hui, X.; Wang, Z.; Drijber, R.A.; Liu, J. Changes in soil microbial communities after 10 years of winter wheat cultivation versus fallow in an organic-poor soil in the Loess Plateau of China. PLoS ONE 2017, 12, e0184223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yurui, L.; Xuanchang, Z.; Zhi, C.; Zhengjia, L.; Zhi, L.; Yansui, L. Towards the progress of ecological restoration and economic development in China’s Loess Plateau and strategy for more sustainable development. Sci. Total. Environ. 2021, 756, 143676. [Google Scholar] [CrossRef]
- Tarjuelo, R.; Margalida, A.; Mougeot, F. Changing the fallow paradigm: A win–win strategy for the post-2020 Common Agricultural Policy to halt farmland bird declines. J. Appl. Ecol. 2020, 57, 642–649. [Google Scholar] [CrossRef]
- Hauchhum, R.; Tripathi, S.K. Impact of Rhizosphere Microbes of Three Early Colonizing Annual Plants on Improving Soil Fertility During Vegetation Establishment Under Different Fallow Periods Following Shifting Cultivation. Agric. Res. 2019, 9, 213–221. [Google Scholar] [CrossRef]
- Joslin, A.; Markewitz, D.; Morris, L.A.; Oliveira, F.D.A.; Kato, O. Improved fallow: Growth and nitrogen accumulation of five native tree species in Brazil. Nutr. Cycl. Agroecosystems 2016, 106, 1–15. [Google Scholar] [CrossRef]
- Sun, B.; Peng, Y.; Yang, H.; Li, Z.; Gao, Y.; Wang, C.; Yan, Y.; Liu, Y. Alfalfa (Medicago sativa L.)/Maize (Zea mays L.) Intercropping Provides a Feasible Way to Improve Yield and Economic Incomes in Farming and Pastoral Areas of Northeast China. PLoS ONE 2014, 9, e110556. [Google Scholar] [CrossRef]
- Antil, R.S.; Gerzabek, M.H.; Haberhauer, G.; Eder, G. Long-term effects of croppedvs. fallow and fertilizer amendments on soil organic matter I. Organic carbon. J. Plant Nutr. Soil Sci. 2005, 168, 108–116. [Google Scholar] [CrossRef]
- Shirani, H.; Hajabbasi, M.; Afyuni, M.; Hemmat, A. Effects of farmyard manure and tillage systems on soil physical properties and corn yield in central Iran. Soil Tillage Res. 2002, 68, 101–108. [Google Scholar] [CrossRef]
- McDaniel, M.D.; Tiemann, L.K.; Grandy, A.S. Does agricultural crop diversity enhance soil microbial biomass and organic matter dynamics? A meta-analysis. Ecol. Appl. 2014, 24, 560–570. [Google Scholar] [CrossRef] [Green Version]
- Li, G.; Wu, C.; Gao, W. Effects of short-term fallow managements on soil microbial properties: A case study in China. Appl. Soil Ecol. 2018, 125, 128–137. [Google Scholar] [CrossRef]
- Lammel, D.R.; Feigl, B.J.; Cerri, C.C.; Nüsslein, K. Specific microbial gene abundances and soil parameters contribute to C, N, and greenhouse gas process rates after land use change in Southern Amazonian Soils. Front. Microbiol. 2015, 6, 1057. [Google Scholar] [CrossRef] [Green Version]
- Bossio, D.; Girvan, M.S.; Verchot, L.; Bullimore, J.; Borelli, T.; Albrecht, A.; Scow, K.; Ball, A.; Pretty, J.; Osborn, A.M. Soil Microbial Community Response to Land Use Change in an Agricultural Landscape of Western Kenya. Microb. Ecol. 2005, 49, 50–62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muchane, M.N.; Jama, B.; Othieno, C.; Okalebo, R.; Odee, D.; Machua, J.; Jansa, J. Influence of improved fallow systems and phosphorus application on arbuscular mycorrhizal fungi symbiosis in maize grown in western Kenya. Agrofor. Syst. 2010, 78, 139–150. [Google Scholar] [CrossRef]
- Moghimian, N.; Hosseini, S.M.; Kooch, Y.; Darki, B.Z. Impacts of changes in land use/cover on soil microbial and enzyme activities. CATENA 2017, 157, 407–414. [Google Scholar] [CrossRef]
- Lv, K. Spatial and Temporal Variation of Soil Nutrients and Its Influencing Factors—A Case Study of Wudi County. Master’s Thesis, Zhejiang University, Hangzhou, China, 2018. [Google Scholar]
- Dick, R.P.; Breakwell, D.P.; Turco, R.F. Soil Enzyme Activities and Biodiversity Measurements as Integrative Microbiological Indicators. In Methods for Assessing Soil Quality; Doran, J.W., Jones, A.J., Eds.; the Soil Science Society of America, Inc.: Madison, WI, USA, 2015; pp. 247–271. [Google Scholar] [CrossRef]
- Hoffmann, G.; Teicher, K. Ein kolorimetrisches Verfahren zur Bestimmung der Ureaseaktivität in Böden. J. Plant Nutr. Soil Sci. 1961, 95, 55–63. [Google Scholar] [CrossRef]
- Angenent, L.T.; Kelley, S.T.; Amand, A.S.; Pace, N.R.; Hernandez, M.T. Molecular identification of potential pathogens in water and air of a hospital therapy pool. Proc. Natl. Acad. Sci. USA 2005, 102, 4860–4865. [Google Scholar] [CrossRef] [Green Version]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, I.J.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naїve Bayesian Classifier for Rapid Assignment of rRNA Sequences into the New Bacterial Taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abarenkov, K.; Nilsson, H.; Larsson, K.; Alexander, I.J.; Eberhardt, U.; Erland, S.; Høiland, K.; Kjøller, R.; Larsson, E.; Pennanen, T.; et al. The UNITE database for molecular identification of fungi—Recent updates and future perspectives. New Phytol. 2010, 186, 281–285. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019. [Google Scholar]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [Green Version]
- Liaw, A.; Wiener, M. Classification and Regression by RandomForest. R News 2002, 2, 18–22. [Google Scholar]
- Sánchez-Marañón, M.; Miralles, I.; Garrido, J.F.A.; Anguita-Maeso, M.; Millán, V.; Ortega, R.; García-Salcedo, J.A.; Martínez-Abarca, F.; Soriano, M. Changes in the soil bacterial community along a pedogenic gradient. Sci. Rep. 2017, 7, 14593. [Google Scholar] [CrossRef]
- Tan, G.; Liu, Y.; Peng, S.; Yin, H.; Meng, D.; Tao, J.; Gu, Y.; Li, J.; Yang, S.; Xiao, N.; et al. Soil potentials to resist continuous cropping obstacle: Three field cases. Environ. Res. 2021, 200, 111319. [Google Scholar] [CrossRef]
- Garrido-Oter, R.; Nakano, R.T.; Dombrowski, N.; Ma, K.-W.; McHardy, A.C.; Schulze-Lefert, P. Modular Traits of the Rhizobiales Root Microbiota and Their Evolutionary Relationship with Symbiotic Rhizobia. Cell Host Microbe 2018, 24, 155–167. [Google Scholar] [CrossRef] [Green Version]
- Bhatti, A.A.; Haq, S.; Bhat, R.A. Actinomycetes benefaction role in soil and plant health. Microb. Pathog. 2017, 111, 458–467. [Google Scholar] [CrossRef]
- Sathya, A.; Vijayabharathi, R.; Gopalakrishnan, S. Plant growth-promoting actinobacteria: A new strategy for enhancing sustainable production and protection of grain legumes. 3 Biotech 2017, 7, 102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Y.; Zheng, C.; Liang, L.; Yi, Z.; Xue, S. Quantitative assessment of the potential for soil improvement by planting Miscanthus on saline-alkaline soil and the underlying microbial mechanism. GCB Bioenergy 2021, 13, 1191–1205. [Google Scholar] [CrossRef]
- Abdulla, H.M.; El Shatoury, S. Actinomycetes in rice straw decomposition. Waste Manag. 2007, 27, 850–853. [Google Scholar] [CrossRef]
- Pedrinho, A.; Mendes, L.W.; Merloti, L.F.; Andreote, F.D.; Tsai, S.M. The natural recovery of soil microbial community and nitrogen functions after pasture abandonment in the Amazon region. FEMS Microbiol. Ecol. 2020, 96, fiaa149. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Zhang, M.; Wu, C. Short-term fallow practices drive soil bacterial community changes: A case study from China. Appl. Soil Ecol. 2021, 165, 103988. [Google Scholar] [CrossRef]
- Wolińska, A.; Kuźniar, A.; Zielenkiewicz, U.; Banach, A.; Błaszczyk, M. Indicators of arable soils fatigue—Bacterial families and genera: A metagenomic approach. Ecol. Indic. 2018, 93, 490–500. [Google Scholar] [CrossRef]
- Safdarian, M.; Askari, H.; Nematzadeh, G. Transcriptional responses of wheat roots inoculated with Arthrobacter nitroguajacolicus to salt stress. Sci. Rep. 2019, 9, 1792. [Google Scholar] [CrossRef]
- Tang, X.; Zhong, R.; Jiang, J.; He, L.; Huang, Z.; Shi, G.; Wu, H.; Liu, J.; Xiong, F.; Han, Z.; et al. Cassava/peanut intercropping improves soil quality via rhizospheric microbes increased available nitrogen contents. BMC Biotechnol. 2020, 20, 13. [Google Scholar] [CrossRef] [Green Version]
- Intra, B.; Mungsuntisuk, I.; Nihira, T.; Igarashi, Y.; Panbangred, W. Identification of actinomycetes from plant rhizospheric soils with inhibitory activity against Colletotrichum spp., the causative agent of anthracnose disease. BMC Res. Notes 2011, 4, 98. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Yu, H.; Zhou, X.; Wu, F. Cucumber (Cucumis sativus L.) Seedling Rhizosphere Trichoderma and Fusarium spp. Communities Altered by Vanillic Acid. Front. Microbiol. 2018, 9, 2195. [Google Scholar] [CrossRef] [Green Version]
- Garrido-Jurado, I.; Fernández-Bravo, M.; Campos, C.; Quesada-Moraga, E. Diversity of entomopathogenic Hypocreales in soil and phylloplanes of five Mediterranean cropping systems. J. Invertebr. Pathol. 2015, 130, 97–106. [Google Scholar] [CrossRef] [PubMed]
- Aziz, A.A.; Lee, K.; Park, B.; Park, H.; Park, K.; Choi, I.-G.; Chang, I.S. Comparative study of the airborne microbial communities and their functional composition in fine particulate matter (PM2.5) under non-extreme and extreme PM2.5 conditions. Atmospheric Environ. 2018, 194, 82–92. [Google Scholar] [CrossRef]
- Jansa, J.; Mozafar, A.; Anken, T.; Ruh, R.; Sanders, I.R.; Frossard, E. Diversity and structure of AMF communities as affected by tillage in a temperate soil. Mycorrhiza 2002, 12, 225–234. [Google Scholar] [CrossRef]
- Banerjee, S.; Walder, F.; Büchi, L.; Meyer, M.; Held, A.Y.; Gattinger, A.; Keller, T.; Charles, R.; van der Heijden, M.G.A. Agricultural intensification reduces microbial network complexity and the abundance of keystone taxa in roots. ISME J. 2019, 13, 1722–1736. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Saixi, Y.; Yi, R.; Baoyin, T. Characterization of soil microbes associated with a grazing-tolerant grass species, Stipa breviflora, in the Inner Mongolian desert steppe. Ecol. Evol. 2020, 10, 10607–10618. [Google Scholar] [CrossRef]
- Wang, J.; Wang, C.; Li, J.; Bai, P.; Li, Q.; Shen, M.; Li, R.; Li, T.; Zhao, J. Comparative Genomics of Degradative Novosphingobium Strains with Special Reference to Microcystin-Degrading Novosphingobium sp. THN1. Front. Microbiol. 2018, 9, 2238. [Google Scholar] [CrossRef] [Green Version]
- Yagi, J.M.; Sims, D.; Brettin, T.; Bruce, D.; Madsen, E.L. The genome of Polaromonas naphthalenivorans strain CJ2, isolated from coal tar-contaminated sediment, reveals physiological and metabolic versatility and evolution through extensive horizontal gene transfer. Environ. Microbiol. 2009, 11, 2253–2270. [Google Scholar] [CrossRef] [PubMed]
- Mackelprang, R.; Grube, A.M.; Lamendella, R.; Jesus, E.D.C.; Copeland, A.; Liang, C.; Jackson, R.D.; Rice, C.W.; Kapucija, S.; Parsa, B.; et al. Microbial Community Structure and Functional Potential in Cultivated and Native Tallgrass Prairie Soils of the Midwestern United States. Front. Microbiol. 2018, 9, 1775. [Google Scholar] [CrossRef] [Green Version]
- Yao, R.; Yang, J.; Wang, X.; Xie, W.; Zheng, F.; Li, H.; Tang, C.; Zhu, H. Response of soil characteristics and bacterial communities to nitrogen fertilization gradients in a coastal salt-affected agroecosystem. Land Degrad. Dev. 2021, 32, 338–353. [Google Scholar] [CrossRef]
- Mujahid, M.; Sasikala, C.; Ramana, C.V. Production of indole-3-acetic acid and related indole derivatives from L-tryptophan by Rubrivivax benzoatilyticus JA2. Appl. Microbiol. Biotechnol. 2011, 89, 1001–1008. [Google Scholar] [CrossRef]
- Hrynkiewicz, K.; Haug, I.; Baum, C. Ectomycorrhizal community structure under willows at former ore mining sites. Eur. J. Soil Biol. 2008, 44, 37–44. [Google Scholar] [CrossRef]
- Huang, X.; Zhou, X.; Zhang, J.; Cai, Z. Highly connected taxa located in the microbial network are prevalent in the rhizosphere soil of healthy plant. Biol. Fertil. Soils 2019, 55, 299–312. [Google Scholar] [CrossRef]
- Abbott, S.P.; Sigler, L.; Currah, R.S. Microascus brevicaulis sp. nov., the teleomorph of Scopulariopsis brevicaulis, supports placement of Scopulariopsiswith the Microascaceae. Mycologia 1998, 90, 297–302. [Google Scholar] [CrossRef]
- Vitale, A.; Aiello, D.; Guarnaccia, V.; Perrone, G.; Stea, G.; Polizzi, G. First Report of Root Rot Caused by Ilyonectria (=Neonectria) macrodidyma on Avocado (Persea americana) in Italy. J. Phytopathol. 2012, 160, 156–159. [Google Scholar] [CrossRef]
- Hernández-Restrepo, M.; Bezerra, J.D.P.; Tan, Y.P.; Wiederhold, N.; Crous, P.; Guarro, J.; Gené, J. Re-evaluation of Mycoleptodiscus species and morphologically similar fungi. Persoonia Mol. Phylogeny Evol. Fungi 2019, 42, 205–227. [Google Scholar] [CrossRef] [Green Version]
- Hou, C.-L.; Piepenbring, M. New species and new records of Rhytismatales from Panama. Mycologia 2009, 101, 565–572. [Google Scholar] [CrossRef] [PubMed]
- Pattarasaikul, W.; Soytong, K.; Poeaim, S. Biological control of anthracnose disease on banana var ‘Namwa Mali-Ong’ by Neosartorya species. Int. J. Agric. Technol. 2018, 14, 1589–1598. [Google Scholar]
- Li, H.; Cai, X.; Gong, J.; Xu, T.; Ding, G.-C.; Li, J. Long-Term Organic Farming Manipulated Rhizospheric Microbiome and Bacillus Antagonism Against Pepper Blight (Phytophthora capsici). Front. Microbiol. 2019, 10, 342. [Google Scholar] [CrossRef] [PubMed]
- Ding, P.; Chu, L.; Wang, J. Biological treatment of actual petrochemical wastewater using anaerobic/anoxic/oxic process and the microbial diversity analysis. Appl. Microbiol. Biotechnol. 2016, 100, 10193–10202. [Google Scholar] [CrossRef]
- Mason, L.; Eagar, A.; Patel, P.; Blackwood, C.; DeForest, J. Potential microbial bioindicators of phosphorus mining in a temperate deciduous forest. J. Appl. Microbiol. 2021, 130, 109–122. [Google Scholar] [CrossRef]
- Wiggins, B.E.; Kinkel, L.L. Green manures and crop sequences influence alfalfa root rot and pathogen inhibitory activity among soil-borne streptomycetes. Plant Soil 2005, 268, 271–283. [Google Scholar] [CrossRef]
- Burns, A.S.; Padilla, C.C.; Pratte, Z.A.; Gilde, K.; Regensburger, M.; Hall, E.; Dove, A.D.M.; Stewart, F.J. Broad Phylogenetic Diversity Associated with Nitrogen Loss through Sulfur Oxidation in a Large Public Marine Aquarium. Appl. Environ. Microbiol. 2018, 84, e01250–e01218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, Y.; Wang, X.; Li, X.; Xu, M.; Li, Y.; Zheng, H.; Luo, Y.; Smith, P. Impacts of land use and salinization on soil inorganic and organic carbon in the middle-lower Yellow River Delta. Pedosphere 2021, 31, 839–848. [Google Scholar] [CrossRef]
- Huhe; Borjigin, S.; Cheng, Y.; Nomura, N.; Nakajima, T.; Nakamura, T.; Uchiyama, H. Effect of Abandonment on Diversity and Abundance of Free-Living Nitrogen-Fixing Bacteria and Total Bacteria in the Cropland Soils of Hulun Buir, Inner Mongolia. PLoS ONE 2014, 9, e106714. [Google Scholar] [CrossRef] [Green Version]
- Koulouri, M.; Giourga, C. Land abandonment and slope gradient as key factors of soil erosion in Mediterranean terraced lands. CATENA 2007, 69, 274–281. [Google Scholar] [CrossRef]
- Liu, M.; Han, G.; Zhang, Q. Effects of agricultural abandonment on soil aggregation, soil organic carbon storage and stabilization: Results from observation in a small karst catchment, Southwest China. Agric. Ecosyst. Environ. 2020, 288, 106719. [Google Scholar] [CrossRef]
- Taniguchi, T.; Yuzawa, T.; HuiPing, M.; Yamamoto, F.; Yamanaka, N. Plantation soil inoculation combined with straw checkerboard barriers enhances ectomycorrhizal colonization and subsequent growth of nursery grown Pinus tabulaeformis seedlings in a dryland. Ecol. Eng. 2021, 163, 106191. [Google Scholar] [CrossRef]
- Aburto-Medina, A.; Adetutu, E.M.; Aleer, S.; Weber, J.; Patil, S.S.; Sheppard, P.J.; Ball, A.S.; Juhasz, A.L. Comparison of indigenous and exogenous microbial populations during slurry phase biodegradation of long-term hydrocarbon-contaminated soil. Biodegradation 2012, 23, 813–822. [Google Scholar] [CrossRef]
- Thammannagowda, S.; Magnusson, L.; Jo, J.H.; Maness, P.C.; Seibert, M. Renewable Hydrogen from Biomass. In Encyclopedia of Biological Chemistry, 2nd ed.; Lennarz, W.J., Lane, M.D., Eds.; Academic Press: Waltham, MA, USA, 2013; pp. 72–75. [Google Scholar] [CrossRef]
- Raggi, L.; García-Guevara, F.; Godoy-Lozano, E.E.; Martínez-Santana, A.; Escobar-Zepeda, A.; Gutierrez-Rios, R.M.; Loza, A.; Merino, E.; Sanchez-Flores, A.; Licea-Navarro, A.; et al. Metagenomic Profiling and Microbial Metabolic Potential of Perdido Fold Belt (NW) and Campeche Knolls (SE) in the Gulf of Mexico. Front. Microbiol. 2020, 11, 1825. [Google Scholar] [CrossRef]
- Zak, D.R.; Argiroff, W.; Freedman, Z.B.; Upchurch, R.A.; Entwistle, E.M.; Romanowicz, K. Anthropogenic N deposition, fungal gene expression, and an increasing soil carbon sink in the Northern Hemisphere. Ecology 2019, 100, e02804. [Google Scholar] [CrossRef]
- Ma, X.; Geng, Q.; Zhang, H.; Bian, C.; Chen, H.Y.H.; Jiang, D.; Xu, X. Global negative effects of nutrient enrichment on arbuscular mycorrhizal fungi, plant diversity and ecosystem multifunctionality. New Phytol. 2021, 229, 2957–2969. [Google Scholar] [CrossRef] [PubMed]
- Bosso, L.; Lacatena, F.; Varlese, R.; Nocerino, S.; Cristinzio, G.; Russo, D. Plant pathogens but not antagonists change in soil fungal communities across a land abandonment gradient in a Mediterranean landscape. Acta Oecologica 2017, 78, 1–6. [Google Scholar] [CrossRef]
- Umaerus, V.R.; Scholte, K.; Turkensteen, L.J. Crop Rotation and the Occurrence of Fungal Diseases in Potatoes. Effects of Crop Rotation on Potato Production in the Temperate Zones. In Proceedings of the International Conference on Effects of Crop Rotation on Potato Production in the Temperate Zones, Wageningen, The Netherlands, 14–19 August 1988; Vos, J., Van Loon, C.D., Bollen, G.J., Eds.; Springer: Dordrecht, The Netherlands, 1989; pp. 171–189. [Google Scholar] [CrossRef]
- Xiao, G.; Zhu, X.; Hou, C.; Xia, X. Extraction and analysis of abandoned farmland: A case study of Qingyun and Wudi counties in Shandong Province. J. Geogr. Sci. 2019, 29, 581–597. [Google Scholar] [CrossRef] [Green Version]
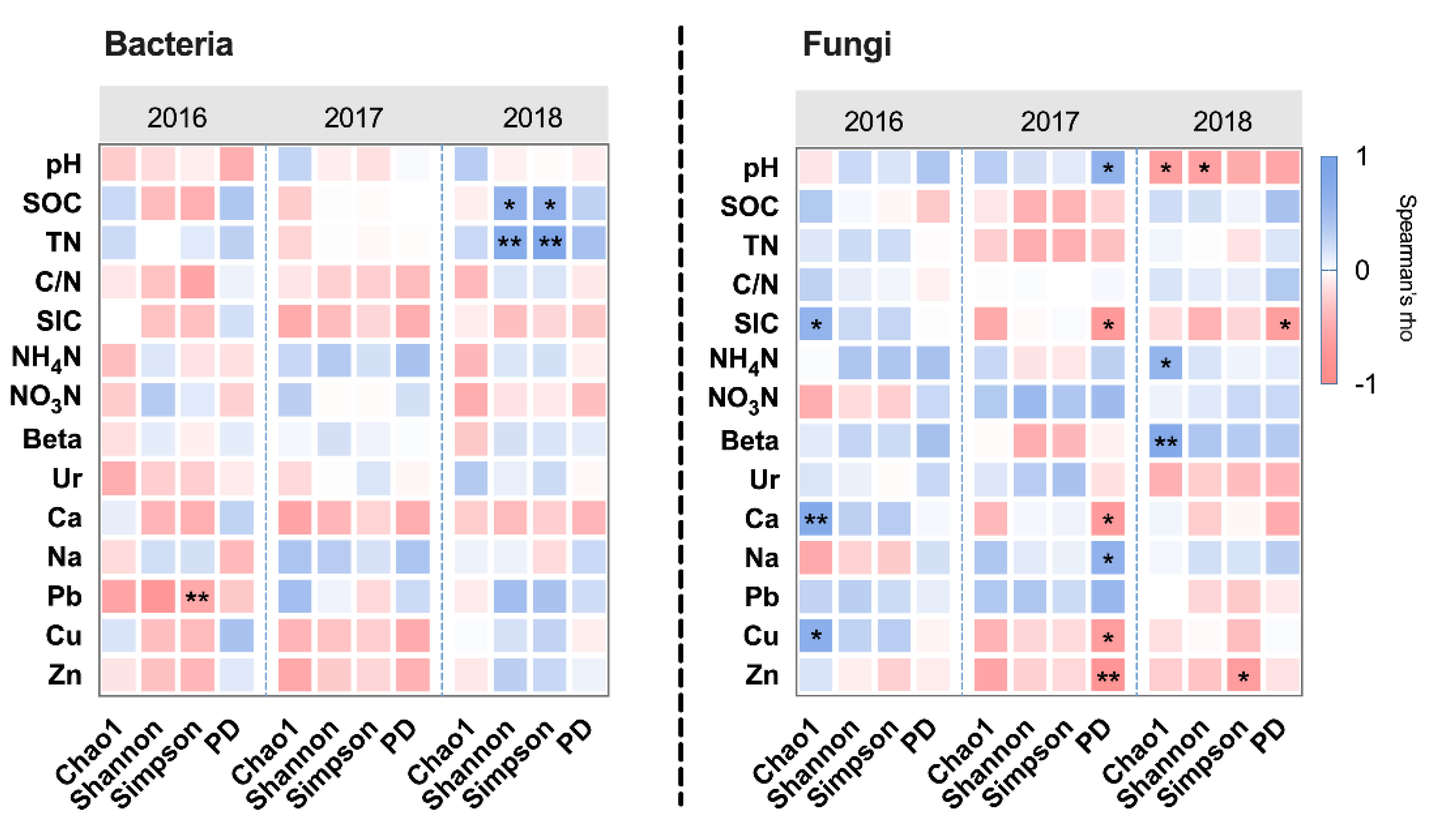


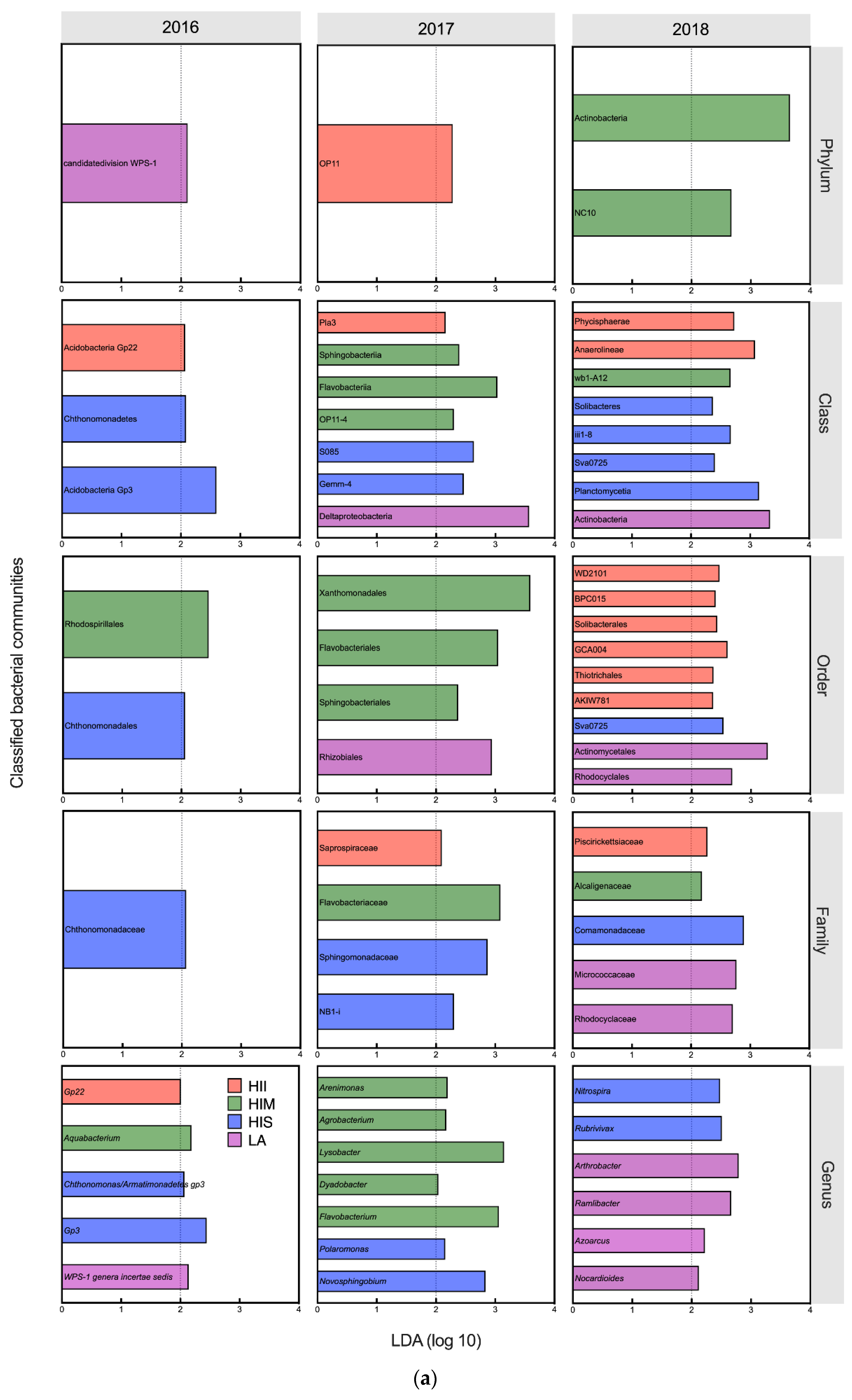



| Edaphic Properties | LA (n = 3) | HIS (n = 3) | HIM (n = 3) | HII (n = 3) | K-W Statistic (Significance) |
|---|---|---|---|---|---|
| 2016 | |||||
| pH | 8.40 ± 0.04 a | 8.42 ± 0.08 a | 8.56 ± 0.10 a | 8.58 ± 0.14 a | 4.66, ns |
| Ca (g kg−1) | 50.45 ± 3.40 a | 49.25 ± 2.64 a | 43.25 ± 1.69 b | 43.58 ± 0.91 b | 8.33, * |
| Na (g kg−1) | 12.58 ± 0.46 b | 13.18 ± 0.37 ab | 14.07 ± 0.08 a | 14.35 ± 0.42 a | 9.26, ** |
| SIC (g kg−1) | 13.03 ± 1.23 a | 12.78 ± 0.61 a | 11.41 ± 0.52 b | 11.69 ± 0.52 ab | 8.69, ** |
| SOC (g kg−1) | 6.68 ± 0.91 a | 6.47 ± 0.77 a | 6.27 ± 0.39 a | 5.46 ± 0.52 a | 4.44, ns |
| TN (g kg−1) | 1.08 ± 0.13 a | 0.71 ± 0.19 a | 0.84 ± 0.06 a | 0.86 ± 0.02 a | 6.15, ns |
| C/N | 6.20 ± 0.18 b | 9.40 ± 1.95 a | 7.45 ± 0.45 ab | 6.35 ± 0.52 b | 8.95, ** |
| NH4N (mg kg−1) | 0.93 ± 0.35 a | 0.47 ± 0.41 a | 1.68 ± 0.85 a | 0.90 ± 0.14 a | 4.51, ns |
| NO3N (mg kg−1) | 2.37 ± 0.18 b | 6.67 ± 4.07 ab | 8.91 ± 1.46 ab | 13.65 ± 2.85 a | 8.74, ** |
| Beta (PNP mg g−1) | 33.99 ± 2.94 a | 33.04 ± 11.40 a | 38.01 ± 14.08 a | 33.59 ± 14.23 a | 0.54, ns |
| Ur (NH4N mg g−1) | 0.43 ± 0.06 a | 0.47 ± 0.05 a | 0.48 ± 0.21 a | 0.42 ± 0.05 a | 1.77, ns |
| Pb (mg kg−1) | 19.03 ± 3.84 a | 25.56 ± 8.36 a | 17.07 ± 0.53 a | 16.91 ± 1.02 a | 0.90, ns |
| Cu (mg kg−1) | 42.16 ± 2.77 a | 32.33 ± 0.54 ab | 30.36 ± 0.77 b | 28.63 ± 1.77 b | 9.67, *** |
| Zn (mg kg−1) | 87.37 ± 7.95 a | 82.7 ± 8.24 a | 81.1 ± 1.64 a | 82.35 ± 2.73 a | 1.56, ns |
| 2017 | |||||
| pH | 8.98 ± 0.01 b | 9.11 ± 0.01 ab | 9.13 ± 0.11 ab | 9.29 ± 0.05 a | 9.23, ** |
| Ca (g kg−1) | 48.39 ± 6.83 ab | 56.73 ± 3.05 a | 38.45 ± 0.38 b | 38.00 ± 0.91 b | 9.05, ** |
| Na (g kg−1) | 11.03 ± 0.20 ab | 10.71 ± 0.76 a | 13.55 ± 0.45 b | 13.58 ± 0.88 b | 8.44, * |
| SIC (g kg−1) | 13.71 ± 0.51 ab | 14.92 ± 0.98 a | 11.87 ± 0.12 b | 12.17 ± 0.62 b | 9.05, ** |
| SOC (g kg−1) | 6.27 ± 0.38 a | 5.56 ± 0.10 ab | 6.33 ± 0.64 a | 4.96 ± 0.57 b | 8.64, ** |
| TN (g kg−1) | 0.98 ± 0.06 a | 0.73 ± 0.04 b | 0.84 ± 0.08 ab | 0.69 ± 0.04 b | 9.46, ** |
| C/N | 6.39 ± 0.41 b | 7.66 ± 0.26 a | 7.56 ± 0.15 a | 7.13 ± 0.42 ab | 7.82, * |
| NH4N (mg kg−1) | 4.50 ± 1.09 a | 0.82 ± 0.73 b | 6.08 ± 2.06 a | 3.44 ± 0.56 ab | 8.44, * |
| NO3N (mg kg−1) | 2.51 ± 0.83 b | 9.09 ± 5.64 a | 8.51 ± 2.55 ab | 14.96 ± 1.84 a | 9.46, ** |
| Beta (PNP mg g−1) | 37.74 ± 17.18 a | 26.60 ± 12.03 a | 51.07 ± 16.87 a | 21.93 ± 5.58 a | 6.08, ns |
| Ur (NH4N mg g−1) | 0.33 ± 0.05 a | 0.46 ± 0.11 a | 0.37 ± 0.06 a | 0.32 ± 0.05 a | 5.97, ns |
| Pb (mg kg−1) | 19.09 ± 2.25 a | 19.29 ± 1.54 a | 20.76 ± 3.59 a | 21.25 ± 2.30 a | 2.48, ns |
| Cu (mg kg−1) | 32.00 ± 0.48 ab | 35.13 ± 0.80 a | 29.37 ± 0.92 b | 28.51 ± 1.99 b | 9.05, ** |
| Zn (mg kg−1) | 89.1 ± 1.71 ab | 94.37 ± 1.49 a | 83.16 ± 2.06 b | 80.83 ± 4.73 b | 9.46, ** |
| 2018 | |||||
| pH | 8.97 ± 0.04 a | 9.06 ± 0.11 a | 8.77 ± 0.10 a | 8.91 ± 0.12 a | 6.73, ns |
| Ca (g kg−1) | 45.40 ± 1.55 ab | 48.25 ± 2.19 a | 36.57 ± 0.60 b | 34.55 ± 4.26 b | 9.51, ** |
| Na (g kg−1) | 11.52 ± 0.18 b | 11.82 ± 0.54 ab | 12.56 ± 0.21 a | 12.81 ± 0.96 a | 7.62, * |
| SIC (g kg−1) | 12.29 ± 0.22 ab | 12.69 ± 0.38 a | 10.67 ± 0.74 c | 11.18 ± 0.78 bc | 8.95, ** |
| SOC (g kg−1) | 4.69 ± 1.79 a | 4.66 ± 1.05 a | 6.50 ± 2.29 a | 5.34 ± 1.49 a | 1.36, ns |
| TN (g kg−1) | 0.80 ± 0.06 a | 0.65 ± 0.10 a | 0.80 ± 0.21 a | 0.68 ± 0.15 a | 2.74, ns |
| C/N | 5.84 ± 2.13 a | 7.13 ± 0.59 a | 7.98 ± 0.83 a | 7.76 ± 0.69 a | 4.33, ns |
| NH4N (mg kg−1) | 3.29 ± 0.80 ab | 2.56 ± 0.38 b | 5.39 ± 1.75 a | 1.84 ± 1.10 b | 7.40, * |
| NO3N (mg kg−1) | 4.06 ± 1.74 a | 15.61 ± 4.09 a | 12.02 ± 4.16 a | 15.45 ± 4.11 a | 6.90, ns |
| Beta (PNP mg g−1) | 40.45 ± 5.18 ab | 25.12 ± 0.62 b | 59.23 ± 25.91 a | 24.37 ± 0.90 b | 9.05, ** |
| Ur (NH4N mg g−1) | 0.43 ± 0.07 a | 0.50 ± 0.20 a | 0.36 ± 0.07 a | 0.34 ± 0.13 a | 2.69, ns |
| Pb (mg kg−1) | 8.36 ± 2.01 a | 10.64 ± 2.75 a | 17.15 ± 8.17 a | 13.36 ± 4.70 a | 5.36, ns |
| Cu (mg kg−1) | 29.20 ± 0.82 a | 28.48 ± 2.97 a | 26.88 ± 0.88 a | 25.87 ± 4.81 a | 2.59, ns |
| Zn (mg kg−1) | 78.57 ± 5.48 a | 79.98 ± 7.17 a | 79.25 ± 3.69 a | 79.08 ± 13.10 a | 0.54, ns |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, G.; He, T.; Zhang, M.; Wu, C. Could Land Abandonment with Human Intervention Benefit Cropland Restoration? From the Perspective of Soil Microbiota. Land 2021, 10, 1049. https://doi.org/10.3390/land10101049
Li G, He T, Zhang M, Wu C. Could Land Abandonment with Human Intervention Benefit Cropland Restoration? From the Perspective of Soil Microbiota. Land. 2021; 10(10):1049. https://doi.org/10.3390/land10101049
Chicago/Turabian StyleLi, Guangyu, Tingting He, Maoxin Zhang, and Cifang Wu. 2021. "Could Land Abandonment with Human Intervention Benefit Cropland Restoration? From the Perspective of Soil Microbiota" Land 10, no. 10: 1049. https://doi.org/10.3390/land10101049
APA StyleLi, G., He, T., Zhang, M., & Wu, C. (2021). Could Land Abandonment with Human Intervention Benefit Cropland Restoration? From the Perspective of Soil Microbiota. Land, 10(10), 1049. https://doi.org/10.3390/land10101049







