The Exudation of Surplus Products Links Plant Functional Traits and Plant-Microbial Stoichiometry
Abstract
:1. Introduction
2. Materials and Methods
2.1. Exudates
2.2. Plant Properties and Functional Traits (PFTs)
2.3. Physico-Chemical Properties of Soil and Soil Solution
2.4. Soil Microbial Biomass and Soil Activities
2.5. Statistical Analyses
3. Results
3.1. C, N, and P Content and Stoichiometry in All Pools
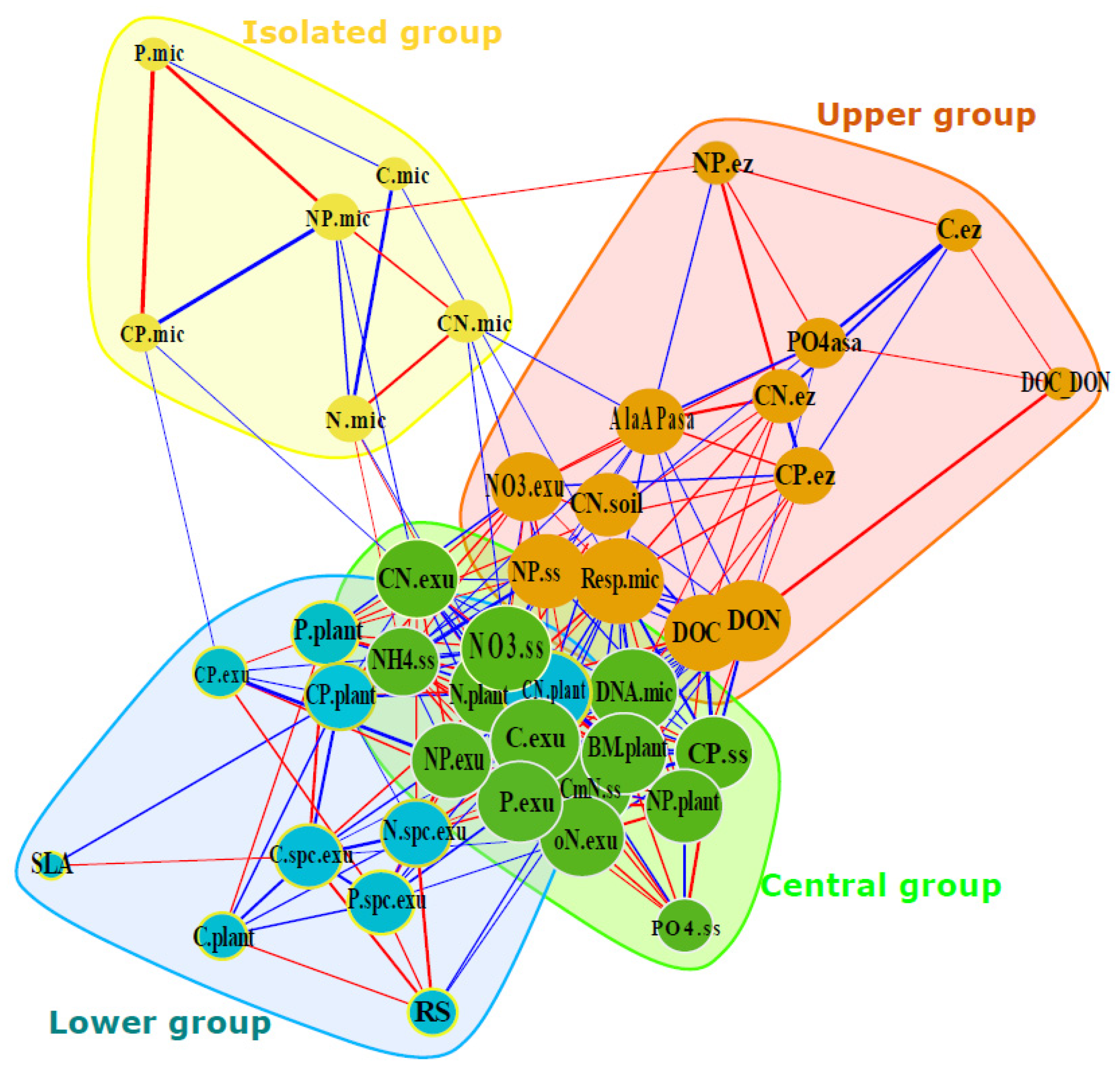
3.2. Variable Clustering and Correlation
3.2.1. Central Group: Close Link to Plant Biomass
3.2.2. Upper Group: DOC, Microbial Respiration, and Enzymatic Activity
3.2.3. Lower Group: Plant and Exudates Stoichiometry
3.2.4. Isolated Group: Microbial Biomass and Stoichiometry
4. Discussion
4.1. Larger Plants Optimised Their Utilisation of N, the Most Limiting Element
4.2. Root Exudation Served to Get Rid of Surplus Elements
4.3. Fast Growth Implied an Adjustment of PFT to Depletion of Mineral N
4.4. Plant Microbe Links Were Mediated through the Competition for N and Exudation Stoichiometry
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- De Graaff, M.A.; Classen, A.T.; Castro, H.F.; Schadt, C.W. Labile soil carbon inputs mediate the soil microbial community composition and plant residue decomposition rates. New Phytol. 2010, 188, 1055–1064. [Google Scholar] [CrossRef] [Green Version]
- Orwin, K.H.; Buckland, S.M.; Johnson, D.; Turner, B.L.; Smart, S.; Oakley, S.; Bardgett, R.D. Linkages of plant traits to soil properties and the functioning of temperate grassland. J. Ecol. 2010, 98, 1074–1083. [Google Scholar] [CrossRef] [Green Version]
- Bai, Z.; Liang, C.; Bodé, S.; Huygens, D.; Boeckx, P. Phospholipid 13C stable isotopic probing during decomposition of wheat residues. Appl. Soil Ecol. 2016, 98, 65–74. [Google Scholar] [CrossRef]
- Cheng, W.; Zhang, Q.; Coleman, D.C.; Carroll, C.R.; Hoffman, C.A. Is available carbon limiting microbial respiration in the rhizosphere? Soil Biol. Biochem. 1996, 28, 1283–1288. [Google Scholar] [CrossRef]
- Blagodatskaya, E.; Blagodatsky, S.; Anderson, T.H.; Kuzyakov, Y. Microbial growth and carbon use efficiency in the rhizosphere and root-free soil. PLoS ONE 2014, 9, e93282. [Google Scholar] [CrossRef]
- Kuzyakov, Y. Review: Factors affecting rhizosphere priming effects. J. Plant Nutr. Soil Sci. 2002, 165, 382–396. [Google Scholar] [CrossRef]
- Hunter, P. Plant microbiomes and sustainable agriculture: Deciphering the plant microbiome and its role in nutrient supply and plant immunity has great potential to reduce the use of fertilizers and biocides in agriculture. EMBO Rep. 2016, 17, 1696–1699. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, T.; Ke, M.; Lavoie, M.; Jin, Y.; Fan, X.; Zhang, Z.; Fu, Z.; Sun, L.; Gillings, M.; Peñuelas, J.; et al. Rhizosphere microorganisms can influence the timing of plant flowering. Microbiome 2018, 6, 231. [Google Scholar] [CrossRef] [PubMed]
- Senthil Kumar, C.M.; Jacob, T.K.; Devasahayam, S.; Thomas, S.; Geethu, C. Multifarious plant growth promotion by an entomopathogenic fungus Lecanicillium psalliotae. Microbiol. Res. 2018, 207, 153–160. [Google Scholar] [CrossRef]
- Čapek, P.; Manzoni, S.; Kaštovská, E.; Wild, B.; Diáková, K.; Bárta, J.; Schnecker, J.; Biasi, C.; Martikainen, P.J.; Alves, R.J.E.; et al. A plant–microbe interaction framework explaining nutrient effects on primary production. Nat. Ecol. Evol. 2018, 2, 1588–1596. [Google Scholar] [CrossRef] [Green Version]
- Manzoni, S. Flexible carbon-use efficiency across litter types and during decomposition partly compensates nutrient imbalances-results from analytical stoichiometric models. Front. Microbiol. 2017, 8, 661. [Google Scholar] [CrossRef] [Green Version]
- Manzoni, S.; Trofymow, J.A.; Jackson, R.B.; Porporato, A. Stoichiometric controls on carbon, nitrogen, and phosphorus dynamics in decomposing litter. Ecol. Monogr. 2010, 80, 89–106. [Google Scholar] [CrossRef]
- Manzoni, S.; Taylor, P.; Richter, A.; Porporato, A.; Ågren, G.I. Environmental and stoichiometric controls on microbial carbon-use efficiency in soils. New Phytol. 2012, 196, 79–91. [Google Scholar] [CrossRef] [PubMed]
- Zechmeister-Boltenstern, S.; Keiblinger, K.M.; Mooshammer, M.; Peñuelas, J.; Richter, A.; Sardans, J.; Wanek, W. The application of ecological stoichiometry to plant-microbial-soil organic matter transformations. Ecol. Monogr. 2015, 85, 133–155. [Google Scholar] [CrossRef] [Green Version]
- Henneron, L.; Kardol, P.; Wardle, D.A.; Cros, C.; Fontaine, S. Rhizosphere control of soil nitrogen cycling: A key component of plant economic strategies. New Phytol. 2020, 228, 1269–1282. [Google Scholar] [CrossRef]
- Henneron, L.; Cros, C.; Picon-Cochard, C.; Rahimian, V.; Fontaine, S. Plant economic strategies of grassland species control soil carbon dynamics through rhizodeposition. J. Ecol. 2020, 108, 528–545. [Google Scholar] [CrossRef]
- Craine, J.M.; Morrow, C.; Fierer, N. Microbial nitrogen limitation increases decomposition. Ecology 2007, 88, 2105–2113. [Google Scholar] [CrossRef]
- Guyonnet, J.P.; Cantarel, A.A.M.; Simon, L.; Haichar, F.Z. Root exudation rate as functional trait involved in plant nutrient-use strategy classification. Ecol. Evol. 2018, 8, 8573–8581. [Google Scholar] [CrossRef] [Green Version]
- Ordoñez, J.C.; Van Bodegom, P.M.; Witte, J.P.M.; Wright, I.J.; Reich, P.B.; Aerts, R. A global study of relationships between leaf traits, climate and soil measures of nutrient fertility. Glob. Ecol. Biogeogr. 2009, 18, 137–149. [Google Scholar] [CrossRef]
- Carrillo, Y.; Bell, C.; Koyama, A.; Canarini, A.; Boot, C.M.; Wallenstein, M.; Pendall, E. Plant traits, stoichiometry and microbes as drivers of decomposition in the rhizosphere in a temperate grassland. J. Ecol. 2017, 105, 1750–1765. [Google Scholar] [CrossRef] [Green Version]
- Freschet, G.T.; Cornelissen, J.H.C.; van Logtestijn, R.S.P.; Aerts, R. Substantial nutrient resorption from leaves, stems and roots in a subarctic flora: What is the link with other resource economics traits? New Phytol. 2010, 186, 879–889. [Google Scholar] [CrossRef]
- de Vries, F.T.; Bardgett, R.D. Plant community controls on short-term ecosystem nitrogen retention. New Phytol. 2016, 210, 861–874. [Google Scholar] [CrossRef] [Green Version]
- De Deyn, G.B.; Cornelissen, J.H.C.; Bardgett, R.D. Plant functional traits and soil carbon sequestration in contrasting biomes. Ecol. Lett. 2008, 11, 516–531. [Google Scholar] [CrossRef]
- Meunier, C.L.; Boersma, M.; El-Sabaawi, R.; Halvorson, H.M.; Herstoff, E.M.; Van de Waal, D.B.; Vogt, R.J.; Litchman, E. From elements to function: Toward unifying ecological stoichiometry and trait-based ecology. Front. Environ. Sci. 2017, 5, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Edwards, K.R.; Kaštovská, E.; Borovec, J.; Šantrůčková, H.; Picek, T. Species effects and seasonal trends on plant efflux quantity and quality in a spruce swamp forest. Plant Soil 2018, 426, 179–196. [Google Scholar] [CrossRef]
- Canarini, A.; Kaiser, C.; Merchant, A.; Richter, A.; Wanek, W. Root exudation of primary metabolites: Mechanisms and their roles in plant responses to environmental stimuli. Front. Plant Sci. 2019, 10, 157. [Google Scholar] [CrossRef] [Green Version]
- Prescott, C.E.; Grayston, S.J.; Helmisaari, H.S.; Kaštovská, E.; Körner, C.; Lambers, H.; Meier, I.C.; Millard, P.; Ostonen, I. Surplus Carbon Drives Allocation and Plant–Soil Interactions. Trends Ecol. Evol. 2020, 35, 1110–1118. [Google Scholar] [CrossRef]
- Baptist, F.; Aranjuelo, I.; Legay, N.; Lopez-Sangil, L.; Molero, G.; Rovira, P.; Nogués, S. Rhizodeposition of organic carbon by plants with contrasting traits for resource acquisition: Responses to different fertility regimes. Plant Soil 2015, 394, 391–406. [Google Scholar] [CrossRef] [Green Version]
- Kaštovská, E.; Edwards, K.; Šantrůčková, H. Rhizodeposition flux of competitive versus conservative graminoid: Contribution of exudates and root lysates as affected by N loading. Plant Soil 2017, 412, 331–344. [Google Scholar] [CrossRef]
- Bengtson, P.; Barker, J.; Grayston, S.J. Evidence of a strong coupling between root exudation, C and N availability, and stimulated SOM decomposition caused by rhizosphere priming effects. Ecol. Evol. 2012, 2, 1843–1852. [Google Scholar] [CrossRef] [PubMed]
- Drake, J.E.; Darby, B.A.; Giasson, M.A.; Kramer, M.A.; Phillips, R.P.; Finzi, A.C. Stoichiometry constrains microbial response to root exudation-insights from a model and a field experiment in a temperate forest. Biogeosciences 2013, 10, 821–838. [Google Scholar] [CrossRef] [Green Version]
- Oburger, E.; Jones, D.L. Sampling root exudates—Mission impossible? Rhizosphere 2018, 6, 116–133. [Google Scholar] [CrossRef]
- Gougherty, S.W.; Bauer, J.E.; Pohlman, J.W. Exudation rates and δ13C signatures of tree root soluble organic carbon in a riparian forest. Biogeochemistry 2018, 137, 235–252. [Google Scholar] [CrossRef]
- Kolaczyk, E.D.; Csardi, G. Statistical Analysis of Network Data with R Introduction; Springer: Berlin, Germany, 2014; Volume 65. [Google Scholar]
- Miles, J.; Grime, J.P.; Hodgson, J.G.; Hunt, R.; Davy, A.J. Comparative Plant Ecology. A Functional Approach to Common British Species; Springer: Berlin, Germany, 1988. [Google Scholar] [CrossRef]
- Reischke, S.; Rousk, J.; Bååth, E. The effects of glucose loading rates on bacterial and fungal growth insoil. Soil Biol. Biochem. 2014, 70, 88–95. [Google Scholar] [CrossRef]
- Reischke, S.; Kumar, M.G.K.; Bååth, E. Threshold concentration of glucose for bacterial growth in soil. Soil Biol. Biochem. 2015, 80, 218–223. [Google Scholar] [CrossRef]
- Knee, E.M.; Gong, F.C.; Gao, M.; Teplitski, M.; Jones, A.R.; Foxworthy, A.; Mort, A.J.; Bauer, W.D. Root mucilage from pea and its utilization by rhizosphere bacteria as a sole carbon source. Mol. Plant-Microbe Interact. 2001, 14, 775–784. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaboud, A.; Rougier, M. Identification and Localization of Sugar Components of Rice (Oryza sativa L.) Root Cap Mucilage. J. Plant Physiol. 1984, 116, 323–330. [Google Scholar] [CrossRef]
- Bacic, A.; Moody, S.F.; Clarke, A.E. Structural Analysis of Secreted Root Slime from Maize ( Zea mays L.). Plant Physiol. 1986, 80, 771–777. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moody, S.F.; Clarke, A.E.; Bacic, A. Structural analysis of secreted slime from wheat and cowpea roots. Phytochemistry 1988, 27, 2857–2861. [Google Scholar] [CrossRef]
- Song, T.; Zhang, W.; Wei, C.; Jiang, T.; Xu, H.; Cao, Y.; Cao, Y.; Qiao, D. Isolation and characterization of agar-degrading endophytic bacteria from plants. Curr. Microbiol. 2015, 70, 275–281. [Google Scholar] [CrossRef]
- Song, T.; Cao, Y.; Xu, H.; Zhang, W.; Fei, B.; Qiao, D.; Cao, Y. Purification and characterization of a novel β-agarase of Paenibacillus sp. SSG-1 isolated from soil. J. Biosci. Bioeng. 2014, 118, 125–129. [Google Scholar] [CrossRef]
- Hosoda, A.; Sakai, M.; Kanazawa, S. Isolation and Characterization of Agar-degrading Paenibacillus spp. Associated with the Rhizosphere of Spinach. Biosci. Biotechnol. Biochem. 2003, 67, 1048–1055. [Google Scholar] [CrossRef] [Green Version]
- Sakai, M.; Hosoda, A.; Ogura, K.; Ikenaga, M. The growth of steroidobacter agariperforans sp. nov., a novel agar-degrading bacterium isolated from soil, is enhanced by the diffusible metabolites produced by bacteria belonging to rhizobiales. Microbes Environ. 2014, 29, 89–95. [Google Scholar] [CrossRef] [Green Version]
- Canarini, A.; Merchant, A.; Dijkstra, F.A. Drought effects on Helianthus annuus and Glycine max metabolites: From phloem to root exudates. Rhizosphere 2016, 2, 85–97. [Google Scholar] [CrossRef]
- Kopáček, J.; Borovec, J.; Hejzlar, J.; Porcal, P. Spectrophotometric determination of iron, aluminum, and phosphorus in soil and sediment extracts after their nitric and perchloric acid digestion. Commun. Soil Sci. Plant Anal. 2001, 32, 1431–1443. [Google Scholar] [CrossRef]
- Brookes, P.C.; Landman, A.; Pruden, G.; Jenkinson, D.S. Chloroform fumigation and the release of soil nitrogen: A rapid direct extraction method to measure microbial biomass nitrogen in soil. Soil Biol. Biochem. 1985, 17, 837–842. [Google Scholar] [CrossRef]
- Vance, E.D.; Brookes, P.C.; Jenkinson, D.S. An extraction method for measuring soil microbial biomass C. Soil Biol. Biochem. 1987, 19, 703–707. [Google Scholar] [CrossRef]
- Brookes, P.C.; Powlson, D.S.; Jenkinson, D.S. Measurement of microbial biomass phosphorus in soil. Soil Biol. Biochem. 1982, 14, 319–329. [Google Scholar] [CrossRef]
- Sparling, G.P.; Shepherd, T.G.; Schipper, L.A. Topsoil characteristics of three contrasting New Zealand soils under four long-term land uses. N. Z. J. Agric. Res. 2000, 43, 569–583. [Google Scholar] [CrossRef]
- Chen, X.; Daniell, T.J.; Neilson, R.; O’Flaherty, V.; Griffiths, B.S. Microbial and microfaunal communities in phosphorus limited, grazed grassland change composition but maintain homeostatic nutrient stoichiometry. Soil Biol. Biochem. 2014, 75, 94–101. [Google Scholar] [CrossRef]
- Sinsabaugh, R.L.; Hill, B.H.; Follstad Shah, J.J. Ecoenzymatic stoichiometry of microbial organic nutrient acquisition in soil and sediment. Nature 2009, 462, 795–798. [Google Scholar] [CrossRef]
- Marx, M.C.; Wood, M.; Jarvis, S.C. A microplate fluorimetric assay for the study of enzyme diversity in soils. Soil Biol. Biochem. 2001, 33, 1633–1640. [Google Scholar] [CrossRef]
- Bárta, J.; Šlajsová, P.; Tahovská, K.; Picek, T.; Šantrůčková, H. Different temperature sensitivity and kinetics of soil enzymes indicate seasonal shifts in C, N and P nutrient stoichiometry in acid forest soil. Biogeochemistry 2014, 117, 525–537. [Google Scholar] [CrossRef]
- Burns, R.G.; DeForest, J.L.; Marxsen, J.; Sinsabaugh, R.L.; Stromberger, M.E.; Wallenstein, M.D.; Weintraub, M.N.; Zoppini, A. Soil enzymes in a changing environment: Current knowledge and future directions. Soil Biol. Biochem. 2013, 58, 216–234. [Google Scholar] [CrossRef]
- Chavent, M.; Kuentz-Simonet, V.; Liquet, B.; Saracco, J. ClustOfVar: An R package for the clustering of variables. J. Stat. Softw. 2012, 50. [Google Scholar] [CrossRef] [Green Version]
- Box, G.E.P.; Cox, D.R. An Analysis of Transformations. J. R. Stat. Soc. Ser. B 1964, 26, 211–243. [Google Scholar] [CrossRef]
- Brandes, U.; Delling, D.; Gaertler, M.; Görke, R.; Hoefer, M.; Nikoloski, Z.; Wagner, D. On modularity clustering. IEEE Trans. Knowl. Data Eng. 2008, 20, 172–188. [Google Scholar] [CrossRef] [Green Version]
- Biernath, C.; Fischer, H.; Kuzyakov, Y. Root uptake of N-containing and N-free low molecular weight organic substances by maize: A 14C/15N tracer study. Soil Biol. Biochem. 2008, 40, 2237–2245. [Google Scholar] [CrossRef]
- Harrison, K.A.; Bol, R.; Bardgett, R.D. Preferences for different nitrogen forms by coexisting plant species and soil microbes. Ecology 2007, 88, 989–999. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, J.; Sauheitl, L.; Eriksen, J.; Kuzyakov, Y. Plant uptake of dual-labeled organic N biased by inorganic C uptake: Results of a triple labeling study. Soil Biol. Biochem. 2010, 42, 524–527. [Google Scholar] [CrossRef]
- Xu, X.; Stange, C.F.; Richter, A.; Wanek, W.; Kuzyakov, Y. Light affects competition for inorganic and organic nitrogen between maize and rhizosphere microorganisms. Plant Soil 2008, 304, 59–72. [Google Scholar] [CrossRef]
- Tinker, P.B.; Nye, P. Solute Movement in the Rhizosphere; Oxford University Press (OUP): Oxford, UK, 2000. [Google Scholar]
- Bélanger, G.; Gastal, F.; Warembourg, F.R. Carbon Balance of Tall Fescue (Festuca arundinacea Schreb.): Effects of Nitrogen Fertilization and the Growing Season. Ann. Bot. 1994, 74, 653–659. [Google Scholar] [CrossRef]
- Brouwer, R. Distribution of dry matter in the plant. Neth. J. Agric. Sci. 1962, 10, 361–376. [Google Scholar] [CrossRef]
- Jílková, V.; Sim, A.; Thornton, B.; Jandová, K.; Cajthaml, T.; Paterson, E. Impact of plant species and atmospheric CO2 concentration on rhizodeposition and soil microbial activity and community composition. J. Plant Nutr. Soil Sci. 2020, 183, 327–337. [Google Scholar] [CrossRef]
- Kaštovská, E.; Edwards, K.; Picek, T.; Šantrůčková, H. A larger investment into exudation by competitive versus conservative plants is connected to more coupled plant–microbe N cycling. Biogeochemistry 2015, 122, 47–59. [Google Scholar] [CrossRef]
- Wang, Q.; Zhu, Y.; Zou, X.; Li, F.; Zhang, J.; Kang, Z.; Li, X.; Yin, C.; Lin, Y. Nitrogen Deficiency-Induced Decrease in Cytokinins Content Promotes Rice Seminal Root Growth by Promoting Root Meristem Cell Proliferation and Cell Elongation. Cells 2020, 9, 916. [Google Scholar] [CrossRef] [Green Version]
- Foito, A.; Byrne, S.L.; Hackett, C.A.; Hancock, R.D.; Stewart, D.; Barth, S. Short-term response in leaf metabolism of perennial ryegrass (Lolium perenne) to alterations in nitrogen supply. Metabolomics 2013, 9, 145–156. [Google Scholar] [CrossRef]
- Liang, Z.; Bao, A.; Li, H.; Cai, H. The effect of nitrogen level on rice growth, carbon-nitrogen metabolism and gene expression. Biologia 2015, 70, 1340–1350. [Google Scholar] [CrossRef]
- Cramer, M.D.; Lewis, O.A.M. The influence of NO3- and NH4+ nutrition on the carbon and nitrogen partitioning characteristics of wheat (Triticum aestivum L.) and maize (Zea mays L.) plants. Plant Soil 1993, 154, 289–300. [Google Scholar] [CrossRef]
- Schjoerring, J.K.; Husted, S.; Mäck, G.; Mattsson, M. The regulation of ammonium translocation in plants. J. Exp. Bot. 2002, 53, 883–890. [Google Scholar] [CrossRef] [Green Version]
- Wright, I.J.; Reich, P.B.; Westoby, M.; Ackerly, D.D.; Baruch, Z.; Bongers, F.; Cavender-Bares, J.; Chapin, T.; Cornellssen, J.H.C.; Diemer, M.; et al. The worldwide leaf economics spectrum. Nature 2004, 428, 821–827. [Google Scholar] [CrossRef]
- Withington, J.M.; Reich, P.B.; Oleksyn, J.; Eissenstat, D.M. Comparisons of structure and life span in roots and leaves among temperate trees. Ecol. Monogr. 2006, 76, 381–397. [Google Scholar] [CrossRef]
- German, D.P.; Chacon, S.S.; Allison, S.D. Substrate concentration and enzyme allocation can affect rates of microbial decomposition. Ecology 2011, 92, 1471–1480. [Google Scholar] [CrossRef] [PubMed]
- Khalid, M.; Soleman, N.; Jones, D.L. Grassland plants affect dissolved organic carbon and nitrogen dynamics in soil. Soil Biol. Biochem. 2007, 39, 378–381. [Google Scholar] [CrossRef]
- Kuzyakov, Y.; Biryukova, O.V.; Kuznetzova, T.V.; Molter, K.; Kandeler, E.; Stahr, K. Carbon partitioning in plant and soil, carbon dioxide fluxes and enzyme activities as affected by cutting ryegrass. Biol. Fertil. Soils 2002, 35, 348–358. [Google Scholar] [CrossRef]
- Tian, P.; Razavi, B.S.; Zhang, X.; Wang, Q.; Blagodatskaya, E. Microbial growth and enzyme kinetics in rhizosphere hotspots are modulated by soil organics and nutrient availability. Soil Biol. Biochem. 2020, 141, 107662. [Google Scholar] [CrossRef]
- Cleveland, C.C.; Liptzin, D. C:N:P stoichiometry in soil: Is there a “Redfield ratio” for the microbial biomass? Biogeochemistry 2007, 85, 235–252. [Google Scholar] [CrossRef]
- Fujita, K.; Miyabara, Y.; Kunito, T. Microbial biomass and ecoenzymatic stoichiometries vary in response to nutrient availability in an arable soil. Eur. J. Soil Biol. 2019, 91, 1–8. [Google Scholar] [CrossRef]
- Kirkby, C.A.; Kirkegaard, J.A.; Richardson, A.E.; Wade, L.J.; Blanchard, C.; Batten, G. Stable soil organic matter: A comparison of C:N:P:S ratios in Australian and other world soils. Geoderma 2011, 163, 197–208. [Google Scholar] [CrossRef]
- Chen, J.; Seven, J.; Zilla, T.; Dippold, M.A.; Blagodatskaya, E.; Kuzyakov, Y. Microbial C:N:P stoichiometry and turnover depend on nutrients availability in soil: A 14 C, 15 N and 33 P triple labelling study. Soil Biol. Biochem. 2019, 131, 206–216. [Google Scholar] [CrossRef]
- Bünemann, E.K.; Oberson, A.; Liebisch, F.; Keller, F.; Annaheim, K.E.; Huguenin-Elie, O.; Frossard, E. Rapid microbial phosphorus immobilization dominates gross phosphorus fluxes in a grassland soil with low inorganic phosphorus availability. Soil Biol. Biochem. 2012, 51, 84–95. [Google Scholar] [CrossRef]
- Bünemann, E.K.; Prusisz, B.; Ehlers, K. Characterization of Phosphorus Forms in Soil Microorganisms. In Phosphorus in Action; Soil Biology; Springer: Berlin/Heidelberg, Germany, 2011. [Google Scholar]
- Kuzyakov, Y.; Xu, X. Competition between roots and microorganisms for nitrogen: Mechanisms and ecological relevance. New Phytol. 2013, 198, 656–669. [Google Scholar] [CrossRef]
- Azam, F.; Ifzal, M. Microbial populations immobilizing NH4+-N and NO3--N differ in their sensitivity to sodium chloride salinity in soil. Soil Biol. Biochem. 2006, 38, 2491–2494. [Google Scholar] [CrossRef]
- Rice, C.W.; Tiedje, J.M. Regulation of nitrate assimilation by ammonium in soils and in isolated soil microorganisms. Soil Biol. Biochem. 1989, 21, 597–602. [Google Scholar] [CrossRef]
- Liu, M.; Wang, Z.; Li, S.; Lü, X.; Wang, X.; Han, X. Changes in specific leaf area of dominant plants in temperate grasslands along a 2500-km transect in northern China. Sci. Rep. 2017, 7, 10780. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhu, B.; Zhang, J.; Müller, C.; Cai, Z. Mechanisms of soil N dynamics following long-term application of organic fertilizers to subtropical rain-fed purple soil in China. Soil Biol. Biochem. 2015, 91, 222–231. [Google Scholar] [CrossRef]
- Allison, S.D.; Vitousek, P.M. Responses of extracellular enzymes to simple and complex nutrient inputs. Soil Biol. Biochem. 2005, 37, 937–944. [Google Scholar] [CrossRef]
- Ekenler, M.; Tabatabai, M.A. β-glucosaminidase activity of soils: Effect of cropping systems and its relationship to nitrogen mineralization. Biol. Fertil. Soils 2002, 36, 367–376. [Google Scholar] [CrossRef]
- Jezierska-Tys, S.; Frac, M. Impact of dairy sewage sludge on enzymatic activity and inorganic nitrogen concentrations in the soils. Int. Agrophysics 2009, 23, 31–37. [Google Scholar]
- Michel, K.; Matzner, E. Response of enzyme activities to nitrogen addition in forest floors of different C-to-N ratios. Biol. Fertil. Soils 2003, 38, 102–109. [Google Scholar] [CrossRef]



| Mean | Coefficient of Variance (%) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| POOL | Unit | C | N | P | C:N | C:P | N:P | C | N | P | C:N | C:P | N:P |
| Plant biomass | mg·g−1 | 414.6 | 19.6 | 4.0 | 24.7 | 266.5 | 10.8 | 2.1 | 31.9 | 18.3 | 30.9 | 21.9 | 29.2 |
| Root exudates | µg·plant−1·h−1 | 793.6 | 40.2 | 29.6 | 22.6 | 88.0 | 4.2 | 61.1 | 56.3 | 74.7 | 31.4 | 63.7 | 57.6 |
| Spec. Exudation | µg·g−1 root·h−1 | 1073.9 | 59.2 | 41.6 | 42.0 | 46.3 | 60.1 | ||||||
| Soil solution | µg·g−1 | 22.0 | 2.9 | 1.1 | 9.2 | 53.4 | 6.0 | 22.0 | 26.7 | 17.9 | 30.6 | 30.9 | 24.5 |
| Soil microbes | µg·g−1 | 200.4 | 29.6 | 77.9 | 7.9 | 6.6 | 0.8 | 20.6 | 28.4 | 42.6 | 20.6 | 40.5 | 51.4 |
| Enzym. activity | nM·g−1·h−1 | 48.5 | 16.1 | 29.5 | 3.0 | 1.7 | 0.6 | 18.2 | 15.5 | 16.7 | 16.7 | 12.9 | 11.8 |
| Total soil | mg·g−1 | 14.2 | 1.3 | 0.2 | 12.9 | 173.4 | 13.6 | 9.4 | 7.4 | 19.0 | 6.5 | 12.2 | 13.5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cardenas, J.; Santa, F.; Kaštovská, E. The Exudation of Surplus Products Links Plant Functional Traits and Plant-Microbial Stoichiometry. Land 2021, 10, 840. https://doi.org/10.3390/land10080840
Cardenas J, Santa F, Kaštovská E. The Exudation of Surplus Products Links Plant Functional Traits and Plant-Microbial Stoichiometry. Land. 2021; 10(8):840. https://doi.org/10.3390/land10080840
Chicago/Turabian StyleCardenas, Julian, Fernando Santa, and Eva Kaštovská. 2021. "The Exudation of Surplus Products Links Plant Functional Traits and Plant-Microbial Stoichiometry" Land 10, no. 8: 840. https://doi.org/10.3390/land10080840






