NK Cytotoxicity Mediated by NK-92 Cell Lines Expressing Combinations of Two Allelic Variants for FCGR3
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Construction of Plasmids Carrying FCGR3A Allelic Variants and NK-92 Transfectant Cell Lines
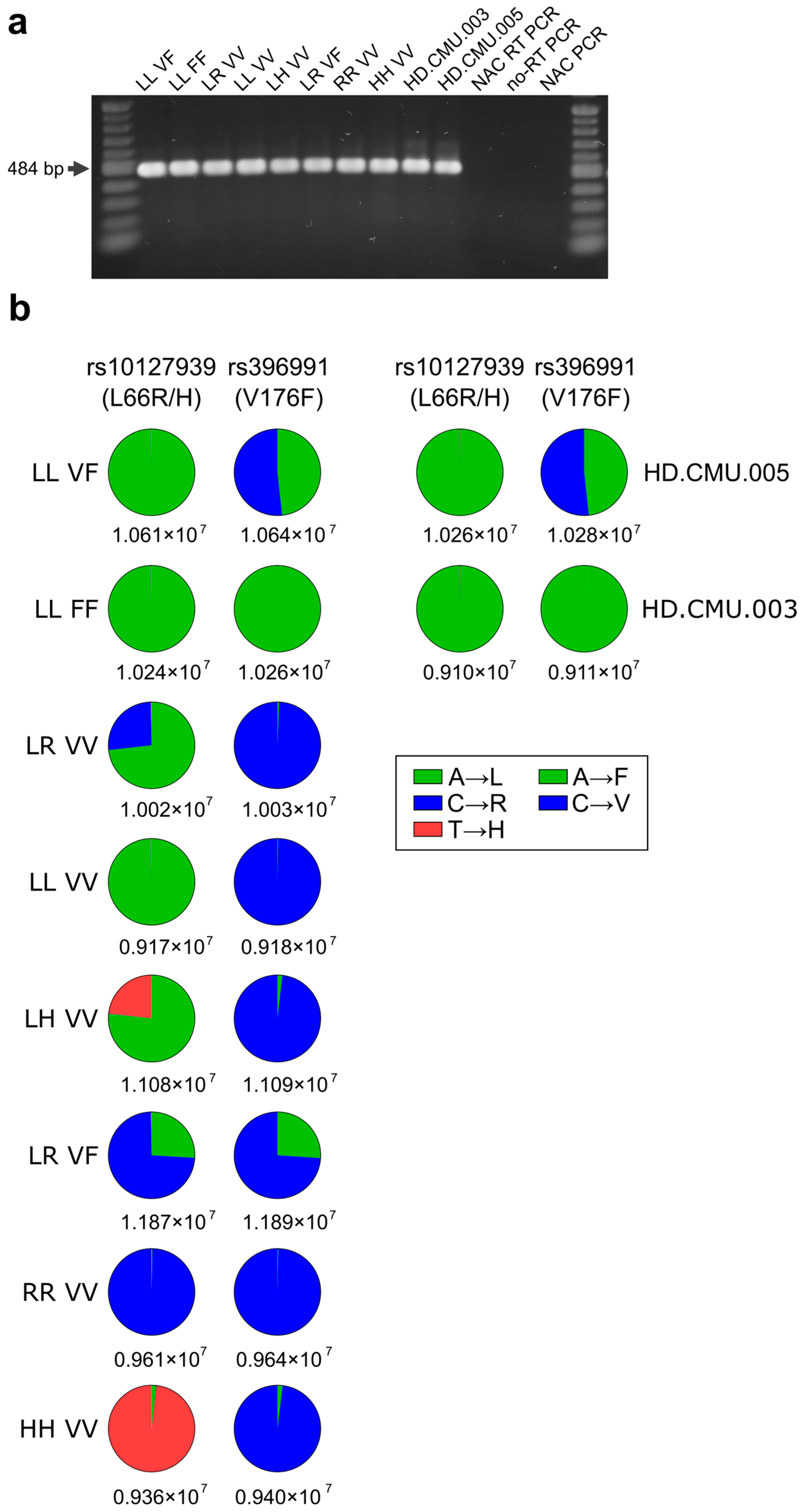
2.3. Determination of FCGR3A Variants in NK-92 Transfectants from Messenger RNA Followed by New-Generation Sequencing (NGS) Quantification
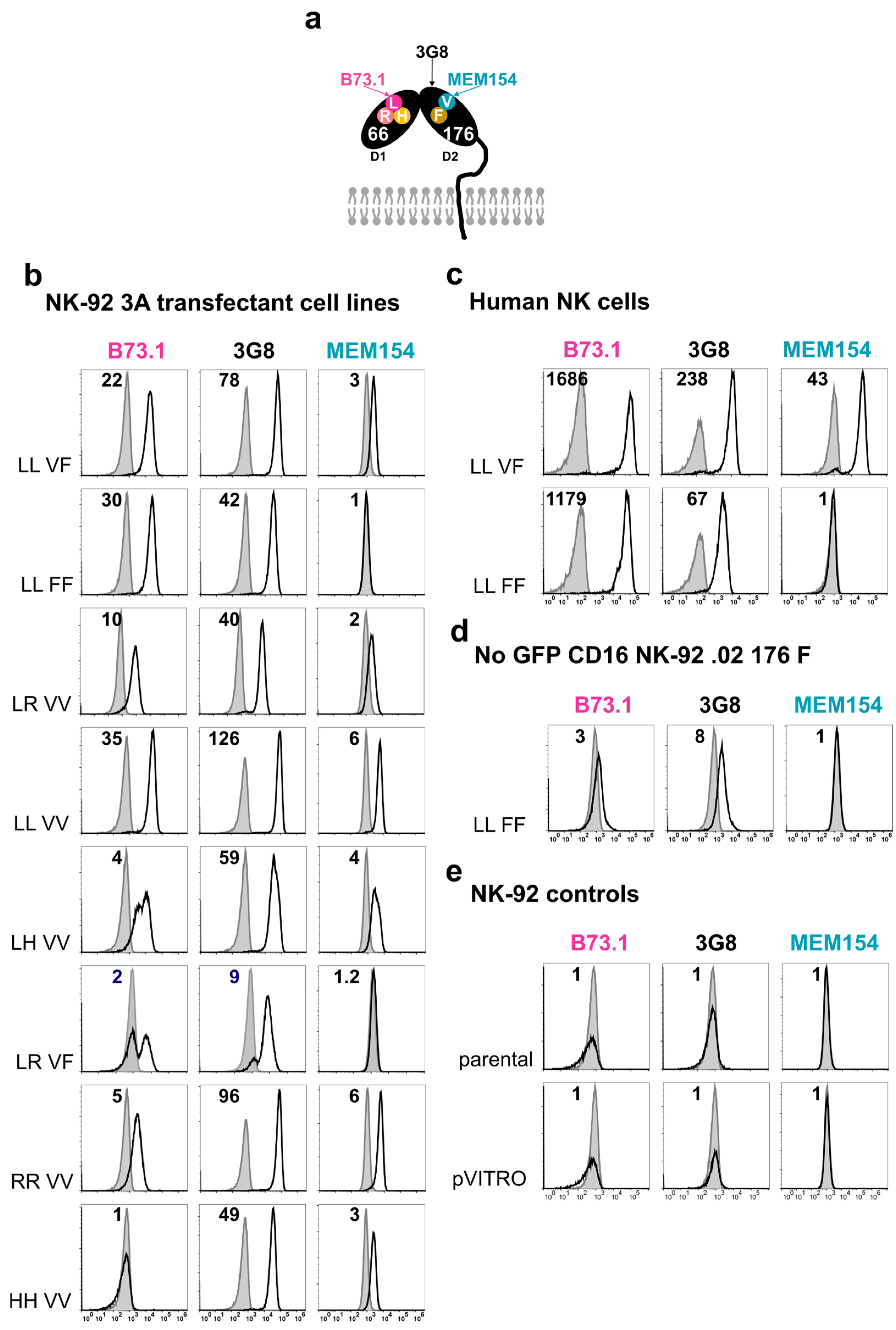
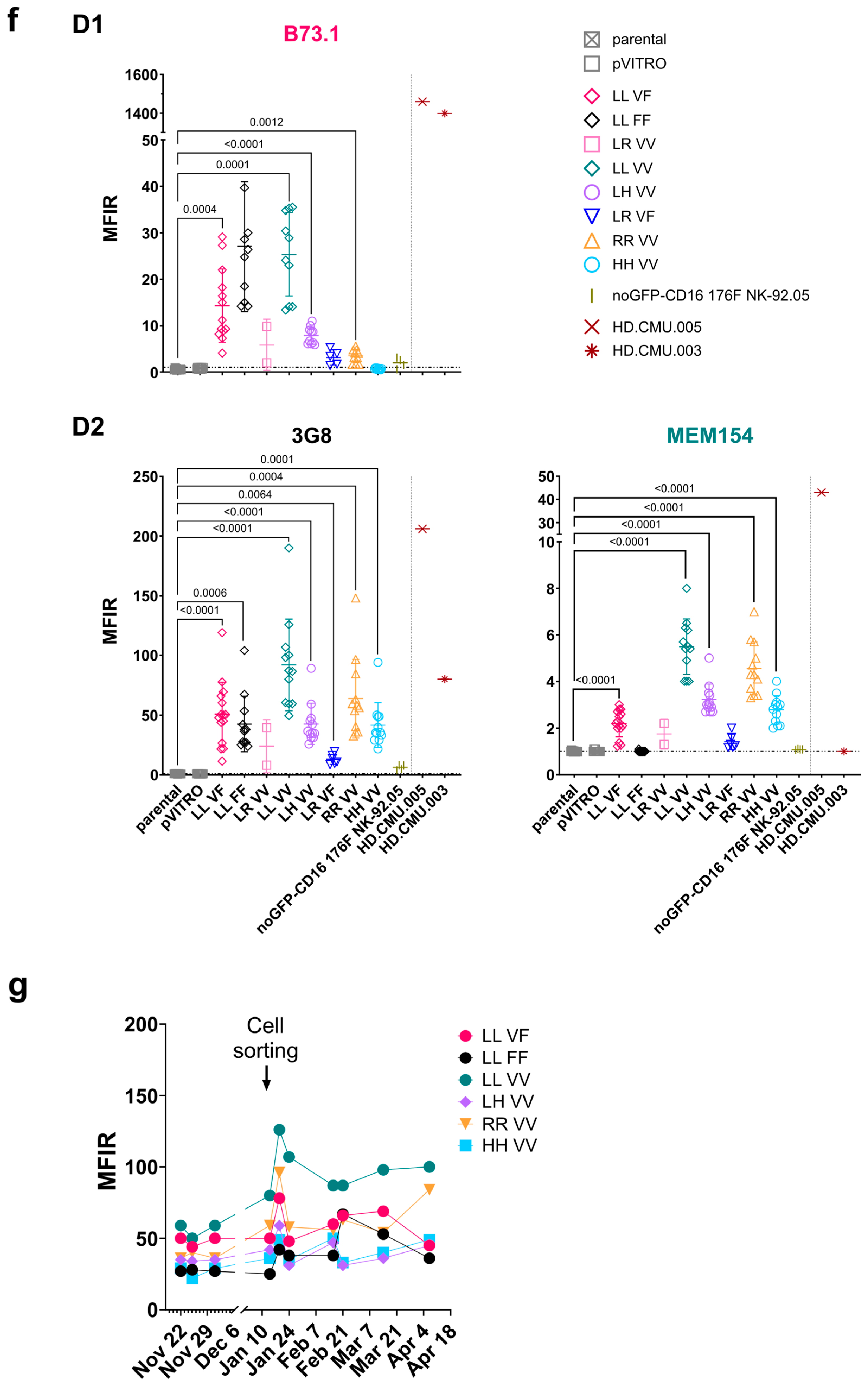
2.4. Surface Marker Expression Using Flow Cytometry Analysis, Cell Sorting, and Cytotoxicity Assays with NK-92 Cell Lines
2.5. Cytotoxicity Assays by NK-92 Cell Lines
2.6. Statistical Analysis
3. Results
3.1. Expression of mRNA of FCGR3A SNPs rs10127939 and rs396991 in Transfected NK-92 Cell Lines
3.2. Stable FcγRIIIa/CD16 Cell Surface Expression of Different Combinations of the FCGR3A SNPs rs10127939 and rs396991
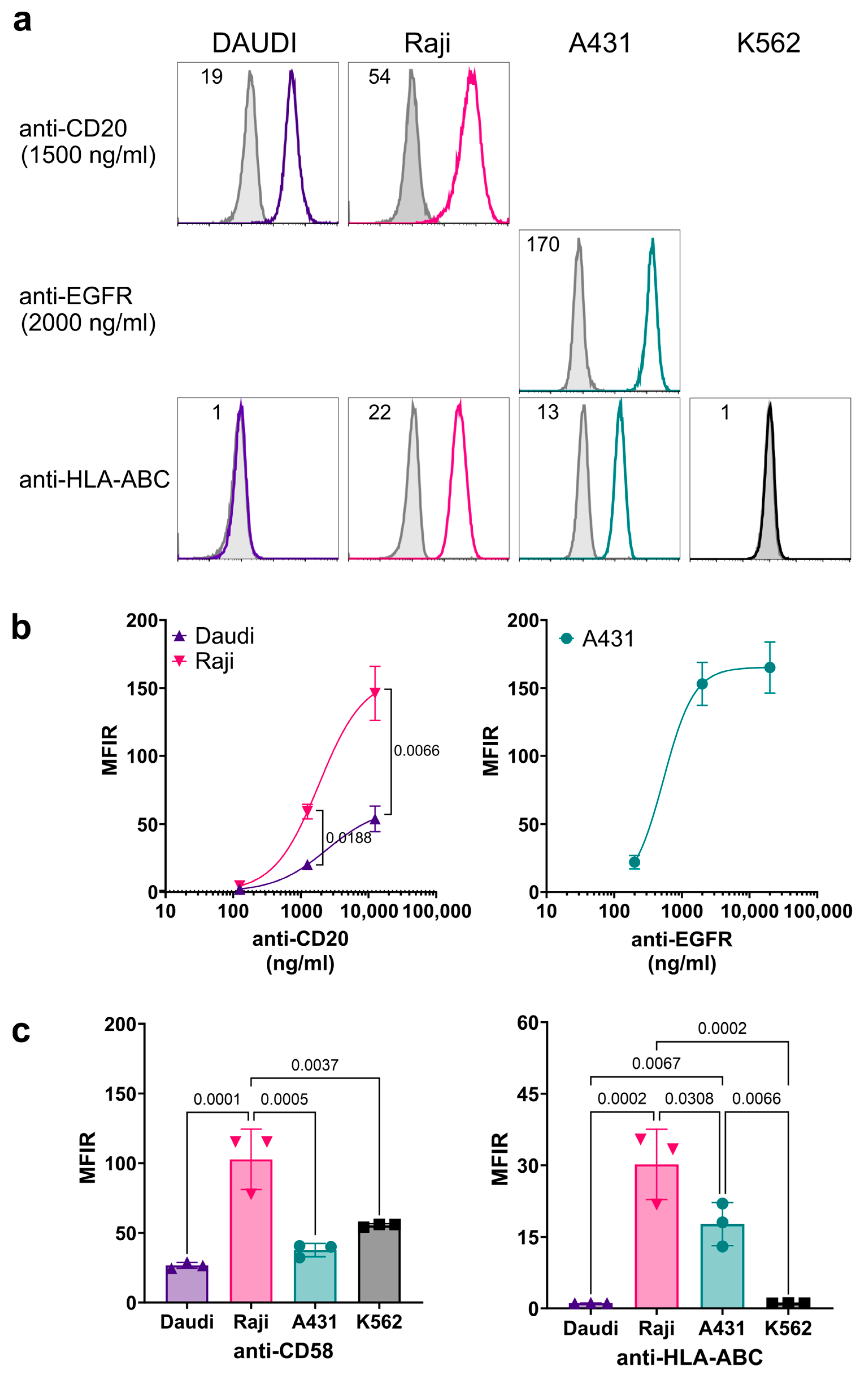
3.3. Characterization of Cell Surface Antigen Expression on Target Cells
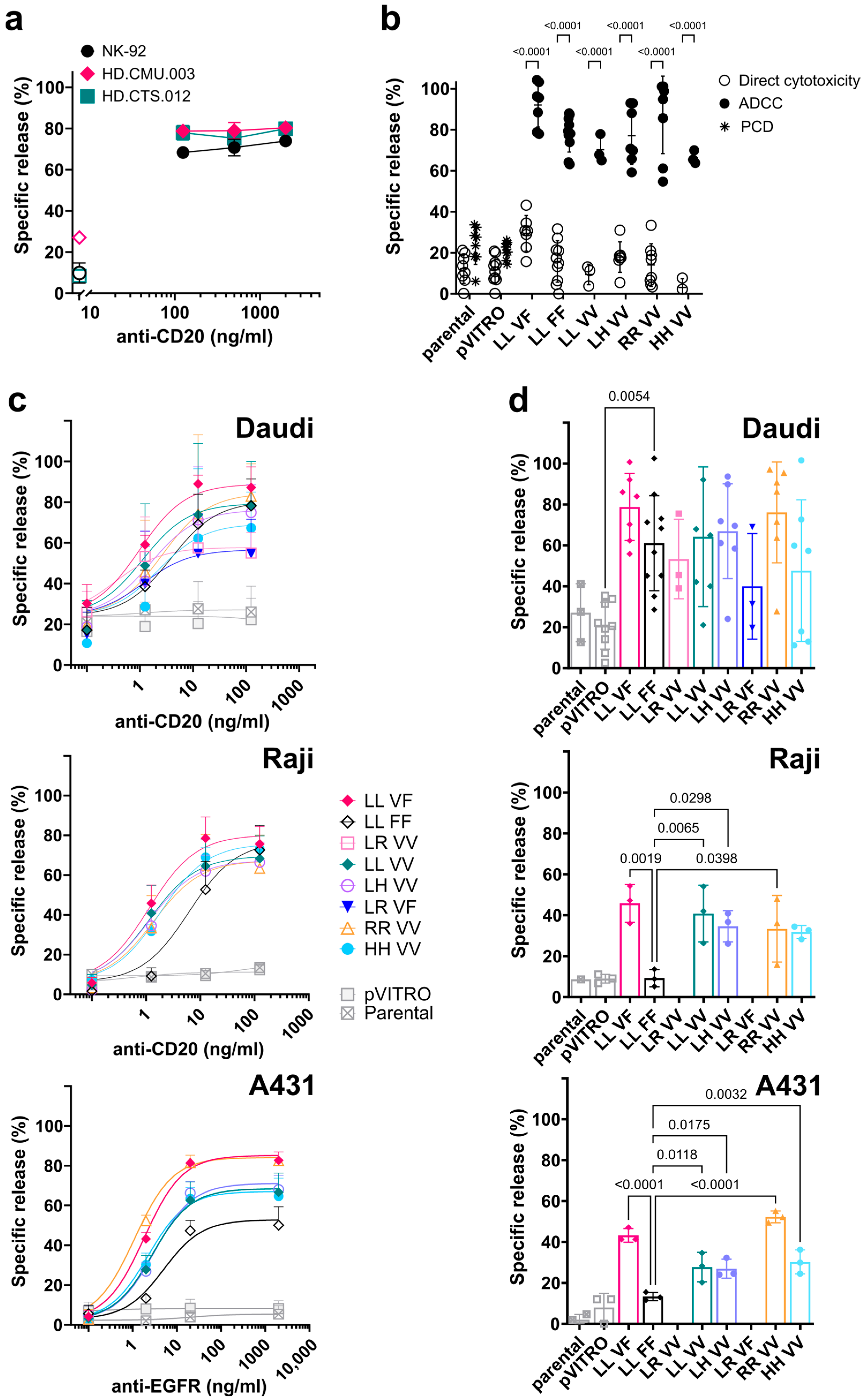
3.4. FcγRIIIa/CD16-Transfected NK-92 Cell Lines Induce Antibody-Dependent Cell-Mediated Cytotoxicity (ADCC)
3.5. Direct Cytotoxicity against K562 Target Cells Mediated by FcγRIIIa/CD16-Transfected NK-92 Cell Lines
4. Discussion
4.1. Expression of FcγRIIIa/CD16
4.2. ADCC Mediated by NK-92 Expressing FcγRIIIa/CD16
4.3. Direct or Natural Cytotoxicity of NK-92 Transfectants
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Del Zotto, G.; Marcenaro, E.; Vacca, P.; Sivori, S.; Pende, D.; Della Chiesa, M.; Moretta, F.; Ingegnere, T.; Mingari, M.C.; Moretta, A.; et al. Markers and Function of Human Nk cells in Normal and Pathological Conditions. Cytom. Part B Clin. Cytom. 2017, 92, 100–114. [Google Scholar] [CrossRef] [PubMed]
- Krabbendam, L.; Bernink, J.H.; Spits, H. Innate Lymphoid Cells: From Helper to Killer. Curr. Opin. Immunol. 2020, 68, 28–33. [Google Scholar] [CrossRef] [PubMed]
- Seillet, C.; Brossay, L.; Vivier, E. Natural Killers or Ilc1s? That is the Question. Curr. Opin. Immunol. 2020, 68, 48–53. [Google Scholar] [CrossRef] [PubMed]
- Vivier, E.; Raulet, D.H.; Moretta, A.; Caligiuri, M.A.; Zitvogel, L.; Lanier, L.L.; Yokoyama, W.M.; Ugolini, S. Innate or Adaptive Immunity? The Example of Natural Killer Cells. Science 2011, 331, 44–49. [Google Scholar] [CrossRef] [PubMed]
- Del Zotto, G.; Antonini, F.; Pesce, S.; Moretta, F.; Moretta, L.; Marcenaro, E. Comprehensive Phenotyping of Human Pb Nk Cells by Flow Cytometry. Cytom. A 2020, 97, 891–899. [Google Scholar] [CrossRef] [PubMed]
- Nimmerjahn, F.; Ravetch, J.V. Fcgamma Receptors as Regulators of Immune Responses. Nat. Rev. Immunol. 2008, 8, 34–47. [Google Scholar] [CrossRef] [PubMed]
- Caaveiro, J.M.; Kiyoshi, M.; Tsumoto, K. Structural Analysis of Fc/Fcgammar Complexes: A Blueprint for Antibody Design. Immunol. Rev. 2015, 268, 201–221. [Google Scholar] [CrossRef] [PubMed]
- Canfield, S.M.; Morrison, S.L. The Binding Affinity of Human Igg for Its High Affinity Fc Receptor Is Determined by Multiple Amino Acids in the Ch2 Domain and Is Modulated by the Hinge Region. J. Exp. Med. 1991, 173, 1483–1491. [Google Scholar] [CrossRef] [PubMed]
- Hogarth, P.M.; Pietersz, G.A. Fc Receptor-Targeted Therapies for the Treatment of Inflammation, Cancer and Beyond. Nat. Rev. Drug Discov. 2012, 11, 311–331. [Google Scholar] [CrossRef] [PubMed]
- Bournazos, S.; Wang, T.T.; Ravetch, J.V. The Role and Function of Fcgamma Receptors on Myeloid Cells. Microbiol. Spectr. 2016, 4, MCHD-0045-2016. [Google Scholar] [CrossRef]
- Bournazos, S.; Woof, J.M.; Hart, S.P.; Dransfield, I. Functional and Clinical Consequences of Fc Receptor Polymorphic and Copy Number Variants. Clin. Exp. Immunol. 2009, 157, 244–254. [Google Scholar] [CrossRef] [PubMed]
- Shamir, J.; Slomp, A.; Boross, P.; Leussen, J.H.W. Fc Receptors. eLS. 2013. Available online: www.els.net (accessed on 1 March 2013).
- Lajoie, L.; Congy-Jolivet, N.; Bolzec, A.; Gouilleux-Gruart, V.; Sicard, E.; Sung, H.C.; Peiretti, F.; Moreau, T.; Vie, H.; Clemenceau, B.; et al. Adam17-Mediated Shedding of Fcgammariiia on Human Nk Cells: Identification of the Cleavage Site and Relationship with Activation. J. Immunol. 2014, 192, 741–751. [Google Scholar] [CrossRef]
- Romee, R.; Foley, B.; Lenvik, T.; Wang, Y.; Zhang, B.; Ankarlo, D.; Luo, X.; Cooley, S.; Verneris, M.; Walcheck, B.; et al. Nk Cell Cd16 Surface Expression and Function Is Regulated by a Disintegrin and Metalloprotease-17 (Adam17). Blood 2013, 121, 3599–3608. [Google Scholar] [CrossRef] [PubMed]
- Koene, H.R.; Kleijer, M.; Algra, J.; Roos, D.; von dem Borne, A.E.; de Haas, M. Fc Gammariiia-158v/F Polymorphism In-fluences the Binding of Igg by Natural Killer Cell Fc Gammariiia, Independently of the Fc Gammariiia-48l/R/H Phenotype. Blood 1997, 90, 1109–1114. [Google Scholar] [CrossRef] [PubMed]
- Ravetch, J.V.; Perussia, B. Alternative Membrane Forms of Fc Gamma Riii(CD16) on Human Natural Killer Cells and Neutrophils. Cell Type-Specific Expression of Two Genes That Differ in Single Nucleotide Substitutions. J. Exp. Med. 1989, 170, 481–497. [Google Scholar] [CrossRef] [PubMed]
- de Haas, M.; Koene, H.R.; Kleijer, M.; de Vries, E.; Simsek, S.; van Tol, M.J.; Roos, D.; Borne, A.E.v.D. A Triallelic Fc Gamma Receptor Type Iiia Polymorphism Influences the Binding of Human IgG by Nk Cell Fc Gamma Riiia. J. Immunol. 1996, 156, 2948–2955. [Google Scholar] [CrossRef]
- de Vries, E.; Koene, H.R.; Vossen, J.M.; Gratama, J.W.; von dem Borne, A.E.; Waaijer, J.L.; Haraldsson, A.; de Haas, M.; van Tol, M.J. Identification of an Unusual Fc Gamma Receptor Iiia (Cd16) on Natural Killer Cells in a Patient with Recurrent Infections. Blood 1996, 88, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Grier, J.T.; Forbes, L.R.; Monaco-Shawver, L.; Oshinsky, J.; Atkinson, T.P.; Moody, C.; Pandey, R.; Campbell, K.S.; Orange, J.S. Human Immunodeficiency-Causing Mutation Defines Cd16 in Spontaneous Nk Cell Cytotoxicity. J. Clin. Investig. 2012, 122, 3769–3780. [Google Scholar] [CrossRef] [PubMed]
- Jawahar, S.; Moody, C.; Chan, M.; Finberg, R.; Geha, R.; Chatila, T. Natural Killer (Nk) Cell Deficiency Associated with An Epitope-Deficient Fc Receptor Type Iiia (Cd16-II). Clin. Exp. Immunol. 1996, 103, 408–413. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Gibson, A.W.; Kimberly, R.P. Human Fcr Polymorphism and Disease. Curr. Top. Microbiol. Immunol. 2014, 382, 275–302. [Google Scholar]
- Orange, J.S. Natural Killer Cell Deficiency. J. Allergy Clin. Immunol. 2013, 132, 515–525. [Google Scholar] [CrossRef] [PubMed]
- Papaserafeim, M.; Jandus, P.; Ferfoglia, R.I.; Nieke, J.P.; Vonarburg, C.; Spirig, R.; Yung, G.L.P.; Seebach, J.D. Effect of Intravenous Igg Therapy on Natural Killer Cell Function Related to Fc Gamma Receptor Gene Expression. J. Allergy Clin. Immunol. 2020, 146, 667–670. [Google Scholar] [CrossRef]
- Pradier, A.; Papaserafeim, M.; Li, N.; Rietveld, A.; Kaestel, C.; Gruaz, L.; Vonarburg, C.; Spirig, R.; Yung, G.L.P.; Seebach, J.D. Small-Molecule Immunosuppressive Drugs and Therapeutic Immunoglobulins Differentially Inhibit Nk Cell Effector Functions in vitro. Front. Immunol. 2019, 10, 556. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, R.A.; Luwor, R.B.; Burgess, A.W. Epidermal Growth Factor Receptor: Structure-function Informing the Design of Anticancer Therapeutics. Exp. Cell Res. 2018, 371, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Pierpont, T.M.; Limper, C.B.; Richards, K.L. Past, Present, and Future of Rituximab-the World’s First Oncology Monoclonal Antibody Therapy. Front. Oncol. 2018, 8, 163. [Google Scholar] [CrossRef]
- Grillo-Lopez, A.; White, C.; Dallaire, B.; Varns, C.; Shen, C.; Wei, A.; Leonard, J.; McClure, A.; Weaver, R.; Cairelli, S.; et al. Rituximab the First Monoclonal Antibody Approved for the Treatment of Lymphoma. Curr. Pharm. Biotechnol. 2000, 1, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Boero, S.; Morabito, A.; Banelli, B.; Cardinali, B.; Dozin, B.; Lunardi, G.; Piccioli, P.; Lastraioli, S.; Carosio, R.; Salvi, S.; et al. Analysis of in Vitro Adcc and Clinical Response to Trastuzumab: Possible Relevance of Fcgammariiia/Fcgammariia Gene Polymorphisms and Her-2 Expression Levels on Breast Cancer Cell Lines. J. Transl. Med. 2015, 13, 324. [Google Scholar] [CrossRef] [PubMed]
- Calemma, R.; Ottaiano, A.; Trotta, A.M.; Nasti, G.; Romano, C.; Napolitano, M.; Galati, D.; Borrelli, P.; Zanotta, S.; Cassata, A.; et al. Fc Gamma Receptor Iiia Polymorphisms in Advanced Colorectal Cancer Patients Correlated with Response to Anti-Egfr Antibodies and Clinical Outcome. J. Transl. Med. 2012, 10, 232. [Google Scholar] [CrossRef]
- Gavin, P.G.; Song, N.; Kim, S.R.; Lipchik, C.; Johnson, N.L.; Bandos, H.; Finnigan, M.; Rastogi, P.; Fehrenbacher, L.; Mamounas, E.P.; et al. Association of Polymorphisms in Fcgr2a and Fcgr3a with Degree of Trastuzumab Benefit in the Adjuvant Treatment of Erbb2/Her2-Positive Breast Cancer: Analysis of the Nsabp B-31 Trial. JAMA Oncol. 2017, 3, 335–341. [Google Scholar] [CrossRef]
- Goulet, D.R.; Atkins, W.M. Considerations for the Design of Antibody-Based Therapeutics. J. Pharm. Sci. 2020, 109, 74–103. [Google Scholar] [CrossRef]
- Guillerey, C.; Huntington, N.D.; Smyth, M.J. Targeting Natural Killer Cells in Cancer Immunotherapy. Nat. Immunol. 2016, 17, 1025–1036. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.S.; Flinn, I.W.; Modali, R.; Lehman, T.A.; Webb, J.; Waymer, S.; Moran, M.E.; Lucas, M.S.; Farag, S.S.; Byrd, J.C. Fcgr3a and Fcgr2a Polymorphisms May Not Correlate with Response to Alemtuzumab in Chronic Lymphocytic Leukemia. Blood 2005, 105, 289–291. [Google Scholar] [CrossRef] [PubMed]
- Mellor, J.D.; Brown, M.P.; Irving, H.R.; Zalcberg, J.R.; Dobrovic, A. A Critical Review of the Role of Fc Gamma Receptor Polymorphisms in the Response to Monoclonal Antibodies in Cancer. J. Hematol. Oncol. 2013, 6, 1. [Google Scholar] [CrossRef] [PubMed]
- Negri, F.V.; Musolino, A.; Naldi, N.; Bortesi, B.; Missale, G.; Laccabue, D.; Zerbini, A.; Camisa, R.; Chernyschova, N.; Bisagni, G.; et al. Role of Immunoglobulin G Fragment C Receptor Polymorphism-Mediated Antibody-Dependant Cellular Cytotoxicity In Colorectal Cancer Treated With Cetuximab Therapy. Pharmacogenomics J. 2014, 14, 14–19. [Google Scholar] [CrossRef] [PubMed]
- Park, S.J.; Hong, Y.S.; Lee, J.L.; Ryu, M.H.; Chang, H.M.; Kim, K.P.; Ahn, Y.C.; Na, Y.S.; Jin, D.H.; Yu, C.S.; et al. Genetic Polymorphisms of Fcgammariia and Fcgammariiia Are Not Predictive of Clinical Outcomes after Ce-tuximab Plus Irinotecan Chemotherapy in Patients with Metastatic Colorectal Cancer. Oncology 2012, 82, 83–89. [Google Scholar] [CrossRef] [PubMed]
- Seidel, U.J.; Schlegel, P.; Lang, P. Natural Killer Cell Mediated Antibody-Dependent Cellular Cytotoxicity in Tumor Immunotherapy with Therapeutic Antibodies. Front. Immunol. 2013, 4, 76. [Google Scholar] [CrossRef] [PubMed]
- Stork, A.C.; Notermans, N.C.; van den Berg, L.H.; Schellevis, R.D.; Niermeijer, J.M.; Nederend, M.; Leusen, J.H.; van der Pol, W.L. Fcgamma Receptor Iiia Genotype Is Associated with Rituximab Response in Antimyelin-Associated Glycoprotein Neuropathy. J. Neurol. Neurosurg. Psychiatry 2014, 85, 918–920. [Google Scholar] [CrossRef] [PubMed]
- Taylor, R.J.; Chan, S.L.; Wood, A.; Voskens, C.J.; Wolf, J.S.; Lin, W.; Chapoval, A.; Schulze, D.H.; Tian, G.; Strome, S.E. Fcgammariiia Polymorphisms and Cetuximab Induced Cytotoxicity in Squamous Cell Carcinoma of the Head and Neck. Cancer Immunol. Immunother. 2009, 58, 997–1006. [Google Scholar] [CrossRef]
- Munde, E.O.; Okeyo, W.A.; Raballah, E.; Anyona, S.B.; Were, T.; Ong’echa, J.M.; Perkins, D.J.; Ouma, C. Association between Fcγ Receptor Iia, Iiia and Iiib Genetic Polymorphisms and Susceptibility to Severe Malaria Anemia in Children in Western Kenya. BMC Infect. Dis. 2017, 17, 289. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, S.; Tanaka, Y.; Tazawa, H.; Verma, S.; Onoe, T.; Ishiyama, K.; Ohira, M.; Ide, K.; Ohdan, H. Fc-Gamma Receptor Polymorphisms Predispose Patients to Infectious Complications after Liver Transplantation. Am. J. Transplant. 2016, 16, 625–633. [Google Scholar] [CrossRef]
- Vietzen, H.; Danklmaier, V.; Zoufaly, A.; Puchhammer-Stockl, E. High-Affinity Fcgammariiia Genetic Variants and Potent Nk Cell-Mediated Antibody-Dependent Cellular Cytotoxicity (Adcc) Responses Contributing to Severe COVID-19. Genet. Med. 2022, 24, 1449–1458. [Google Scholar] [CrossRef]
- Drexler, H.G.; Matsuo, Y. Malignant Hematopoietic Cell Lines: In Vitro Models for the Study of Natural Killer Cell Leukemia-Lymphoma. Leukemia 2000, 14, 777–782. [Google Scholar] [CrossRef]
- Gong, J.H.; Maki, G.; Klingemann, H.G. Characterization of a Human Cell Line (Nk-92) with Phenotypical and Functional Characteristics of Activated Natural Killer Cells. Leukemia 1994, 8, 652–658. [Google Scholar] [PubMed]
- Maki, G.; Klingemann, H.-G.; Martinson, J.A.; Tam, Y.K. Factors Regulating the Cytotoxic Activity of the Human Natural Killer Cell Line, Nk-92. J. Hematotherapy Stem Cell Res. 2001, 10, 369–383. [Google Scholar] [CrossRef]
- Huang, R.-S.; Shih, H.-A.; Lai, M.-C.; Chang, Y.-J.; Lin, S. Enhanced Nk-92 Cytotoxicity by CRISPR Genome Engineering Using Cas9 Ribonucleoproteins. Front. Immunol. 2020, 11, 1008. [Google Scholar] [CrossRef] [PubMed]
- Clémenceau, B.; Vivien, R.; Pellat, C.; Foss, M.; Thibault, G.; Vié, H. The human natural killer cytotoxic cell line Nk-92, Once Armed with A Murine CD16 Receptor, Represents a Convenient Cellular Tool for the Screening of Mouse mAbs According to their ADCC Potential. mAbs 2013, 5, 587–594. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Zhou, Z.; Li, G.; Liu, G.; Lin, S.; Chen, W.; Xiong, S. An Nk Cell Line (Nk92-41bb) Expressing High Levels of Granzyme Is Engineered to Express the High Affinity Chimeric Genes Cd16/Car. Cytotechnology 2021, 73, 539–553. [Google Scholar] [CrossRef] [PubMed]
- Binyamin, L.; Alpaugh, R.K.; Hughes, T.L.; Lutz, C.T.; Campbell, K.S.; Weiner, L.M. Blocking Nk Cell Inhibitory Self-Recognition Promotes Antibody-Dependent Cellular Cytotoxicity in a Model of Anti-Lymphoma Therapy. J. Immunol. 2008, 180, 6392–6401. [Google Scholar] [CrossRef] [PubMed]
- Joseph, S.; Maniatis, T.; Fritsch, E.F. Molecular Cloning: A Laboratory Manual, 2nd ed.; Cold Spring Harbor Laboratory: Cold Spring Harbor, NY, USA, 1989. [Google Scholar]
- Millard, A.-L.; Valli, P.V.; Stussi, G.; Mueller, N.J.; Yung, G.P.; Seebach, J.D. Brief Exercise Increases Peripheral Blood Nk Cell Counts without Immediate Functional Changes, but Impairs their Responses to Ex Vivo Stimulation. Front. Immunol. 2013, 4, 125. [Google Scholar] [CrossRef] [PubMed]
- Freitas Monteiro, M.; Papaserafeim, M.; Real, A.; Yung, G.L.P.; Seebach, J.D. Anti-Cd20 Rituximab Igg1, Igg3, and Igg4 but Not Igg2 Subclass Trigger Ca2+ Mobilization and Cytotoxicity in Human Nk Cells. J. Leukoc. Biol. 2020, 108, 1409–1423. [Google Scholar] [CrossRef]
- Pross, H.F.; Baines, M.G.; Rubin, P.; Shragge, P.; Patterson, M.S. Spontaneous Human Lymphocyte-Mediated Cytotoxicity against Tumor Target Cells. Ix. The Quantitation of Natural Killer Cell Activity. J. Clin. Immunol. 1981, 1, 51–63. [Google Scholar] [CrossRef]
- Garrick, D.; Fiering, S.; Martin, D.I.; Whitelaw, E. Repeat-Induced Gene Silencing in Mammals. Nat. Genet. 1998, 18, 56–59. [Google Scholar] [CrossRef] [PubMed]
- Festenstein, R.; Tolaini, M.; Corbella, P.; Mamalaki, C.; Parrington, J.; Fox, M.; Miliou, A.; Jones, M.; Kioussis, D. Locus Control Region Function and Heterochromatin-Induced Position Effect Variegation. Science 1996, 271, 1123–1125. [Google Scholar] [CrossRef]
- Berg, M.; Lundqvist, A.; McCoy, P.; Samsel, L.; Fan, Y.; Tawab, A.; Childs, R. Clinical-Grade Ex Vivo-Expanded Human Natural Killer Cells Up-Regulate Activating Receptors and Death Receptor Ligands and Have Enhanced Cytolytic Activity against Tumor Cells. Cytotherapy 2009, 11, 341–355. [Google Scholar] [CrossRef]
- Törnroos, H.; Hägerstrand, H.; Lindqvist, C. Culturing the Human Natural Killer Cell Line Nk-92 in Interleukin-2 and Interleukin-15—Implications for Clinical Trials. Anticancer. Res. 2019, 39, 107–112. [Google Scholar] [CrossRef]
- Herter, S.; Herting, F.; Mundigl, O.; Waldhauer, I.; Weinzierl, T.; Fauti, T.; Muth, G.; Ziegler-Landesberger, D.; Van Puijenbroek, E.; Lang, S.; et al. Preclinical Activity of the Type Ii Cd20 Antibody Ga101 (Obinutuzumab) Compared with Rituximab and Ofatumumab in Vitro and in Xenograft Models. Mol. Cancer Ther. 2013, 12, 2031–2042. [Google Scholar] [CrossRef]
- Ivanov, A.; Beers, S.A.; Walshe, C.A.; Honeychurch, J.; Alduaij, W.; Cox, K.L.; Potter, K.N.; Murray, S.; Chan, C.H.; Klymenko, T.; et al. Monoclonal Antibodies Directed to CD20 and Hla-Dr can Elicit Homotypic Adhesion Followed by Lysosome-Mediated Cell Death in Human Lymphoma and Leukemia Cells. J. Clin. Investig. 2009, 119, 2143–2159. [Google Scholar] [CrossRef]
- Meyer, S.; Evers, M.; Jansen, J.H.M.; Buijs, J.; Broek, B.; Reitsma, S.E.; Moerer, P.; Amini, M.; Kretschmer, A.; Broeke, T.T.; et al. New Insights in Type I and II CD20 Antibody Mechanisms-of-Action with a Panel of Novel CD20 Antibodies. Br. J. Haematol. 2018, 180, 808–820. [Google Scholar] [CrossRef]
- Klingemann, H. The Nk-92 cell line—30 years later: Its Impact on Natural Killer Cell Research and Treatment of Cancer. Cytotherapy 2023, 25, 451–457. [Google Scholar] [CrossRef]
- Schnueriger, A.; Grau, R.; Sondermann, P.; Schreitmueller, T.; Marti, S.; Zocher, M. Development of a Quantitative, Cell-Line Based Assay to Measure Adcc Activity Mediated by Therapeutic Antibodies. Mol. Immunol. 2011, 48, 1512–1517. [Google Scholar] [CrossRef]
- Yamashita-Kashima, Y.; Iijima, S.; Yorozu, K.; Furugaki, K.; Kurasawa, M.; Ohta, M.; Fujimoto-Ouchi, K. Pertuzumab in Combination with Trastuzumab Shows Significantly Enhanced Antitumor Activity in Her2-Positive Human Gastric Cancer Xenograft Models. Clin. Cancer Res. 2011, 17, 5060–5070. [Google Scholar] [CrossRef] [PubMed]
- Fabian, K.P.; Hodge, J.W. The Emerging Role of Off-the-Shelf Engineered Natural Killer Cells in Targeted Cancer Immunotherapy. Mol. Ther. Oncolytics 2021, 23, 266–276. [Google Scholar] [CrossRef] [PubMed]
- Fujii, R.; Schlom, J.; Hodge, J.W. A potential Therapy for Chordoma via Antibody-Dependent Cell-Mediated Cytotoxicity Employing Nk or High-Affinity Nk Cells in Combination with Cetuximab. J. Neurosurg. 2018, 128, 1419–1427. [Google Scholar] [CrossRef] [PubMed]
- Jochems, C.; Hodge, J.W.; Fantini, M.; Fujii, R.; Morillon, Y.M.; Greiner, J.W., 2nd; Padget, M.R.; Tritsch, S.R.; Tsang, K.Y.; Campbell, K.S.; et al. An Nk Cell Line (Hank) Expressing High Levels of Granzyme and Engineered to Express the High Affinity Cd16 Allele. Oncotarget 2016, 7, 86359–86373. [Google Scholar] [CrossRef] [PubMed]
- Rataj, F.; Jacobi, S.J.; Stoiber, S.; Asang, F.; Ogonek, J.; Tokarew, N.; Cadilha, B.L.; van Puijenbroek, E.; Heise, C.; Duewell, P.; et al. High-Affinity CD16-Polymorphism and Fc-Engineered Antibodies Enable Activity of CD16-Chimeric Antigen Receptor-Modified T Cells for Cancer Therapy. Br. J. Cancer 2019, 120, 79–87. [Google Scholar] [CrossRef]
- Perez-Romero, C.A.; Sanchez, I.P.; Naranjo-Piedrahita, L.; Orrego-Arango, J.C.; Muskus-Lopez, C.E.; Rojas-Montoya, W.; Restrepo, J.L.F.; Trujillo-Vargas, C.M. Frequency Analysis of the G.7081t>G/a and G.10872t>G Polymorphisms in the Fcgr3a Gene (Cd16a) Using Nested Pcr and Their Functional Specific Effects. Genes Immun. 2019, 20, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Machiela, M.J.; Chanock, S.J. LDlink: A Web-Based Application for Exploring Population-Specific Haplotype Structure and Linking Correlated Alleles of Possible Functional Variants. Bioinformatics 2015, 31, 3555–3557. [Google Scholar] [CrossRef] [PubMed]
- Blázquez-Moreno, A.; Park, S.; Im, W.; Call, M.J.; Call, M.E.; Reyburn, H.T. Transmembrane Features Governing Fc Receptor CD16A Assembly with CD16A Signaling Adaptor Molecules. Proc. Natl. Acad. Sci. USA 2017, 114, E5645–E5654. [Google Scholar] [CrossRef] [PubMed]
- Victor, A.R.; Weigel, C.; Scoville, S.D.; Chan, W.K.; Chatman, K.; Nemer, M.M.; Mao, C.; Young, K.A.; Zhang, J.; Yu, J.; et al. Epigenetic and Posttranscriptional Regulation of CD16 Expression during Human Nk Cell Development. J. Immunol. 2018, 200, 565–572. [Google Scholar] [CrossRef]
- Li, W.; Yang, H.; Dimitrov, D.S. Identification of High-Affinity Anti-CD16A allotype-Independent Human Antibody Domains. Exp. Mol. Pathol. 2016, 101, 281–289. [Google Scholar] [CrossRef]
- Dong, C.; Ptacek, T.S.; Redden, D.T.; Zhang, K.; Brown, E.E.; Edberg, J.C.; McGwin, G., Jr.; Alarcon, G.S.; Ramsey-Goldman, R.; Reveille, J.D.; et al. Fcgamma Receptor Iiia Single-Nucleotide Polymorphisms and Haplotypes Affect Human Igg Binding and Are Associated with Lupus Nephritis in African Americans. Arthritis Rheumatol. 2014, 66, 1291–1299. [Google Scholar] [CrossRef]
- Deuse, T.; Hu, X.; Agbor-Enoh, S.; Jang, M.K.; Alawi, M.; Saygi, C.; Gravina, A.; Tediashvili, G.; Nguyen, V.Q.; Liu, Y.; et al. The Sirpalpha-Cd47 Immune Checkpoint in Nk Cells. J. Exp. Med. 2021, 218, e20200839. [Google Scholar] [CrossRef]
- Barta, P.; Andersson, K.; Trejtnar, F.; Buijs, J. Exploring Time-Resolved Characterization of the Heterogeneity and Dynamics of Ligand-Receptor Interactions on Living Cells. J. Anal. Oncol. 2014, 3, 94–104. [Google Scholar] [CrossRef]
- Mandelboim, O.; Malik, P.; Davis, D.M.; Jo, C.H.; Boyson, J.E.; Strominger, J.L. Human CD16 as a Lysis Receptor Mediating Direct Natural Killer Cell Cytotoxicity. Proc. Natl. Acad. Sci. USA 1999, 96, 5640–5644. [Google Scholar] [CrossRef]
- Binder, C.; Cvetkovski, F.; Sellberg, F.; Berg, S.; Visbal, H.P.; Sachs, D.H.; Berglund, E.; Berglund, D. Cd2 Immuno-biology. Front. Immunol. 2020, 11, 1090. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, Q.; Yang, S.; Liao, Q. Cd58 Immunobiology at a Glance. Front. Immunol. 2021, 12, 705260. [Google Scholar] [CrossRef]
- Carlsten, M.; Childs, R.W. Genetic Manipulation of Nk Cells for Cancer Immunotherapy: Techniques and Clinical Implications. Front. Immunol. 2015, 6, 266. [Google Scholar] [CrossRef]
- Izadi, N.; Sun, M.; Mace, E.M.; O’gorman, M.R.G.; Church, J.A. Undetectable Nk Cells Due to the Fcgr3a Variant, l66h, Which May Not Be Directly Disease-Causing. J. Clin. Immunol. 2021, 41, 1957–1959. [Google Scholar] [CrossRef]


| Daudi | Raji | A431 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Transfectant | n | * R2 | ‡ EC50 (ng/mL) | n | R2 | EC50 (ng/mL) | n | R2 | EC50 (ng/mL) |
| NK-92 LL_VF | 2, 5 | 0.9139 | 0.99 | 3 | 0.9268 | 1.1 | 3 | 0.9715 | 1.8 |
| NK-92 LL_FF | 4, 8 | 0.8504 | 2.13 | 3 | 0.9039 | 6.2 | 3 | 0.9011 | 5.0 |
| NK-92 LR_VV | 3 | 0.5515 | 0.02 | ND | ND | ||||
| NK-92 LL_VV | 4 | 0.5578 | 1.02 | 3 | 0.8626 | 1.2 | 3 | 0.9284 | 2.8 |
| NK-92 LH_VV | 2, 4 | 0.7898 | 1.58 | 3 | 0.9106 | 1.1 | 3 | 0.9472 | 3.1 |
| NK-92 LR_VF | 3 | 0.5181 | 0.62 | ND | ND | ||||
| NK-92 RR_VV | 2, 5 | 0.7526 | 1.64 | 3 | 0.9749 | 1.4 | 3 | 0.9689 | 1.1 |
| NK-92 HH_VV | 2, 4 | 0.5679 | 2.23 | 3 | 0.8003 | 1.8 | 3 | 0.9379 | 2.2 |
| Parental | 3, 6 | 0.0476 | -; | 3 | - | 3 | 0.0097 | - | |
| NK-92 pVITRO | 7, 9 | 0.0738 | - | 3 | - | 3 | 0 | - | |
| Name | Goodness of Fit, R2 * | Maximum Specific Lysis (%) | LU30/107 Effectors |
|---|---|---|---|
| NK-92 LL_VF | 0.8095 | 83 | 63 |
| NK-92 LL_FF | 0.8588 | 52 | 18 |
| NK-92 LR_VV | 0.6181 | 71 | 28 |
| NK-92 LL_VV | 0.9251 | 55 | 10 |
| NK-92 LH_VV | 0.8174 | 80 | 18 |
| NK-92 LR_VF | 0.7148 | 100 * | 11 |
| NK-92 RR_VV | 0.3276 | 77 | n/a † |
| NK-92 HH_VV | 0.6209 | 73 | 31 |
| parental | 0.6521 | 91 | 8 |
| NK-92 pVITRO | 0.5487 | 83 | 10 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Freitas Monteiro, M.; Papaserafeim, M.; Andreani, M.; Réal, A.; Kouklas, A.; Reis Galvão, D.; Seebach, J.D.; Puga Yung, G.L. NK Cytotoxicity Mediated by NK-92 Cell Lines Expressing Combinations of Two Allelic Variants for FCGR3. Antibodies 2024, 13, 55. https://doi.org/10.3390/antib13030055
Freitas Monteiro M, Papaserafeim M, Andreani M, Réal A, Kouklas A, Reis Galvão D, Seebach JD, Puga Yung GL. NK Cytotoxicity Mediated by NK-92 Cell Lines Expressing Combinations of Two Allelic Variants for FCGR3. Antibodies. 2024; 13(3):55. https://doi.org/10.3390/antib13030055
Chicago/Turabian StyleFreitas Monteiro, Marta, Maria Papaserafeim, Matteo Andreani, Aline Réal, Athanasios Kouklas, Daniela Reis Galvão, Jörg D. Seebach, and Gisella L. Puga Yung. 2024. "NK Cytotoxicity Mediated by NK-92 Cell Lines Expressing Combinations of Two Allelic Variants for FCGR3" Antibodies 13, no. 3: 55. https://doi.org/10.3390/antib13030055








