Identification of Apple Tree Leaf Diseases Based on Deep Learning Models
Abstract
:1. Introduction
2. Building the Dataset
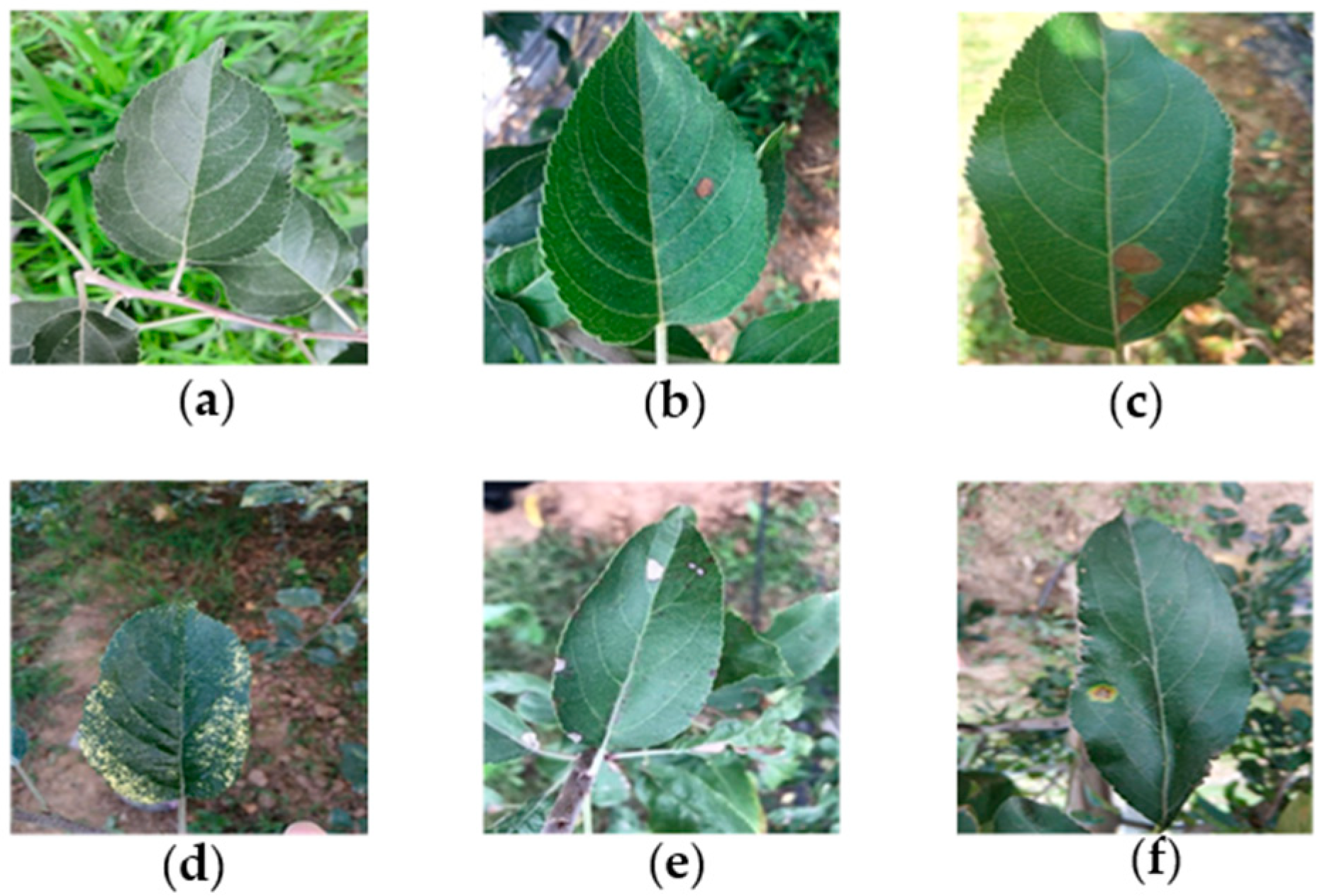
2.1. Collecting the Dataset
2.2. Dataset Image Preprocessing
2.2.1. Data Augmentation
2.2.2. Data Normalization
2.3. Dividing the Dataset
3. Constructing Deep Convolutional Neural Network
3.1. Xception
- The Inception module is replaced by a depthwise separable convolution layer in Xception, and the standard convolution is decomposed into a spatial convolution and a point-by-point convolution. Spatial convolution operations are first performed independently on each channel, followed by point-wise convolution operation, and finally connect the results. The use of depthwise separable convolution can greatly reduce the amount of parameters and calculations with a tiny loss of accuracy. This structure is similar to the conventional convolution operation and can be used to extract features. Compared with the conventional convolution operation, the number of parameters and the calculation cost of depth-wise separable convolution are lower.
- Xception contains 14 modules. Except for the first module and the last module, all modules have added a residual connection mechanism similar to ResNet [32], which significantly accelerates the convergence process of Xception and obtains higher accuracy rate [19]. The structure of Xception network is shown in Figure 3. The front part of the network is mainly used to continuously down sample and reduce the spatial dimension. The middle part is to continuously learn the correlation and optimize the features. The latter part is to summarize and consolidate the features, then Softmax activation function is used to calculate the probability vector of a given input class.
3.2. DenseNet
- The biggest feature of DenseNet is that for each layer, the function maps of all the previous layers are used as inputs, and its own function map is used as the input of all subsequent layers. It clearly distinguishes the information added to the network from the information reused. The connection scheme is shown in Figure 4, which ensures that the information flow between the layers in the network reaches the maximum, and there is no need to re-learn redundant feature mappings. Therefore, the number of parameters is greatly reduced, and the parameter efficiency is improved. The model improves the information flow and gradient of the entire network. Each layer can directly access the gradient from the loss function to the original input signal, thereby achieving an implicit deep monitoring and alleviating the problem of vanishing gradients. Moreover, the dense connection has regularization effect, so it can restrain the over-fitting on a small scale training dataset to some extent.
- Function maps of the same size between any two layers are directly connected, which has good feed-forward characteristics, enhancing the feature propagation and feature reuse.
- DenseNet has a small number of filters per convolution operation. Only a small part of the feature map is added to the network, and the remaining feature maps are kept unchanged. This structure reduces the number of input feature maps and helps to build a deep network architecture.
- The structure of the DenseNet-201 [20] model is shown in Figure 4. Since the output of the dense block connects all the layers in the block, the larger the depth in the dense block is, the larger the size of the feature map becomes, which will increase the calculation costs continuously. Therefore, the transition layer is added between the dense blocks. The transition layer consists of 1 × 1 convolution and 2 × 2 average-pooling. Through the 2 × 2 average pool, the width and height can be halved to improve the computational efficiency [35].
3.3. The Proposed XDNet
4. Experimental Evaluation
4.1. Experimental Device
4.2. ATLDs Detection Process
4.3. Experimental Results and Analysis
4.3.1. Confusion Matrix
4.3.2. Comparative Experiment of Transfer Learning
4.3.3. Experiment on Data Augmentation
4.3.4. Comparison of DCNNs
4.3.5. Importance of Training Images Type
4.3.6. Feature Visualization
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Mahlein, A.; Rumpf, T.; Welke, P.; Dehne, H.W.; Plumer, L.; Steiner, U.; Oerke, E. Development of spectral indices for detecting and identifying plant diseases. Remote Sens. Environ. 2013, 128, 21–30. [Google Scholar] [CrossRef]
- Yuan, L.; Huang, Y.; Loraamm, R.W.; Nie, C.; Wang, J.; Zhang, J. Spectral analysis of winter wheat leaves for detection and differentiation of diseases and insects. Field Crop. Res. 2014, 156, 199–207. [Google Scholar] [CrossRef] [Green Version]
- Qin, F.; Liu, D.; Sun, B.; Ruan, L.; Ma, Z.; Wang, H. Identification of Alfalfa Leaf Diseases Using Image Recognition Technology. PLoS ONE 2016, 11, e0168274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, S.; Zhang, Q.; LI, P. Apple disease identification based on improved deep convolutional neural network. J. For. Eng. 2019, 4, 107–112. [Google Scholar]
- Liu, W.; Wang, Z.; Liu, X.; Zeng, N.; Liu, Y.; Alsaadi, F.E. A survey of deep neural network architectures and their applications. Neurocomputing 2017, 234, 11–26. [Google Scholar] [CrossRef]
- Lee, S.H.; Chan, C.S.; Wilkin, P.; Remagnino, P. Deep-plant: Plant identification with convolutional neural networks. In Proceedings of the 2015 IEEE International Conference on Image Processing (ICIP), Quebec City, QC, Canada, 27–30 September 2015; pp. 452–456. [Google Scholar]
- Kawasaki, Y.; Uga, H.; Kagiwada, S.; Iyatomi, H. Basic Study of Automated Diagnosis of Viral Plant Diseases Using Convolutional Neural Networks. In Proceedings of the International Symposium on Visual Computing, Las Vegas, NV, USA, 14–16 December 2015; pp. 638–645. [Google Scholar]
- Sladojevic, S.; Arsenovic, M.; Anderla, A.; Stefanović, D. Deep Neural Networks Based Recognition of Plant Diseases by Leaf Image Classification. Comput. Intell. Neurosci. 2016, 2016, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Mohanty, S.; Hughes, D.; Salathe, M. Using Deep Learning for Image-Based Plant Disease Detection. Front. Plant Sci. 2016, 7, 1419. [Google Scholar] [CrossRef] [Green Version]
- Ferentinos, K. Deep learning models for plant disease detection and diagnosis. Comput. Electron. Agric. 2018, 145, 311–318. [Google Scholar] [CrossRef]
- Long, M.; Ouyang, C.; Liu, H.; Fu, Q. Image recognition of Camellia oleifera diseases based on convolutional neural network & transfer learning. Nongye Gongcheng Xuebao/Trans. Chin. Soc. Agric. Eng. 2018, 34, 194–201. [Google Scholar]
- Zhang, C.; Zhang, S.; Yang, J.; Shi, Y.; Chen, J. Apple leaf disease identification using genetic algorithm and correlation based feature selection method. Int. J. Agric. Biol. Eng. 2017, 10, 74–83. [Google Scholar]
- Liu, B.; Zhang, Y.; He, D.; Li, Y. Identification of Apple Leaf Diseases Based on Deep Convolutional Neural Networks. Symmetry 2017, 10, 11. [Google Scholar] [CrossRef] [Green Version]
- Baranwal, S.; Khandelwal, S.; Arora, A. Deep Learning Convolutional Neural Network for Apple Leaves Disease Detection. In Proceedings of the International Conference on Sustainable Computing in Science, Technology and Management (SUSCOM), Amity University Rajasthan, Jaipur, India, 26–28 February 2019; p. 8. [Google Scholar]
- Jiang, P.; Chen, Y.; Bin, L.; He, D.; Liang, C. Real-Time Detection of Apple Leaf Diseases Using Deep Learning Approach Based on Improved Convolutional Neural Networks. IEEE Access 2019, 7, 59069–59080. [Google Scholar] [CrossRef]
- Zhong, Y.; Zhao, M. Research on deep learning in apple leaf disease recognition. Comput. Electron. Agric. 2020, 168, 105146. [Google Scholar] [CrossRef]
- Yu, H.; Son, C.; Lee, D. Apple Leaf Disease Identification Through Region-of-Interest-Aware Deep Convolutional Neural Network. J. Imaging Sci. Technol. 2020, 64, 20507-1–20507-10. [Google Scholar] [CrossRef] [Green Version]
- Albayati, J.S.H.; Ustundag, B.B. Evolutionary Feature Optimization for Plant Leaf Disease Detection by Deep Neural Networks. Int. J. Comput. Intell. Syst. 2020, 13, 12–23. [Google Scholar] [CrossRef] [Green Version]
- Chollet, F. Xception: Deep Learning with Depthwise Separable Convolutions. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 1800–1807. [Google Scholar]
- Huang, G.; Liu, Z.; Der Maaten, L.V.; Weinberger, K.Q. Densely Connected Convolutional Networks. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 2261–2269. [Google Scholar]
- Wang, D.; Chai, X. Application of machine learning in plant diseases recognition. J. Chin. Agric. Mech. 2019, 40, 171–180. [Google Scholar] [CrossRef]
- Sweet, J.B.; Barbara, D.J. A yellow mosaic disease of horse chestnut (Aesculus spp.) caused by apple mosaic virus. Ann. Appl. Biol. 2008, 92, 335–341. [Google Scholar] [CrossRef]
- Boulent, J.; Foucher, S.; Théau, J.; St-Charles, P.-L. Convolutional Neural Networks for the Automatic Identification of Plant Diseases. Front. Plant Sci. 2019, 10, 941. [Google Scholar] [CrossRef] [Green Version]
- Dutot, M.; Nelson, L.M.; Tyson, R.C. Predicting the spread of postharvest disease in stored fruit, with application to apples. Postharvest Biol. Technol. 2013, 85, 45–56. [Google Scholar] [CrossRef]
- Joharestani, M.; Cao, C.; Ni, X.; Bashir, B.; Talebiesfandarani, S. PM2.5 Prediction Based on Random Forest, XGBoost, and Deep Learning Using Multisource Remote Sensing Data. Atmosphere 2019, 10, 373. [Google Scholar] [CrossRef] [Green Version]
- Nagasubramanian, K.; Jones, S.; Singh, A.K.; Sarkar, S.; Singh, A.; Ganapathysubramanian, B. Plant disease identification using explainable 3D deep learning on hyperspectral images. Plant Methods 2019, 15, 98. [Google Scholar] [CrossRef] [PubMed]
- Lecun, Y.; Bottou, L.; Bengio, Y.; Haffner, P. Gradient-Based Learning Applied to Document Recognition. Proc. IEEE 1998, 86, 2278–2324. [Google Scholar] [CrossRef] [Green Version]
- Krizhevsky, A.; Sutskever, I.; Hinton, G. ImageNet Classification with Deep Convolutional Neural Networks. Neural Inf. Process. Syst. 2012, 25, 1097–1105. [Google Scholar] [CrossRef]
- Zeiler, M.D.; Fergus, R. Visualizing and Understanding Convolutional Networks. In Proceedings of the European Conference on Computer Vision, Zurich, Switzerland, 6–12 September 2014; pp. 818–833. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. In Proceedings of the Computer Vision and Pattern Recognition, Zurich, Switzerland, 6–12 September 2014. [Google Scholar]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Pleiss, G.; Chen, D.; Huang, G.; Li, T.; van der Maaten, L.; Weinberger, K. Memory-Efficient Implementation of DenseNets. arXiv 2017, arXiv:1707.06990. [Google Scholar]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z.B. Rethinking the Inception Architecture for Computer Vision. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 2818–2826. [Google Scholar]
- Song, J.; Kim, W.; Park, K. Finger-vein Recognition Based on Deep DenseNet Using Composite Image. IEEE Access 2019, 7, 66845–66863. [Google Scholar] [CrossRef]
- Nogueira, K.; Penatti, O.; dos Santos, J. Towards Better Exploiting Convolutional Neural Networks for Remote Sensing Scene Classification. Pattern Recognit. 2017, 61, 539–556. [Google Scholar] [CrossRef] [Green Version]
- Mahdianpari, M.; Salehi, B.; Rezaee, M.; Mohammadimanesh, F.; Zhang, Y. Very Deep Convolutional Neural Networks for Complex Land Cover Mapping Using Multispectral Remote Sensing Imagery. Remote Sens. 2018, 10, 1119. [Google Scholar] [CrossRef] [Green Version]
- Ioffe, S.; Szegedy, C. Batch Normalization: Accelerating Deep Network Training by Reducing Internal Covariate Shift. arXiv 2015, arXiv:1502.03167. [Google Scholar]
- Amara, J.; Bouaziz, B.; Algergawy, A. A Deep Learning-based Approach for Banana Leaf Diseases Classification. In Proceedings of the BTW Workshop, Stuttgart, Germany, 6–10 March 2017; pp. 79–88. [Google Scholar]
- Hasan, M.; Tanawala, B.; Patel, K. Deep Learning Precision Farming: Tomato Leaf Disease Detection by Transfer Learning. SSRN Electron. J. 2019. [Google Scholar] [CrossRef]
- Vioix, J.; Douzals, J.P.; Truchetet, F. Development of a Spatial Method for Weed Detection and Localization. In Proceedings of the International Conference on Robotics and Automation, New Orleans, LA, USA, 26 April–1 May 2004; pp. 169–178. [Google Scholar]
- Tang, Y. Deep Learning using Linear Support Vector Machines. arXiv 2013, arXiv:1306.0239. [Google Scholar]
- Kingma, D.; Ba, J. Adam: A Method for Stochastic Optimization. In Proceedings of the International Conference on Learning Representations (ICLR), San Diego, CA, USA, 7–9 May 2015; p. 13. [Google Scholar]
- Dyson, J.; Mancini, A.; Frontoni, E.; Zingaretti, P. Deep Learning for Soil and Crop Segmentation from Remotely Sensed Data. Remote Sens. 2019, 11, 1859. [Google Scholar] [CrossRef] [Green Version]
- Yosinski, J.; Clune, J.; Bengio, Y.; Lipson, H. How transferable are features in deep neural networks. In Proceedings of the 27th International Conference on Neural Information Processing Systems, Montreal, QC, Canada, 8–13 December 2014; pp. 3320–3328. [Google Scholar]
- Wang, Y.; Shi, D.; Li, Y.; Cai, S. Studies on identification of microscopic images of chinese Chinese medicinal materials powder based on squeezenet deep network. J. Chin. Electron Microsc. Soc. 2019, 38, 130–138. [Google Scholar]
- Howard, A.; Zhu, M.; Chen, B.; Kalenichenko, D.; Wang, W.; Weyand, T.; Andreetto, M.; Adam, H. MobileNets: Efficient Convolutional Neural Networks for Mobile Vision Applications. arXiv 2017, arXiv:1704.04861. [Google Scholar]










| Disease Types | Original Images | Training Images (After Augmentation) | Validation Images | Testing Images |
|---|---|---|---|---|
| Alternaria leaf spot | 279 | 2366 | 55 | 55 |
| Brown spot | 540 | 4536 | 108 | 108 |
| Mosaic | 879 | 7378 | 176 | 176 |
| Grey spot | 430 | 3612 | 86 | 86 |
| Rust | 430 | 3612 | 86 | 86 |
| Heathy | 412 | 3472 | 82 | 82 |
| Sum | 2970 | 24,976 | 593 | 593 |
| Type | Patch Size/Stride | Output Size |
|---|---|---|
| Input | - | 224 × 224 × 3 |
| Convolution | 3 × 3/2 | 111 × 111 × 32 |
| Batch Normalization | - | 111 × 111 × 32 |
| ReLU | - | 111×111 × 32 |
| Convolution | 3×3/1 | 109 × 109 × 64 |
| Batch Normalization | - | 109 × 109 × 64 |
| ReLU | - | 109 × 109 × 64 |
| Block1 × 3 | - | 14 × 14 × 728 |
| Block2 × 5 | - | 14 × 14 × 728 |
| Batch Normalization | - | 14 × 14 × 728 |
| ReLU | - | 14 × 14 × 728 |
| Block3×6 | - | 14 × 14 × 800 |
| Batch Normalization | - | 14 × 14 × 800 |
| ReLU | - | 14 × 14 × 800 |
| Convolution | 1 × 1/1 | 14 × 14 × 400 |
| Pool/Average | 2 × 2/2 | 7 × 7 × 400 |
| Block3 × 12 | - | 7 × 7 × 544 |
| Batch Normalization | - | 7 × 7 × 544 |
| Global Average Pooling | - | 544 |
| SVM | - | 6 |
| Configuration Item | Value |
|---|---|
| CPU | CPU Intel(R) Xeon(R) CPU E5-2620 v4 @ 2.10GHz |
| GPU | NVIDIA TITAN V 12 GB |
| RAM | 128 GB |
| Hard Disk | 4 TB |
| Operating System | Ubuntu 16.04.1 LTS (64-bits) |
| Model | Input Size | Batch Size | Training Time/min | Parameters | Best Accuracy/% | Average Accuracy/% |
|---|---|---|---|---|---|---|
| Inception-v3 | 299 | 16 | 76.85 | 21,815,078 | 97.13 | 96.59 |
| DenseNet-201 | 224 | 16 | 111.17 | 18,333,510 | 98.31 | 97.50 |
| MobileNet | 224 | 16 | 30.42 | 3,235,014 | 97.47 | 96.52 |
| VGG-16 | 224 | 16 | 31.72 | 134,285,126 | 96.29 | 95.85 |
| Xception | 299 | 16 | 133.43 | 20,873,774 | 98.48 | 97.77 |
| VGG-INCEP | 224 | 32 | 144.27 | 148,714,950 | 97.64 | 97.20 |
| XDNet (Our work) | 224 | 16 | 86.67 | 10,159,518 | 98.82 | 98.35 |
| Model | Training: Laboratory | Training: Field |
|---|---|---|
| Accuracy | Accuracy | |
| XDNet | 76.96% | 90.79% |
| Xception | 74.19% | 88.62% |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chao, X.; Sun, G.; Zhao, H.; Li, M.; He, D. Identification of Apple Tree Leaf Diseases Based on Deep Learning Models. Symmetry 2020, 12, 1065. https://doi.org/10.3390/sym12071065
Chao X, Sun G, Zhao H, Li M, He D. Identification of Apple Tree Leaf Diseases Based on Deep Learning Models. Symmetry. 2020; 12(7):1065. https://doi.org/10.3390/sym12071065
Chicago/Turabian StyleChao, Xiaofei, Guoying Sun, Hongke Zhao, Min Li, and Dongjian He. 2020. "Identification of Apple Tree Leaf Diseases Based on Deep Learning Models" Symmetry 12, no. 7: 1065. https://doi.org/10.3390/sym12071065
APA StyleChao, X., Sun, G., Zhao, H., Li, M., & He, D. (2020). Identification of Apple Tree Leaf Diseases Based on Deep Learning Models. Symmetry, 12(7), 1065. https://doi.org/10.3390/sym12071065






