Synchronized Oscillations in Double-Helix B-DNA Molecules with Mirror-Symmetric Codons
Abstract
:1. Introduction
2. Dynamical DNA Model Hamiltonian
3. Dynamical Equations of Motion
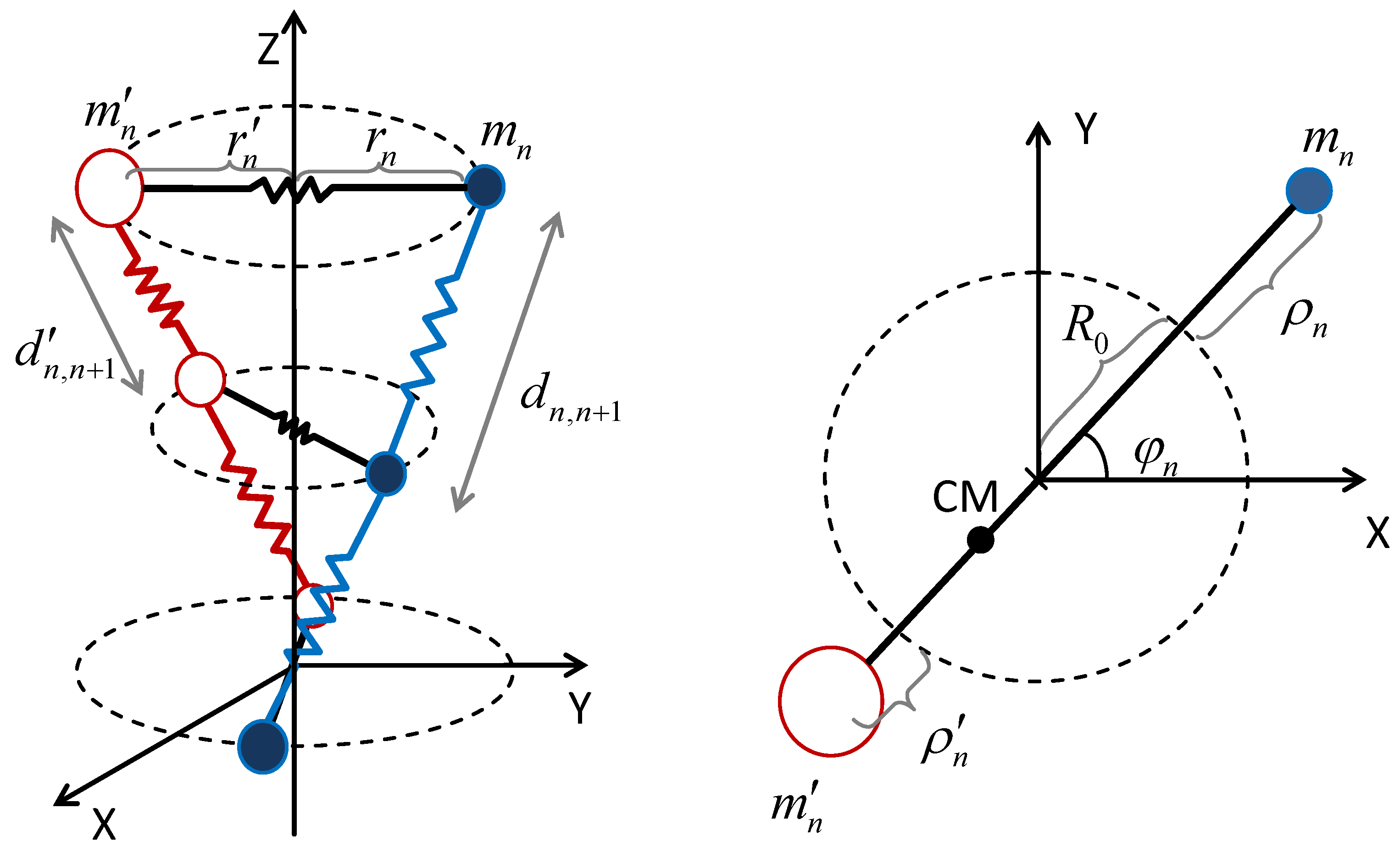
3.1. General Expressions
3.2. Dynamics of Homopolymer dsDNA Chains
3.3. Dynamics of Poly(XX’)-Poly(X’X) dsDNA Molecules
3.4. Dynamics of Poly(XYX)-Poly(X’Y’X’) dsDNA Molecules
4. Helical Waves Related Dispersion Relations
5. Orchestrated Codon Oscillations
6. Environmental Effects in the DNA Dynamics
7. Conclusions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| dsDNA | double-stranded DNA |
| bp | base pair |
| A | adenine |
| C | cytosine |
| G | guanine |
| T | thymine |
| PDB | Peyrard–Dauxois–Bishop |
References
- Chakarborty, T. (Ed.) Charge Migration in DNA: Perspectives from Physics, Chemistry and Biology; Springer: Berlin, Germany, 2007. [Google Scholar]
- Treadway, C.; Hill, M.G.; Barton, J.K. Charge transport through a molecular pi-stack: Double helical DNA. Chem. Phys. 2002, 281, 409–428. [Google Scholar] [CrossRef]
- Zwang, T.J.; Tse, E.C.M.; Barton, J.K. Sensing DNA through DNA Charge Transport. ACS Chem. Biol. 2018, 13, 1799–1809. [Google Scholar] [CrossRef] [PubMed]
- Starikov, E.B.; Fujita, T.; Wanatabe, H.; Sengoku, Y.; Tanaka, S.; Wenzel, W. Effects of molecular motion on charge transfer/transport through DNA duplexes with and without base pair mismatch. Mol. Simul. 2006, 32, 759–764. [Google Scholar] [CrossRef]
- Berlin, Y.A.; Grozema, F.C.; Siebbeles, L.D.A.; Ratner, M.A. Charge transfer in donor-bridge-acceptor systems: Static disorder, dynamic fluctuations, and complex kinetics. J. Phys. Chem. C 2008, 112, 10988–11000. [Google Scholar] [CrossRef]
- Starikov, E.B.; Quintilla, A.; Nganou, C.; Lee, K.H.; Cuniberti, G.; Wenzel, W. Single-molecule DNA conductance in water solutions: Role of DNA low-frequency dynamics. Chem. Phys. Lett. 2009, 467, 369–374. [Google Scholar] [CrossRef]
- Bruinsma, R.; Grüner, G.; D’Orsogna, M.R.; Rudnick, J. Fluctuation-facilitated charge migration along DNA. Phys. Rev. Lett. 2000, 85, 4393–4396. [Google Scholar] [CrossRef] [Green Version]
- Maciá, E. Base-Pairs’ correlated oscillation effects on the charge transfer in double-helix B-DNA molecules. Materials 2020, 13, 5119. [Google Scholar] [CrossRef]
- Peyrard, M. Nonlinear dynamics and statistical physics of DNA. Nonlinearity 2004, 17, R1–R40. [Google Scholar] [CrossRef]
- Peyrard, M.; Bishop, A.R. Statistical mechanics of a nonlinear model for DNA denaturation. Phys. Rev. Lett. 1989, 62, 2755–2758. [Google Scholar] [CrossRef]
- Dauxois, T.; Peyrard, M. Entropy-driven transition in a one-dimensional system. Phys. Rev. E 1995, 51, 4027–4040. [Google Scholar] [CrossRef]
- Peyrard, M.; Cuesta-López, S.; Angelov, D. Experimental and theoretical studies of sequence effects on the fluctuation and melting of short DNA molecule. J. Phys. Condens. Matter 2009, 21, 034103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shinwari, W.; Deen, M.J.; Starikov, E.B.; Cuniberti, G. Electrical Conductance in Biological Molecules. Adv. Funct. Mater. 2010, 20, 1865–1883. [Google Scholar] [CrossRef] [Green Version]
- Barbi, M.; Cocco, S.; Peyrard, M. Helicoidal model for DNA opening. Phys. Lett. A 1999, 253, 358–369. [Google Scholar] [CrossRef]
- Cocco, S.; Monasson, R. Statistical mechanics of torque induced denaturation of DNA. Phys Rev. Lett. 1999, 83, 5178–5181. [Google Scholar] [CrossRef] [Green Version]
- Agarwal, J.D.; Hennig, D. Breather solutions of a nonlinear DNA model including a longitudinal degree of freedom. Physica A 2003, 323, 519–533. [Google Scholar] [CrossRef]
- Krisch, M.; Mermet, A.; Grimm, H.; Forsyth, V.T.; Rupprecht, A. Phonon dispersion of oriented DNA by inelastic X-ray scattering. Phys. Rev. E 2006, 73, 061909. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Torrellas, G.; Maciá, E. Twist–radial normal mode analysis in double-stranded DNA chains. Phys. Lett. A 2012, 376, 3407–3410. [Google Scholar] [CrossRef]
- Maciá, E. Electrical conductance in duplex DNA: Helical effects and low-frequency vibrational coupling. Phys. Rev. B 2007, 76, 245123. [Google Scholar] [CrossRef] [Green Version]
- Dauxois, T.; Peyrard, M.; Bishop, A.R. Entropy-driven DNA denaturation. Phys. Rev. E 1993, 47, R44–R47. [Google Scholar] [CrossRef]
- Cocco, S.; Monasson, R. Theoretical study of collective modes in DNA at ambient temperature. J. Chem. Phys. 2000, 112, 10017–10033. [Google Scholar] [CrossRef] [Green Version]
- Campa, A.; Giansanti, A. Experimental tests of the Peyrard-Bishop model applied to the melting of very short DNA chains. Phys. Rev. E 1998, 58, 3585–3588. [Google Scholar] [CrossRef] [Green Version]
- Cule, D.; Hwa, T. Denaturation of Heterogeneous DNA. Phys. Rev. Lett. 1997, 79, 2375–2378. [Google Scholar] [CrossRef] [Green Version]
- Lee, O.; Jeon, J.H.; Sung, W. How double-stranded DNA breathing enhances its flexibility and instability on short length scales. Phys. Rev. E 2010, 81, 021906. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michoel, T.; Van de Peer, Y. Helicoidal transfer matrix model for inhomogeneous DNA melting. Phys. Rev. E 2006, 73, 011908. [Google Scholar] [CrossRef] [Green Version]
- Okonogi, T.M.; Alley, S.C.; Harwood, E.A.; Hopkins, P.B.; Robinson, B.H. Phosphate backbone neutralization increases duplex DNA flexibility. Proc. Natl. Acad. Sci. USA 2002, 99, 4156–4160. [Google Scholar] [CrossRef] [Green Version]
- Duduială, C.I.; Wattis, J.A.D.; Dryden, I.L.; Laughton, C.A. Nonlinear breathing modes at a defect site in DNA. Phys. Rev. E 2009, 80, 061906. [Google Scholar] [CrossRef] [Green Version]
- Zdravković, S.; Satarić, M.V. Single-molecule unzippering experiments on DNA and Peyrard-Bishop-Dauxois model. Phys. Rev. E 2006, 73, 021905. [Google Scholar] [CrossRef]
- Ghorbani, M.; Rafiee, F.M. Geometrical correlations in the nucleosomal DNA conformation and the role of the covalent bonds rigidity. Nucleic Acids Res. 2011, 39, 1220–1230. [Google Scholar] [CrossRef] [Green Version]
- Barbi, M.; Lepri, S.; Peyrard, M.; Theodorakopoulos, N. Thermal denaturation of a helicoidal DNA model. Phys. Rev. E 2003, 68, 061909. [Google Scholar] [CrossRef] [Green Version]
- Leal, M.R.; Weber, G. Sharp DNA denaturation in a helicoidal mesoscopic model. Chem. Phys. Lett. 2020, 755, 137781. [Google Scholar] [CrossRef]
- Maciá-Barber, E. Aperiodic Structures in Condensed Matter: Fundamentals and Applications; Taylor and Francis, CRC Press: Boca Raton, FL, USA, 2009; pp. 209–297. [Google Scholar]
- Maciá, E. Charge transfer in DNA: Effective Hamiltonian approaches. Z. Kristallogr. 2009, 224, 91–95. [Google Scholar] [CrossRef] [Green Version]
- Mantela, M.; Lambropoulos, K.; Theodorakou, M.; Simserides, C. Quasi-Periodic and Fractal Polymers: Energy Structure and Carrier Transfer. Materials 2019, 12, 2177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lambropoulos, K.; Simserides, C. Periodic, quasiperiodic, fractal, Kolakoski, and random binary polymers: Energy structure and carrier transport. Phys. Rev. E 2019, 99, 032415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lambropoulos, K.; Simserides, C. Tight-binding modeling of nucleic acid sequences: Interplay between various types of order or disorder and charge transport. Symmetry 2019, 11, 968. [Google Scholar] [CrossRef] [Green Version]
- Landi, A.; Borrelli, R.; Capobianco, A.; Peluso, A. Transient and enduring electronic resonances drive coherent long distance charge transport in molecular wires. J. Phys. Chem. Lett. 2019, 10, 1845–1851. [Google Scholar] [CrossRef]
- Behnia, S.; Fathizadeh, S.; Javanshour, E.; Nemati, F. Light-driven modulation of electrical current through DNA sequences: Engineering of a molecular optical switch. J. Phys. Chem. B 2020, 124, 3261–3270. [Google Scholar] [CrossRef] [PubMed]
- Maciá, E. π-π orbital resonance in twisting duplex DNA: Dynamical phyllotaxis and electronic structure effects. Phys. Rev. B 2009, 80, 125102. [Google Scholar] [CrossRef] [Green Version]
- Young, M.A.; Ravishanker, G.; Beveridge, D.L. A 5-nanosecond molecular dynamics trajectory for B-DNA: Analysis of structure, motions, and solvation. Biophys. J. 1997, 73, 2313–2336. [Google Scholar] [CrossRef] [Green Version]
- Okaly, J.B.; Mvogo, A.; Woulaché, R.L.; Kofané, T.C. Nonlinear dynamics of damped DNA systems with long-range interactions. Commun. Nonlinear Sci. Numer. Simulat. 2018, 55, 183–193. [Google Scholar] [CrossRef] [Green Version]
- Okaly, J.B.; Mvogo, A.; Tabi, C.B.; Ekobena Fouda, H.P.; Kofané, T.C. Base pair opening in a damped helicoidal Joyeux-Buyukdagli model of DNA in an external force field. Phys. Rev. E 2020, 102, 062402. [Google Scholar] [CrossRef]
- Harris, S.A.; Laughton, C.A. A simple physical description of DNA dynamics: Quasi-harmonic analysis as a route to the configurational entropy. J. Phys. Condens. Matter 2007, 19, 076103. [Google Scholar] [CrossRef] [PubMed]
- Fischer, B.M.; Walther, M.; Uhd Jepsen, P. Far-infrared vibrational modes of DNA components studied by terahertz time-domain spectroscopy. Phys. Med. Biol. 2002, 47, 3807. [Google Scholar] [CrossRef] [PubMed]

| Geometrical | Dynamical | Potential |
|---|---|---|
| rad | amu | Å [15,30] |
| nm | Å [14,30,31] | |
| nm | eV [21,30,31] | |
| nm | amu | eV Å [21] |
| nm | amu | eV Å [30] |
| Oscillation | Codon | ( rad s) | (THz) | (ps) | |
|---|---|---|---|---|---|
| Twist | |||||
| Stacking | |||||
| Stretching | |||||
| Twist-Stacking | X’XX’ | ||||
| YXY | |||||
| XXX | |||||
| XYX | |||||
| XX’X | |||||
| Twist-Stretching | XXX | ||||
| Twist-Stacking-Stretching | XX’X | ||||
| XYX | |||||
| YXY | |||||
| X’XX’ | |||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maciá, E. Synchronized Oscillations in Double-Helix B-DNA Molecules with Mirror-Symmetric Codons. Symmetry 2021, 13, 241. https://doi.org/10.3390/sym13020241
Maciá E. Synchronized Oscillations in Double-Helix B-DNA Molecules with Mirror-Symmetric Codons. Symmetry. 2021; 13(2):241. https://doi.org/10.3390/sym13020241
Chicago/Turabian StyleMaciá, Enrique. 2021. "Synchronized Oscillations in Double-Helix B-DNA Molecules with Mirror-Symmetric Codons" Symmetry 13, no. 2: 241. https://doi.org/10.3390/sym13020241






