1. Introduction
Clustering analysis is a main technique used to solve the problems that exist in data mining. It aims to classify data points into several groups called clusters so that similar elements are clustered into one group, while different elements are separated from each other [
1]. Because clustering can be used to deeply mine internal and possible knowledge, rules, and patterns, it has been applied to many practical fields, including data mining, pattern recognition, machine learning, information retrieval, and image analysis [
2,
3,
4]. Clustering has been widely and intensely studied by the data mining community in recent years. Many clustering algorithms have been proposed: K-means [
5], fuzzy C-means [
6], affinity propagation [
7], Gaussian mixtures [
8], etc. These algorithms perform well on many occasions, but they cannot deal with datasets with arbitrary shapes, and the determination of hyperparameters and clustering centers is also difficult for clustering algorithms.
Density-based clustering methods (such as DBSCAN) [
9,
10] consider clusters as high-density regions separated by low-density regions. DBSCAN can process datasets with arbitrary shapes. However, it is sensitive to the density radius parameter, and small parameter changes can lead to widely different clustering results. In 2014, Alex and Rodriguez proposed the density peak clustering (DPC) algorithm [
11] in
Science, a novel fast density peak clustering algorithm that can recognize clusters with arbitrary shapes without setting the number of clusters in advance. Compared to a traditional clustering algorithm, DPC can quickly identify cluster centers, needs few parameters, has a fast clustering speed, and can be used with datasets with different shapes. Therefore, several research studies [
12,
13,
14,
15,
16,
17,
18] have been performed focusing on this method.
Although the DPC algorithm and its improved version have achieved great success and proved their high performance in practical applications, they can only be used to process complete data. However, in many practical applications, we may encounter missing features for various reasons (such as sensor failure, measurement error, unreliable features, etc.).
Researchers have expended considerable effort to study the problem of incomplete data. Some methods have been proposed for supervised learning and unsupervised learning [
19,
20,
21,
22,
23]. A simple way is to discard those missing values by eliminating instances that contain missing values before clustering. More commonly, the missing data are imputed by an imputation algorithm (such as zero imputation, mean imputation, KNN imputation, or EM imputation [
24,
25,
26]) and then clustered by the original clustering algorithm. However, the clustering performance on incomplete data is much lower. Recently, imputation and clustering were considered in the same framework [
23,
27] to improve the clustering performance on incomplete data, but most of these methods cannot handle incomplete data with arbitrary shapes well. In addition, the imputation-based method does not work well in the DPC algorithm because of the aggregation phenomenon of imputed points. The cluster centers in DPC have a higher density than their neighbors, and the different cluster centers are far away from each other. The example in
Figure 1 illustrates the aggregation phenomenon. The five-cluster dataset is a two-dimensional synthetic dataset that contains five clusters. We can accurately obtain five clusters and their centers (the cross mark in
Figure 1a) by executing the DPC algorithm on the five-cluster dataset; the results are shown in
Figure 1a. For comparison, we can generate missing data by randomly deleting 25% of all features in the five-cluster dataset. Then, the missing feature data points are imputed by the mean imputation method and the DPC algorithm is executed on the missing feature data. The clustering results are shown in
Figure 1b. As shown in
Figure 1b, the two straight lines in the middle area are the missing feature points imputed by mean imputation, and these imputed data points form some high-density areas, which breaks the assumption of the DPC algorithm. Thus, applying DPC on incomplete data can lead to the wrong centers being chosen (the cross marks in
Figure 1b) and to a very poor clustering result. Other advanced imputation algorithms (such as KNN imputation) are also likely to break the assumption of the DPC algorithm and lead to the aggregation phenomenon of imputed data points.
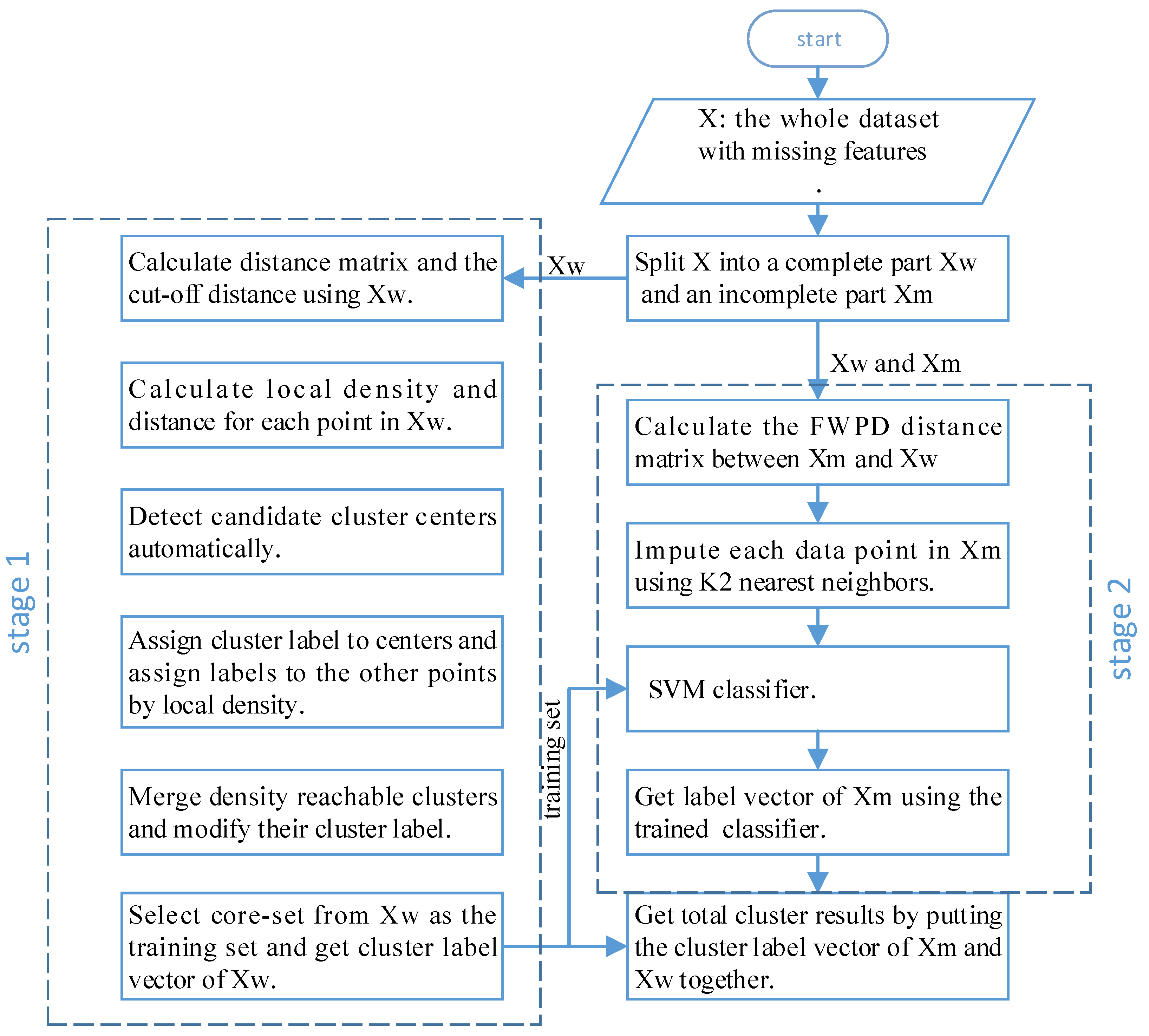
Classification algorithms are highly accurate and can accurately learn the distribution of data, but they need labeled training data. Clustering algorithms do not need data with labels, but the accuracy of the learned model will not be particularly high. In addition, during the label assignment process of DPC, a point is always assigned the same label as the nearest neighbor with a higher density, and errors propagated in the subsequent assignment process. Incomplete points with many missing features are more likely to be assigned a wrong label and propagate errors. Therefore, to avoid the above problems and improve the clustering performance when features of data points are missing, in this paper, we propose the use of a novel two-stage clustering algorithm based on DPC. In stage 1, based on the idea of training a classification model using clustering results, a batch of labeled data is first obtained by density peak clustering, then some representative points are selected. In stage 2, a powerful classifier is trained using these representative points. Then, a symmetrical FWPD distance matrix for incomplete data points is computed, after that, the missing feature data are imputed by the FWPD distance matrix and classified by the classifier. Our algorithm works well with incomplete data while maintaining the advantages of the DPC algorithm. The main diagram of the proposed method is shown in
Figure 2. We conducted extensive experiments on three synthetic datasets and six UCI benchmark datasets; the experimental results showed the effectiveness of our proposed algorithm.
The main contributions of this article are summarized as follows:
(1) To the best of our knowledge, this is the first work about clustering on incomplete data of arbitrary shapes. To solve this problem, we propose combining clustering with classification. The classifier has a high accuracy but it needs labeled data. In stage 1, we use the density clustering method to obtain labeled data, then select some high-quality labeled data to train the classifier in stage 2. Stage 2 relies on the samples from stage 1. Although the original DPC can deal with irregular data, we introduced some improvements, such as improving the local density formula, making it automatically select centers, and merging clusters. Therefore, the clustering performance in stage 1 has obvious advantages. The classifier in stage 2 retains advantages and improves the clustering performance.
(2) Based on the above ideas, we propose a density peak clustering method for incomplete data named DPC-INCOM. The traditional DPC method fails when handling incomplete data due to the aggregation phenomenon. DPC-INCOM can deal with this kind of situation well.
(3) We conducted extensive experiments on three synthetic datasets and six UCI benchmark datasets. Our algorithm achieves superior performance compared to other imputation-based methods.
The rest of this paper is organized as follows:
Section 2 describes DPC and reviews related work. Then, we propose the DPC-INCOM method in
Section 3.
Section 4 discusses the experimental results. We draw conclusions in
Section 5.
3. Density Peak Clustering with Incomplete Data
3.1. The Problem of Incomplete Data Clustering
The problem of clustering with incomplete data has been the subject of several prior studies [
10,
29].
Table 1 provides the list of notations used in this paper. Given a set of objects
represented by a numerical data matrix
,
is the
kth data vector that describes the object
by specifying values for M particular features. The
jth component of the data vector
is denoted by
. In practice, some features of certain data points may be missing in some cases. Such data can be called incomplete data. For example,
is incomplete datum, where
and
are missing. Otherwise, the point is called complete datum. Any dataset containing such points (missing values) can be regarded as an incomplete dataset. For convenience of expression, we use
to note that data that no features are missing in the dataset and
to note the incomplete part of
.
Problem Statement. Given a dataset with N unlabeled points, each point has M dimensions (features). Based on the features, the clustering algorithm aims to find a set of clusters , where each can mapped to one cluster and . The points in the same cluster should have similar features, while the points far away should never appear in the same cluster. Accordingly, we denote as the cluster label of sample . After, a cluster label vector can be used to represent the clustering result with N elements.
When using the DPC algorithm to process incomplete data, it makes sense to impute the missing features first and then apply the DPC algorithm for imputed data. However, the use of a simple imputation-based method (zero imputation, mean imputation, or median imputation) may lead to the aggregation phenomenon, where some imputed missing feature data points gather and form a high-density area. Thus, their density exceeds that of the original clustering centers, leading to a poor clustering result. Complex imputation methods such as KNN imputation can also suffer from the aggregation phenomenon.
To extend the DPC algorithm to incomplete data, we designed the DPC-INCOM method, which can work well on incomplete data and automatically identify cluster centers without manually setting the number of clusters
k. The proposed method divides the clustering problem into two major stages: Stage 1 uses improved DPC to obtain a batch of labeled data samples. The details of this process can be found in
Section 3.2 and
Section 3.3. In stage 2, the labeled data points with a high local density are selected as a core set, a proper classification model is selected, and the labeled training dataset is fed to train the model. Then, missing feature data samples are imputed with K nearest neighbors. Finally, the trained classifier is used to classify the imputed missing feature data.
Section 3.4 provides a detailed description of Stage 2.
3.2. Improved Density Formula
Ref. [
13] proposes an improved DPC, which overcomes the shortcoming of DPC: the original algorithm cannot effectively identify the center of low-density clusters. The improved local density is calculated by K nearest neighbors. Therefore, not much difference exists between the local density of the center of a low-density cluster and the local density of the center of a high-density cluster, and this improved algorithm no longer needs to set the parameter
. The improved local density
of the
ith data point is defined as:
where
is the number of data points in
and
is the distance between data point
i and its
Kth nearest neighbor, being defined as
.
is the
K nearest neighbors of data point
i.
is the Euclidean distance between data points
i and
j. Our method borrows this improved density formula to improve the performance of Stage 1. Note that any improvement in DPC can be applied in Stage 1.
3.3. Automatic Selection of Centers and Cluster Merging
A decision graph must be established in the original DPC algorithm and then the cluster centers must be manually selected. If the cutoff distance parameter is incorrect, the decision graph may be incorrectly built. In addition, the original algorithm assumes that there is only one density peak in one cluster. If there are multiple density peaks, DPC cannot cluster correctly. The subsequent assignment procedure will propagate errors, but no action is taken in the DPC to fix them. It should also be pointed out that the initial cluster centers are selected manually rather than automatically. However, it is difficult to obtain the correct selection in some datasets. For the same decision graph, different people choose different cluster centers, and the clustering results will therefore different. To solve these problems, we propose a method that involves automatically selecting cluster centers and automatically merging density reachable clusters.
3.3.1. Candidate Centers
According to the local density
and distance
, we set a threshold and then select the candidate centers according to this threshold. Candidate centers are defined as:
where
and
are the local density and distance of the
ith data point
, respectively, and
is the representative data point.
is the product of the local density and distance of
, and
should be larger than the other
of the data points.
is a constant number and can usually be set as 1. Using this threshold, we can select points with a relatively high local density and distance as candidate cluster centers, but there may be more candidate centers than actual centers. We can fix this problem through cluster merging.
3.3.2. Cluster Merging
Usually, the number of candidate cluster centers is greater than the number of actual cluster centers, especially when a cluster contains multiple density peaks, so we need to merge the clustering results. In terms of our requirements, some of the concepts utilized in DBSCAN are redefined below. These concepts defined for objects are extended to clusters.
Definition 1 (Border points between two clusters).
The border points between two clusters and , denoted by , are defined as:where is the Euclidean distance from to . Obviously, . Definition 2 (Border density of two clusters).
The border density between cluster and cluster , denoted by , is defined as:where is the local density of . Obviously, and the amount of computation can be halved by using the symmetry property. Definition 3 (Density directly reachable).
The density of cluster is directly reachable from cluster with respect to border density if:where and are the average local density of clusters and . Definition 4 (Density reachable). Cluster is density reachable by if there is a chain of clusters , such that is density directly reachable from .
The density reachable is symmetric and transitive. To show its veracity, one can apply mathematical induction.
Here is an example of density reachable: As shown in
Figure 3, there are three clusters, A, B, and C. In the enlarged subgraph in the lower-left corner, there are four points, e, f, g, and h, of which e and g are from cluster C, and f and h are from cluster B. It can be seen that f’s circle contains points e and g from cluster C. Therefore, e, g, and f are the border points of cluster B and C. If the average density of all border points of clusters B and C is larger than the average density of cluster B or C, then clusters B and C are density-directly-reachable. If cluster A and B are density-directly-reachable and cluster B and C are density-directly-reachable, then we can say that clusters A, B, and C are density reachable.
Based on the concept of density reachable, we can build a graph model for all cluster centers. The cluster centers are the nodes of the graph. If both two centers are density-directly-reachable, they have a connected edge. We can calculate all connected components using the graph model based on the DFS algorithm and merging the connected clusters. As shown in the example in
Figure 3, three candidate centers are selected using (
7). Let
,
, and
represent the centers of cluster A, B, and C, respectively. Then, we can build a graph model with three nodes,
,
, and
. If clusters B and C are density reachable, then
and
have an edge. If clusters A and B are not density reachable, then
and
have no edge. Then, applying the DFS algorithm on this graph model, we can obtain a connected component with two nodes,
and
. Therefore, clusters B and C can be merged into one.
Based on the above improvements, we can use the improved DPC algorithm to cluster and obtain the cluster label comH.
3.4. Training the Classifier with Core-Set
The advantage of classification algorithms is their high accuracy, but some data points identified by category labels need to be used as a training set. Marking labels manually is a time-consuming task. Clustering algorithms do not need labeled data. However, without providing labels, the learned model will not be accurate enough. The method detailed in
Section 3.2 and
Section 3.3 provides a batch of labeled data. We can use these data to train the classifier, and then the classifier will determine to which cluster each missing feature data point belongs.
3.4.1. Distance Measurement of Incomplete Data Points
To calculate the
K nearest neighbors of the incomplete data point
, we should first know how to calculate the distance between data points with missing features. The partial distance (PDS) of incomplete data was proposed in [
29]. The distance is calculated according to the visible part of the data point and then multiplied by a factor calculated by the number of missing dimensions. Datta proposed the use of the FWPD [
30] distance to directly calculate the distance of incomplete data points and directly applied the FWPD distance to the K-means clustering algorithm to deal with incomplete datasets. However, this method cannot handle incomplete data with an arbitrary shape. FWPD distance is the weighted sum of two terms: the first is the observed distance of two data points, and the second is a penalty term. The penalty term is a weighted sum of the penalty coefficients of all missing dimensions, and the FWPD distance is a weighted sum of the observed distance and the penalty term. It is denoted by
and calculated as follows:
where
is the observed distance of data points
and
and
S is the set of all feature dimensions.
and
are the sets of feature dimensions that can be observed by
and
, respectively.
belongs to
, which is the number of data points in
with the observed values of the
lth feature. Obviously, if a feature observed in most samples is missing in a specific sample, this weighting scheme will impose a greater penalty. On the other hand, if the missing feature is not observed in many other samples, a smaller penalty will be imposed. Utilizing the FWPD distance, we can calculate the K nearest complete neighbors of the missing data point
and then use the mean value of the corresponding dimension of the K neighbors to impute the missing feature.
3.4.2. Training Classifier
Definition 5 (Core set of a cluster).
The core set of a cluster , denoted by , is defined as: The core set of a cluster contains data points that have a relatively higher local density in the cluster, meaning it can better represent the distribution of the cluster. Using these points as a training set can lead to obtaining a higher-accuracy classifier. A core set is selected from the clustering results of , then the core set is fed to a proper classification model (such as SVM). When the trained classifier is obtained, the classifier can be applied to classify the imputed missing feature data points. The final clustering result is obtained by combining the labels of and the labels of the imputed missing data assigned by the classifier.
3.5. Main Steps of DPC-INCOM
Algorithm 1 provides a summary of the proposed DPC-INCOM.
| Algorithm 1 DPC-INCOM. |
Input: data matrix with missing features, constant number , Output: cluster label vector H
- 1:
Divide into and - 2:
Use to cluster complete dataset , and obtain the cluster label vector - 3:
Calculate the FWPD distance matrix of data points in using ( 13) - 4:
Impute the missing features of all the data points in using nearest neighbors by the FWPD distance; obtain imputed denoted by - 5:
Select and from and as training sets using ( 14) - 6:
Training a SVM classifier with and - 7:
Obtain the label vector of denoted by using the trained SVM classifier - 8:
Obtain H by union ,
|
3.6. Complexity Analysis
In this section, we measure the computational complexity of the proposed algorithm.The complexity of the entire process is the sum of the complexities of the two stages: DPC clustering in
and imputation and classification in
. Suppose the dataset has
N points and is divided into a complete part with
points and an incomplete part with
points. In Stage 1, the time complexity of Algorithm 2 depends on the following aspects: (a) calculating the distance between points (
); (b) sorting the distance vector of each point (
); (c) calculating the cutoff distance
(
); (d) calculating the local density
with the
nearest neighbors(
); (e) calculating the distance
for each point (
); (f) selecting initial cluster centers and assigning each remaining point to the nearest point with a higher density (
); (g) calculating the border points of each cluster (
); (h) calculating the border density of each cluster and merge density-reachable clusters (
). Therefore, the complexity of Stage 1 is
, which is same as that of basic DPC [
11]. In Stage 2, the time complexity depends on the following aspects: (a) calculating the FWPD distance between the incomplete points and the complete points (
); (b) imputing all the incomplete points using
K2 nearest neighbors
; (c) selecting a core set from the complete points
; (d) training a SVM [
33] classifier with core set
. When the dataset is large, the complexity can be reduced by switching to an appropriate classifier. The above analysis demonstrates that the overall time complexity of the proposed algorithm is
.
| Algorithm 2AKdensityPeak. |
- 1:
Calculate the cut-off distance using ( 4) - 2:
Calculate the local density using nearest neighbors and distance for each point in using ( 6) and ( 3), respectively - 3:
Select candidate cluster centers automatically using ( 7) - 4:
Assign cluster labels from candidate centers to low-density points - 5:
Calculate border points of clusters for each cluster using ( 8) - 6:
Calculate the border density of clusters for each cluster using ( 9) - 7:
Find all density reachable clusters using the DFS algorithm and merge these clusters - 8:
Return cluster label vector
|