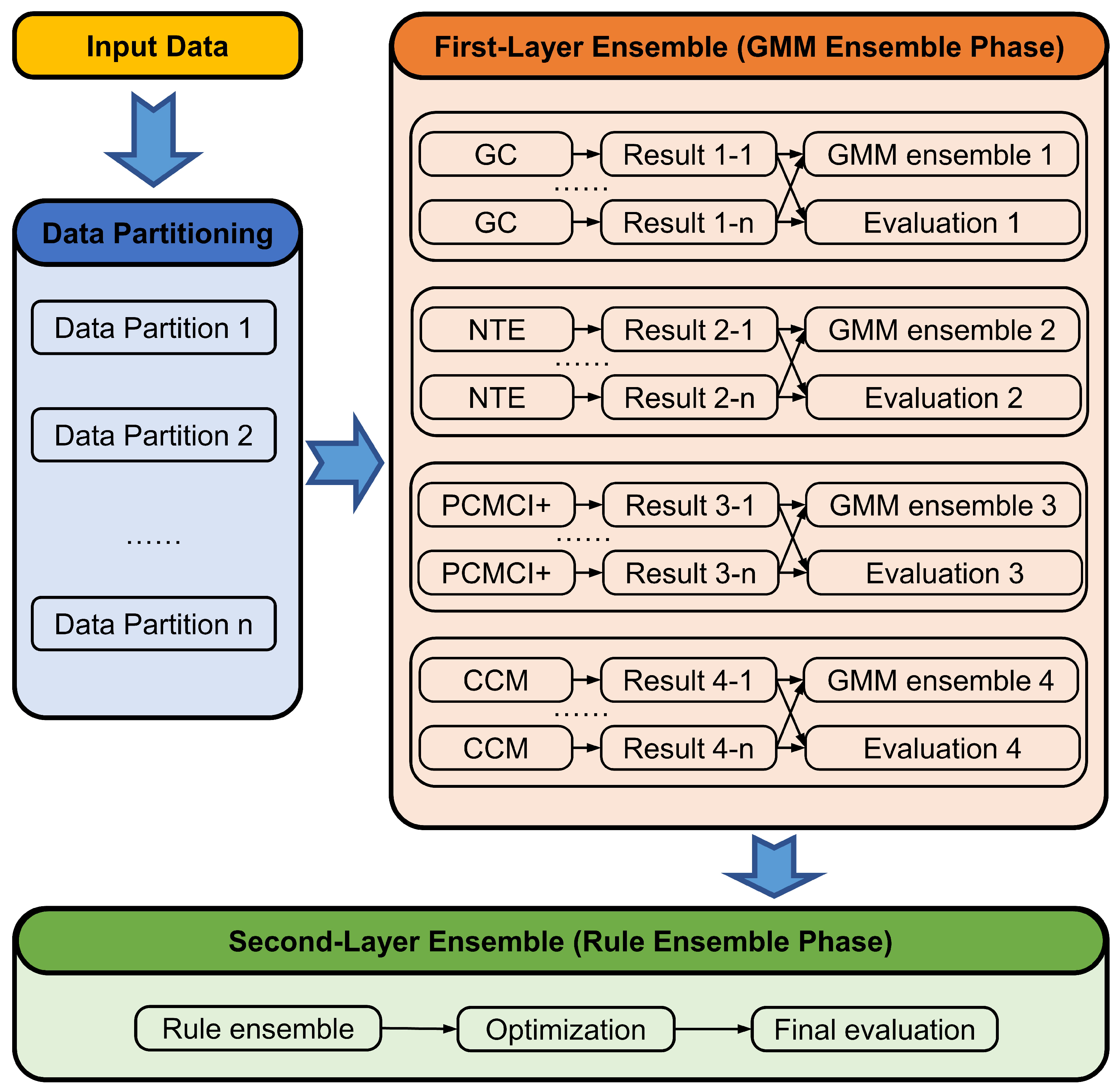
3.1. General Pipeline
To improve the accuracy of existing causal inference approaches, we propose a two-phase multi-split causal ensemble model to derive the full benefit of the four causal inference algorithms (GC, NTE, PCMCI+, and CCM). In many experiments [
38,
39], multiple algorithms often lead to divergent causality conclusions when processing the same dataset, which affects the trustworthiness of causal inference results. The proposed ensemble framework aims to produce more robust results by integrating multiple causal detectors. Sources of noise can often temporarily affect time series, leading to data which are unrepresentative of the underlying causal mechanisms in sections of the time series. This can lead to wrongly inferred causal relationships if the time series is examined as a whole.
To circumvent the mentioned challenges, our model follows a split-and-ensemble approach as
Figure 1 shows, meaning we first partition the given data, apply causal inference methods to each partition, and, finally, combine the results. The split-and-ensemble process makes full use of the data and reduces the impacts of noise in the datasets. In the data partitioning step, the time series are split into many partitions with an overlapping partitioning process. After each base algorithm has been applied to each partition, the first ensemble phase uses a GMM to integrate the causal results of each base algorithm from different data partitions in order to improve the robustness of each base algorithm. This leads to one combined causal inference result for each base algorithm, and the trustworthiness assessment is conducted to check the stability of the result. Therefore, the first-layer ensemble outputs four combined causal inference results associated with the base learners, as well as their evaluation results, which is demonstrated in the GMM ensemble phase of
Figure 1. The second ensemble phase combines these results through three decision-making rules considering the first-layer evaluation results, which take advantage of the strengths of diverse causal inference algorithms and make the results more robust.
Finally, the ensemble result is optimized by removing the indirect causal links and the credibility of the final result is evaluated by a proposed assessment coefficient. The overall process is illustrated in
Figure 1 and formalized in Algorithm 1. The following sections will explain the individual steps of the algorithm which have been briefly introduced above.
3.3. GMM Ensemble Phase
After the partitioning, the four base learners are applied to process each data partition separately and in parallel. After partitioning of the data into
n partitions, each partition is processed by the four individual models, and
causal strength matrices are obtained (see
Figure 1). Next, the results from each individual causal inference method are combined using a Gaussian mixture model (GMM).
When using GMMs for clustering, each Gaussian component represents a cluster, and the corresponding weight represents the probability that a data point belongs to said cluster [
40]. A datum point is assigned to the cluster with the highest probability, which is called the soft assignment. Unlike the k-means algorithm, which can only detect spherical clusters, the GMM can process elliptical or oblong clusters. Moreover, in contrast to the soft assignments of GMM clustering, the k-means algorithm can only perform hard assignments. Since the transformed dataset at this stage is oblong in shape, GMM clustering is the more suitable approach to ensemble causal models. In this ensemble learning framework, the relationships between two variables are divided into two groups, causal and non-causal. Therefore, the number of components of the GMM is set as two.
In this ensemble phase, a two-component GMM is fitted to ensemble the causal strength matrices from the same causal detector but from different data partitions.
Figure 3 visualizes the steps of processing the observations in this phase in a dataset with
n features being split into
K partitions. In this example, the results are
K causal strength matrices with sizes that are all
after the processing of any of the four base learners. The
(
...
)(
...
) represents the causal strength from feature
i to feature
j for the
k-th partition, where
. Before the post-processing, the matrices are filtered in the first step as Equation (
16) shows, because the causal strengths in the four base learners are all based on the correlation coefficient (CC). In order to not be too restrictive too early, two features are considered a low-correlation pair and the causal inference is weakly trustworthy if
. Equation (
16) shows the processing of weakly trustworthy causal links.
In the second step, all the
K matrices are flattened to vectors with size
, where
n self-adjacent values are removed. In other words, the diagonal of the matrix is removed because this model solely analyzes the relationships between different variables. In the time series, the future values of a feature are assumed to be dependent on the past values, so there is a causal relationship within the feature. However, it is not shown in the causality model as the model is the rolled-up version without unrolling in the time dimension. Then, the data points with the same position indices are integrated into one data instance with
K dimensions. Thus, a new dataset
containing
K-dimensional instances is created as the input of the GMM clustering, which is illustrated in
Figure 3 (step 2). The left numbers are the indices of the instance in the
, and the right tuples represent the position of the
where the values are generated. For instance, the first instance in the
is
...
. The
m-th instance in the
is labeled
, where
m is the left index and
refers to the right index in
Figure 3 (step 2).
Next, in the third step, the GMM with two components is fitted to cluster the input data. The Gaussian with the lower mean is denoted as the cluster for the causal strength data from the feature pairs without causal relationships, which is labeled “Group 0”. The Gaussian with the higher mean represents the cluster containing the strength values of causal pairs, which is labeled “Group 1”. All of the
inputs are assigned to the two groups by the fitted GMM. Finally, each clustered
K-dimensional data instance is mapped to a single floating point value. In
Figure 3 (step 3),
...
is the set containing the instances in “Group 0” and
is the corresponding mapping function.
...
is the set including the data points in “Group 1” and the corresponding mapping function is
.
where
is a function to calculate the median of non-zero values in a vector. For instance, assume
...
, and
...
is the set containing the values of the coordinate of the vector
. Define
by removing all elements valued 0.
where
denotes the median of the values in the set
. In this instance, the median is selected instead of the mean because it represents the values’ average information without the influence of outliers.
After the processing, an ensemble causal strength matrix
is produced separately for each base learner, and its size is
, which is shown in
Figure 3 (step 5). The values of
...
are set at 0 because causal detection does not consider self-related pairs as discussed before. The values of
...
rely on the ensemble processing and correspond to the instance
(
Figure 3 (step 2)), which is processed following Equation (
20).
where
is based on Equation (
17) and
is derived from Equation (
18).
It is necessary to evaluate the trustworthiness of the causal relationships from the GMM ensemble phase and make preparations for the next ensemble phase in
Section 3.3. To accomplish that goal, an assessment pipeline is designed, as
Figure 4 illustrates.
The trustworthiness matrix
with the size
is proposed corresponding to the ensemble causal strength matrix
. The elements are then computed by the trustworthiness index in Equations (
21) and (
22). If the value of
is 0, the corresponding element
is set at 0 as well. This is because, under the requirements of the following rule ensemble phase, only causal relationships require assessment. If the value of
is not 0, the value of
is set by Equation (
22).
where
where
is a variable containing
K elements. This variable is transformed from the
K-dimensional data point in the dataset of step 2 in
Figure 4, which is derived from the elements
, where
...
.
denotes the mean of non-zero values in
,
represents the standard deviation of non-zero values in
,
m refers to the number of the non-zero values, and
K is the number of elements in
.
is an extremely small value (e.g.,
) used to avoid a divisor being equal to zero.
The range of any
is
. A value approaching 0 points to barely detectable causal relationships, whereas an extremely high value represents stable and strong causal relationships across all data partitions. In Equation (
22), a high mean represents a strong causal relationship. A high parameter
m indicates a good consistency in the causal detection among different partitions. A low standard deviation indicates the stability of causal inference because the values are distributed over a narrow range. Therefore, a higher value of
indicates greater trustworthiness in the GMM-ensemble processing.
Algorithm 2 outlines the procedure of the GMM ensemble phase.
| Algorithm 2 GMM ensemble phase |
| Input: causal strength matrices with the size (4 refers to the four base learners, K is the number of partitions, ... and ...) |
| Output: GMM-ensemble strength matrices with the size and trustworthiness matrices with the size () |
| 1: for all l in do | ▹ Run in parallel |
| 2: for all k in ... do |
| 3: flatten and remove values (...) to the size |
| 4: end for |
| 5: ← combine all flattened | ▹ the size of is |
| 6: (Group 0, Group 1) ← Run a GMM (number of components) to process |
| 7: for all in do |
| 8: if is in Group 0 then |
| 9: |
| 10: |
| 11: else |
| 12: | ▹ Equation (18) |
| 13: | ▹ Equation (22) |
| 14: end if |
| 15: end for |
| 16: end for |
3.4. Rule Ensemble Phase
The GMM ensemble phase is conducted to process the four base learners separately and in parallel. The outputs of this step are the four ensemble causal strength matrices , , , and , and the corresponding trustworthiness matrices , , , and with sizes that are all . These serve as the inputs of the rule ensemble phase, which is described in the following.
In this phase, the four intermediate ensemble matrices are integrated into one rule-ensemble matrix with the size based on three rules as follows. To determine the element (...), the set is created, and is denoted as the number of non-zero elements in . Before conducting the following rules, the is filtered to improve the credibility of the ensemble processing. Through a number of experiments, for any , if (), the corresponding element in the causal strength matrix is set at 0. This highlights that such detected cause–effect pairs can not be trusted.
Next, three ensemble rules are developed to combine the intermediate ensemble results. The formal definition is as follows. Rule 1 states that there is no causal link if more than half of the intermediate ensemble results indicate non causality. Similarly, as Rule 3 shows, when the majority of the intermediate results specify the causal relationship, the causality is confirmed. Rule 2 expresses the decision-making process without majority. The decisions are made based on the trustworthiness matrices and corresponding thresholds. Furthermore, the quantitative causality result, causal strength, is determined by a weighting strategy.
- Rule 1:
When or 1, set at 0.
- Rule 2:
When , suppose and .
If , set at .
If and , set at as well, otherwise set at 0.
- Rule 3:
When or 4, set at .
The parameters and are the relevant threshold. Based on many experiments, can be selected between 10 and 20, and can be chosen between and .
is the weighted causal strength based on Equation (
23).
where
refers to the normalized weight based on the trustworthiness matrix
(
), which ranges from 0 to 1. Furthermore, the
(
) represents the values in the GMM-ensemble causal strength matrix.
Algorithm 3 demonstrates the procedure of the rule ensemble phase.
| Algorithm 3 Rule ensemble phase |
| Input: GMM-ensemble strength matrices with the size and trustworthiness matrices with the size (...), the threshold and |
| Output: GMM-ensemble strength matrices with the size |
| 1: Initialize ... and representing the number of non-zero elements in |
| 2: for all ......do |
| 3: if then |
| 4: |
| 5: end if |
| 6: end for |
| 7: for all ...do |
| 8: if or 1 then | ▹ Rule 1 |
| 9: |
| 10: else if then | ▹ Rule 2 |
| 11: Select the largest element in and the second largest one |
| 12: if then |
| 13: | ▹ Computing in Equation (23) |
| 14: else if then |
| 15: |
| 16: else |
| 17: |
| 18: end if |
| 19: else | ▹ Rule 3 |
| 20: |
| 21: end if |
| 22: end for |
The three rules constitute the rule ensemble phase, where the causal pairs detected by multiple base learners with high trustworthiness scores are selected and embedded into the final causal strength matrix. The comprehensive evaluation and selection process enhances the reliability of the causal models and is more robust than majority voting and averaging.
3.5. Model Optimization
In the rule-ensemble result
, both direct and indirect causal relationships are present. In the applications, the direct causal relationships are more significant than the indirect ones for analyzing the system’s performance [
41]. Hence, the indirect causal links should be extracted and removed from the
.
Many experiments [
41] demonstrate that the indirect adjacency is weaker than the direct one. As
Figure 5 shows, for instance,
influences
directly, but
can only influence
through
.
denotes the causal strength from
to
. Under the conclusion from the experiments, for instance, the triple
contains the inequalities
and
.
is the weakest value among all causal strengths.
To remove the indirect links of the time series dataset ..., all triples (...) meeting the conditions of and in the rule-ensemble result are analyzed. If and , it indicates that is the indirect adjacency and should be removed from the causal links. In turn, this means that should be set at 0. The filtered matrix from is denoted as .
The optimization step marks the end of the causal discovery procedure. The final causal strength matrix , as well as the corresponding causal graph is then produced. In the next section, an evaluation index is proposed to assess the credibility of the ensemble model.