Human Mitochondrial Pathologies of the Respiratory Chain and ATP Synthase: Contributions from Studies of Saccharomyces cerevisiae
Abstract
1. Introduction
2. Yeast, a Model for Studying Mitochondrial Function and Biogenesis
3. Strategy for Determining the Function of Unknown Mitochondrial Proteins
- (1)
- Expression of one of the mitochondrial gene products of the COX complex;
- (2)
- Assembly of this complex; or
- (3)
- Maturation of the heme active centers of the enzyme.
4. Pathological Mutations in Respiratory Chain and ATP Synthase Human Genes with S. cerevisiae Homologs
4.1. Complex II
4.1.1. Mutations in Complex II Catalytic and Structural Subunits
4.1.2. Mutations in Complex II Assembly Factors
4.1.3. Complex II and Paragangliomas
4.2. Complex III
4.2.1. Mutations in Complex III Catalytic Subunits
4.2.2. Mutations in Complex III Structural Subunits
4.2.3. Mutations in Complex III Assembly Factors
4.3. Complex IV
4.3.1. Mutations in Complex IV Catalytic Subunits
4.3.2. Mutations in Complex IV Structural Subunits
4.3.3. Mutations in Complex IV Assembly Factors
4.4. ATP Synthase
4.4.1. Mutations in ATP Synthase Mitochondrially Encoded Subunits
4.4.2. Mutations in ATP Synthase Nuclear Structural Subunits
4.4.3. Nuclear ATP Synthase Assembly Gene Mutations
4.5. Coenzyme Q
Mutations in COQ Genes
4.6. Cytochrome c
5. Human Pathologies Resulting from Mutations in Genes with No Homologs in S. cerevisiae
5.1. Complex I
5.2. Assembly Factors
6. Concluding Remarks
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Morgenstern, M.; Stiller, S.B.; Lübbert, P.; Peikert, C.D.; Dannenmaier, S.; Drepper, F.; Weill, U.; Höß, P.; Feuerstein, R.; Gebert, M.; et al. Definition of a High-Confidence Mitochondrial Proteome at Quantitative Scale. Cell Rep. 2017, 19, 2836–2852. [Google Scholar] [CrossRef] [PubMed]
- Saccharomyces Genome Database | SGD. Available online: https://www.yeastgenome.org/ (accessed on 23 October 2020).
- Tzagoloff, A.; Dieckmann, C.L. PET genes of Saccharomyces cerevisiae. Microbiol. Rev. 1990, 54, 211–225. [Google Scholar] [CrossRef] [PubMed]
- DiMauro, S. Mitochondrial diseases. Biochim. Biophys. Acta 2004, 1658, 80–88. [Google Scholar] [CrossRef] [PubMed]
- Jonckheere, A.I.; Renkema, G.H.; Bras, M.; van den Heuvel, L.P.; Hoischen, A.; Gilissen, C.; Nabuurs, S.B.; Huynen, M.A.; de Vries, M.C.; Smeitink, J.A.M.; et al. A complex V ATP5A1 defect causes fatal neonatal mitochondrial encephalopathy. Brain J. Neurol. 2013, 136, 1544–1554. [Google Scholar] [CrossRef]
- Mayr, J.A.; Havlícková, V.; Zimmermann, F.; Magler, I.; Kaplanová, V.; Jesina, P.; Pecinová, A.; Nusková, H.; Koch, J.; Sperl, W.; et al. Mitochondrial ATP synthase deficiency due to a mutation in the ATP5E gene for the F1 epsilon subunit. Hum. Mol. Genet. 2010, 19, 3430–3439. [Google Scholar] [CrossRef]
- Meulemans, A.; Seneca, S.; Pribyl, T.; Smet, J.; Alderweirldt, V.; Waeytens, A.; Lissens, W.; Van Coster, R.; De Meirleir, L.; di Rago, J.-P.; et al. Defining the pathogenesis of the human Atp12p W94R mutation using a Saccharomyces cerevisiae yeast model. J. Biol. Chem. 2010, 285, 4099–4109. [Google Scholar] [CrossRef]
- Valnot, I.; von Kleist-Retzow, J.C.; Barrientos, A.; Gorbatyuk, M.; Taanman, J.W.; Mehaye, B.; Rustin, P.; Tzagoloff, A.; Munnich, A.; Rötig, A. A mutation in the human heme A:farnesyltransferase gene (COX10) causes cytochrome c oxidase deficiency. Hum. Mol. Genet. 2000, 9, 1245–1249. [Google Scholar] [CrossRef]
- Quinzii, C.M.; Kattah, A.G.; Naini, A.; Akman, H.O.; Mootha, V.K.; DiMauro, S.; Hirano, M. Coenzyme Q deficiency and cerebellar ataxia associated with an aprataxin mutation. Neurology 2005, 64, 539–541. [Google Scholar] [CrossRef]
- Garcia-Diaz, B.; Barros, M.H.; Sanna-Cherchi, S.; Emmanuele, V.; Akman, H.O.; Ferreiro-Barros, C.C.; Horvath, R.; Tadesse, S.; El Gharaby, N.; DiMauro, S.; et al. Infantile encephaloneuromyopathy and defective mitochondrial translation are due to a homozygous RMND1 mutation. Am. J. Hum. Genet. 2012, 91, 729–736. [Google Scholar] [CrossRef]
- Barrientos, A. Yeast models of human mitochondrial diseases. IUBMB Life 2003, 55, 83–95. [Google Scholar] [CrossRef]
- Foury, F.; Roganti, T.; Lecrenier, N.; Purnelle, B. The complete sequence of the mitochondrial genome of Saccharomyces cerevisiae. FEBS Lett. 1998, 440, 325–331. [Google Scholar] [CrossRef]
- Myers, A.M.; Pape, L.K.; Tzagoloff, A. Mitochondrial protein synthesis is required for maintenance of intact mitochondrial genomes in Saccharomyces cerevisiae. EMBO J. 1985, 4, 2087–2092. [Google Scholar] [CrossRef] [PubMed]
- Rak, M.; Tetaud, E.; Godard, F.; Sagot, I.; Salin, B.; Duvezin-Caubet, S.; Slonimski, P.P.; Rytka, J.; di Rago, J.-P. Yeast cells lacking the mitochondrial gene encoding the ATP synthase subunit 6 exhibit a selective loss of complex IV and unusual mitochondrial morphology. J. Biol. Chem. 2007, 282, 10853–10864. [Google Scholar] [CrossRef] [PubMed]
- Rak, M.; Tzagoloff, A. F1-dependent translation of mitochondrially encoded Atp6p and Atp8p subunits of yeast ATP synthase. Proc. Natl. Acad. Sci. USA 2009, 106, 18509–18514. [Google Scholar] [CrossRef] [PubMed]
- Doeg, K.A.; Krueger, S.; Ziegler, D.M. Studies on the electron transfer system. 29. The isolation and properties of a succinic dehydrogenase-cytochrome b complex from beef-heart mitochondria. Biochim. Biophys. Acta 1960, 41, 491–497. [Google Scholar] [CrossRef]
- Capaldi, R.A.; Sweetland, J.; Merli, A. Polypeptides in the succinate-coenzyme Q reductase segment of the respiratory chain. Biochemistry 1977, 16, 5707–5710. [Google Scholar] [CrossRef]
- Lemire, B.D.; Oyedotun, K.S. The Saccharomyces cerevisiae mitochondrial succinate:ubiquinone oxidoreductase. Biochim. Biophys. Acta 2002, 1553, 102–116. [Google Scholar] [CrossRef]
- Gebert, N.; Gebert, M.; Oeljeklaus, S.; von der Malsburg, K.; Stroud, D.A.; Kulawiak, B.; Wirth, C.; Zahedi, R.P.; Dolezal, P.; Wiese, S.; et al. Dual function of Sdh3 in the respiratory chain and TIM22 protein translocase of the mitochondrial inner membrane. Mol. Cell 2011, 44, 811–818. [Google Scholar] [CrossRef]
- Baginsky, M.L.; Hatefi, Y. Reconstitution of succinate-coenzyme Q reductase (complex II) and succinate oxidase activities by a highly purified, reactivated succinate dehydrogenase. J. Biol. Chem. 1969, 244, 5313–5319. [Google Scholar]
- Davis, K.A.; Hatefi, Y. Resolution and reconstitution of complex II (succinate-ubiquinone reductase) by salts. Arch. Biochem. Biophys. 1972, 149, 505–512. [Google Scholar] [CrossRef]
- Sun, F.; Huo, X.; Zhai, Y.; Wang, A.; Xu, J.; Su, D.; Bartlam, M.; Rao, Z. Crystal structure of mitochondrial respiratory membrane protein complex II. Cell 2005, 121, 1043–1057. [Google Scholar] [CrossRef]
- Kim, H.J.; Jeong, M.-Y.; Na, U.; Winge, D.R. Flavinylation and assembly of succinate dehydrogenase are dependent on the C-terminal tail of the flavoprotein subunit. J. Biol. Chem. 2012, 287, 40670–40679. [Google Scholar] [CrossRef] [PubMed]
- Rivner, M.H.; Shamsnia, M.; Swift, T.R.; Trefz, J.; Roesel, R.A.; Carter, A.L.; Yanamura, W.; Hommes, F.A. Kearns-Sayre syndrome and complex II deficiency. Neurology 1989, 39, 693–696. [Google Scholar] [CrossRef] [PubMed]
- Bourgeron, T.; Rustin, P.; Chretien, D.; Birch-Machin, M.; Bourgeois, M.; Viegas-Péquignot, E.; Munnich, A.; Rötig, A. Mutation of a nuclear succinate dehydrogenase gene results in mitochondrial respiratory chain deficiency. Nat. Genet. 1995, 11, 144–149. [Google Scholar] [CrossRef]
- Leigh, D. Subacute Necrotizing Encephalomyelopathy in an Infant. J. Neurol. Neurosurg. Psychiatry 1951, 14, 216–221. [Google Scholar] [CrossRef]
- Pagnamenta, A.T.; Hargreaves, I.P.; Duncan, A.J.; Taanman, J.-W.; Heales, S.J.; Land, J.M.; Bitner-Glindzicz, M.; Leonard, J.V.; Rahman, S. Phenotypic variability of mitochondrial disease caused by a nuclear mutation in complex II. Mol. Genet. Metab. 2006, 89, 214–221. [Google Scholar] [CrossRef]
- Levitas, A.; Muhammad, E.; Harel, G.; Saada, A.; Caspi, V.C.; Manor, E.; Beck, J.C.; Sheffield, V.; Parvari, R. Familial neonatal isolated cardiomyopathy caused by a mutation in the flavoprotein subunit of succinate dehydrogenase. Eur. J. Hum. Genet. EJHG 2010, 18, 1160–1165. [Google Scholar] [CrossRef]
- Van Coster, R.; Seneca, S.; Smet, J.; Van Hecke, R.; Gerlo, E.; Devreese, B.; Van Beeumen, J.; Leroy, J.G.; De Meirleir, L.; Lissens, W. Homozygous Gly555Glu mutation in the nuclear-encoded 70 kDa flavoprotein gene causes instability of the respiratory chain complex II. Am. J. Med. Genet. Part A 2003, 120, 13–18. [Google Scholar] [CrossRef]
- Saghbini, M.; Broomfield, P.L.; Scheffler, I.E. Studies on the assembly of complex II in yeast mitochondria using chimeric human/yeast genes for the iron-sulfur protein subunit. Biochemistry 1994, 33, 159–165. [Google Scholar] [CrossRef]
- Burgener, A.-V.; Bantug, G.R.; Meyer, B.J.; Higgins, R.; Ghosh, A.; Bignucolo, O.; Ma, E.H.; Loeliger, J.; Unterstab, G.; Geigges, M.; et al. SDHA gain-of-function engages inflammatory mitochondrial retrograde signaling via KEAP1-Nrf2. Nat. Immunol. 2019, 20, 1311–1321. [Google Scholar] [CrossRef]
- Van Vranken, J.G.; Bricker, D.K.; Dephoure, N.; Gygi, S.P.; Cox, J.E.; Thummel, C.S.; Rutter, J. SDHAF4 promotes mitochondrial succinate dehydrogenase activity and prevents neurodegeneration. Cell Metab. 2014, 20, 241–252. [Google Scholar] [CrossRef] [PubMed]
- Moosavi, B.; Berry, E.A.; Zhu, X.-L.; Yang, W.-C.; Yang, G.-F. The assembly of succinate dehydrogenase: A key enzyme in bioenergetics. Cell. Mol. Life Sci. CMLS 2019, 76, 4023–4042. [Google Scholar] [CrossRef] [PubMed]
- Ghezzi, D.; Goffrini, P.; Uziel, G.; Horvath, R.; Klopstock, T.; Lochmüller, H.; D’Adamo, P.; Gasparini, P.; Strom, T.M.; Prokisch, H.; et al. SDHAF1, encoding a LYR complex-II specific assembly factor, is mutated in SDH-defective infantile leukoencephalopathy. Nat. Genet. 2009, 41, 654–656. [Google Scholar] [CrossRef] [PubMed]
- Hao, H.-X.; Khalimonchuk, O.; Schraders, M.; Dephoure, N.; Bayley, J.-P.; Kunst, H.; Devilee, P.; Cremers, C.W.R.J.; Schiffman, J.D.; Bentz, B.G.; et al. SDH5, a gene required for flavination of succinate dehydrogenase, is mutated in paraganglioma. Science 2009, 325, 1139–1142. [Google Scholar] [CrossRef] [PubMed]
- Na, U.; Yu, W.; Cox, J.; Bricker, D.K.; Brockmann, K.; Rutter, J.; Thummel, C.S.; Winge, D.R. The LYR factors SDHAF1 and SDHAF3 mediate maturation of the iron-sulfur subunit of succinate dehydrogenase. Cell Metab. 2014, 20, 253–266. [Google Scholar] [CrossRef]
- Ohlenbusch, A.; Edvardson, S.; Skorpen, J.; Bjornstad, A.; Saada, A.; Elpeleg, O.; Gärtner, J.; Brockmann, K. Leukoencephalopathy with accumulated succinate is indicative of SDHAF1 related complex II deficiency. Orphanet J. Rare Dis. 2012, 7, 69. [Google Scholar] [CrossRef]
- Rustin, P.; Rötig, A. Inborn errors of complex II--unusual human mitochondrial diseases. Biochim. Biophys. Acta 2002, 1553, 117–122. [Google Scholar] [CrossRef]
- Rutter, J.; Winge, D.R.; Schiffman, J.D. Succinate dehydrogenase—Assembly, regulation and role in human disease. Mitochondrion 2010, 10, 393–401. [Google Scholar] [CrossRef]
- Baysal, B.E. Mitochondrial complex II and genomic imprinting in inheritance of paraganglioma tumors. Biochim. Biophys. Acta 2013, 1827, 573–577. [Google Scholar] [CrossRef][Green Version]
- Miettinen, M.; Lasota, J. Succinate dehydrogenase deficient gastrointestinal stromal tumors (GISTs)—A review. Int. J. Biochem. Cell Biol. 2014, 53, 514–519. [Google Scholar] [CrossRef]
- Lussey-Lepoutre, C.; Buffet, A.; Gimenez-Roqueplo, A.-P.; Favier, J. Mitochondrial Deficiencies in the Predisposition to Paraganglioma. Metabolites 2017, 7, 17. [Google Scholar] [CrossRef] [PubMed]
- Nazar, E.; Khatami, F.; Saffar, H.; Tavangar, S.M. The Emerging Role of Succinate Dehyrogenase Genes (SDHx) in Tumorigenesis. Int. J. Hematol. Oncol. Stem Cell Res. 2019, 13, 72–82. [Google Scholar] [CrossRef] [PubMed]
- Bannon, A.E.; Kent, J.; Forquer, I.; Town, A.; Klug, L.R.; McCann, K.; Beadling, C.; Harismendy, O.; Sicklick, J.K.; Corless, C.; et al. Biochemical, Molecular, and Clinical Characterization of Succinate Dehydrogenase Subunit A Variants of Unknown Significance. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2017, 23, 6733–6743. [Google Scholar] [CrossRef] [PubMed]
- Parfait, B.; Chretien, D.; Rötig, A.; Marsac, C.; Munnich, A.; Rustin, P. Compound heterozygous mutations in the flavoprotein gene of the respiratory chain complex II in a patient with Leigh syndrome. Hum. Genet. 2000, 106, 236–243. [Google Scholar] [CrossRef] [PubMed]
- Horváth, R.; Abicht, A.; Holinski-Feder, E.; Laner, A.; Gempel, K.; Prokisch, H.; Lochmüller, H.; Klopstock, T.; Jaksch, M. Leigh syndrome caused by mutations in the flavoprotein (Fp) subunit of succinate dehydrogenase (SDHA). J. Neurol. Neurosurg. Psychiatry 2006, 77, 74–76. [Google Scholar] [CrossRef] [PubMed]
- Birch-Machin, M.A.; Taylor, R.W.; Cochran, B.; Ackrell, B.A.; Turnbull, D.M. Late-onset optic atrophy, ataxia, and myopathy associated with a mutation of a complex II gene. Ann. Neurol. 2000, 48, 330–335. [Google Scholar] [CrossRef]
- Ma, Y.-Y.; Wu, T.-F.; Liu, Y.-P.; Wang, Q.; Li, X.-Y.; Ding, Y.; Song, J.-Q.; Shi, X.-Y.; Zhang, W.-N.; Zhao, M.; et al. Two compound frame-shift mutations in succinate dehydrogenase gene of a Chinese boy with encephalopathy. Brain Dev. 2014, 36, 394–398. [Google Scholar] [CrossRef]
- Alston, C.L.; Davison, J.E.; Meloni, F.; van der Westhuizen, F.H.; He, L.; Hornig-Do, H.-T.; Peet, A.C.; Gissen, P.; Goffrini, P.; Ferrero, I.; et al. Recessive germline SDHA and SDHB mutations causing leukodystrophy and isolated mitochondrial complex II deficiency. J. Med. Genet. 2012, 49, 569–577. [Google Scholar] [CrossRef]
- Courage, C.; Jackson, C.B.; Hahn, D.; Euro, L.; Nuoffer, J.-M.; Gallati, S.; Schaller, A. SDHA mutation with dominant transmission results in complex II deficiency with ocular, cardiac, and neurologic involvement. Am. J. Med. Genet. Part A 2017, 173, 225–230. [Google Scholar] [CrossRef]
- Grønborg, S.; Darin, N.; Miranda, M.J.; Damgaard, B.; Cayuela, J.A.; Oldfors, A.; Kollberg, G.; Hansen, T.V.O.; Ravn, K.; Wibrand, F.; et al. Leukoencephalopathy due to Complex II Deficiency and Bi-Allelic SDHB Mutations: Further Cases and Implications for Genetic Counselling. JIMD Rep. 2017, 33, 69–77. [Google Scholar] [CrossRef]
- Jackson, C.B.; Nuoffer, J.-M.; Hahn, D.; Prokisch, H.; Haberberger, B.; Gautschi, M.; Häberli, A.; Gallati, S.; Schaller, A. Mutations in SDHD lead to autosomal recessive encephalomyopathy and isolated mitochondrial complex II deficiency. J. Med. Genet. 2014, 51, 170–175. [Google Scholar] [CrossRef] [PubMed]
- Hatefi, Y.; Haavik, A.G.; Griffiths, D.E. Studies on the electron transfer system. XLI. Reduced coenzyme Q (QH2)-cytochrome c reductase. J. Biol. Chem. 1962, 237, 1681–1685. [Google Scholar] [PubMed]
- Beckmann, J.D.; Ljungdahl, P.O.; Lopez, J.L.; Trumpower, B.L. Isolation and characterization of the nuclear gene encoding the Rieske iron-sulfur protein (RIP1) from Saccharomyces cerevisiae. J. Biol. Chem. 1987, 262, 8901–8909. [Google Scholar] [PubMed]
- Hunte, C.; Koepke, J.; Lange, C.; Rossmanith, T.; Michel, H. Structure at 2.3 A resolution of the cytochrome bc(1) complex from the yeast Saccharomyces cerevisiae co-crystallized with an antibody Fv fragment. Structure 2000, 8, 669–684. [Google Scholar] [CrossRef]
- Trumpower, B.L. Cytochrome bc1 complexes of microorganisms. Microbiol. Rev. 1990, 54, 101–129. [Google Scholar] [CrossRef] [PubMed]
- Gruschke, S.; Kehrein, K.; Römpler, K.; Gröne, K.; Israel, L.; Imhof, A.; Herrmann, J.M.; Ott, M. Cbp3-Cbp6 interacts with the yeast mitochondrial ribosomal tunnel exit and promotes cytochrome b synthesis and assembly. J. Cell Biol. 2011, 193, 1101–1114. [Google Scholar] [CrossRef]
- Crivellone, M.D. Characterization of CBP4, a new gene essential for the expression of ubiquinol-cytochrome c reductase in Saccharomyces cerevisiae. J. Biol. Chem. 1994, 269, 21284–21292. [Google Scholar]
- Hildenbeutel, M.; Hegg, E.L.; Stephan, K.; Gruschke, S.; Meunier, B.; Ott, M. Assembly factors monitor sequential hemylation of cytochrome b to regulate mitochondrial translation. J. Cell Biol. 2014, 205, 511–524. [Google Scholar] [CrossRef]
- Nobrega, F.G.; Nobrega, M.P.; Tzagoloff, A. BCS1, a novel gene required for the expression of functional Rieske iron-sulfur protein in Saccharomyces cerevisiae. EMBO J. 1992, 11, 3821–3829. [Google Scholar] [CrossRef]
- Cruciat, C.M.; Hell, K.; Fölsch, H.; Neupert, W.; Stuart, R.A. Bcs1p, an AAA-family member, is a chaperone for the assembly of the cytochrome bc(1) complex. EMBO J. 1999, 18, 5226–5233. [Google Scholar] [CrossRef]
- Atkinson, A.; Smith, P.; Fox, J.L.; Cui, T.-Z.; Khalimonchuk, O.; Winge, D.R. The LYR protein Mzm1 functions in the insertion of the Rieske Fe/S protein in yeast mitochondria. Mol. Cell. Biol. 2011, 31, 3988–3996. [Google Scholar] [CrossRef] [PubMed]
- Nobrega, F.G.; Tzagoloff, A. Assembly of the mitochondrial membrane system. DNA sequence and organization of the cytochrome b gene in Saccharomyces cerevisiae D273-10B. J. Biol. Chem. 1980, 255, 9828–9837. [Google Scholar] [PubMed]
- Lazowska, J.; Jacq, C.; Slonimski, P.P. Sequence of introns and flanking exons in wild-type and box3 mutants of cytochrome b reveals an interlaced splicing protein coded by an intron. Cell 1980, 22, 333–348. [Google Scholar] [CrossRef]
- Church, G.M.; Slonimski, P.P.; Gilbert, W. Pleiotropic mutations within two yeast mitochondrial cytochrome genes block mRNA processing. Cell 1979, 18, 1209–1215. [Google Scholar] [CrossRef]
- Gampel, A.; Nishikimi, M.; Tzagoloff, A. CBP2 protein promotes in vitro excision of a yeast mitochondrial group I intron. Mol. Cell. Biol. 1989, 9, 5424–5433. [Google Scholar] [CrossRef]
- Rödel, G. Two yeast nuclear genes, CBS1 and CBS2, are required for translation of mitochondrial transcripts bearing the 5′-untranslated COB leader. Curr. Genet. 1986, 11, 41–45. [Google Scholar] [CrossRef]
- Dieckmann, C.L.; Homison, G.; Tzagoloff, A. Assembly of the mitochondrial membrane system. Nucleotide sequence of a yeast nuclear gene (CBP1) involved in 5′ end processing of cytochrome b pre-mRNA. J. Biol. Chem. 1984, 259, 4732–4738. [Google Scholar]
- WebHome < MITOMAP < Foswiki. Available online: https://www.mitomap.org/foswiki/bin/view/MITOMAP/WebHome (accessed on 19 October 2020).
- Meunier, B.; Fisher, N.; Ransac, S.; Mazat, J.-P.; Brasseur, G. Respiratory complex III dysfunction in humans and the use of yeast as a model organism to study mitochondrial myopathy and associated diseases. Biochim. Biophys. Acta 2013, 1827, 1346–1361. [Google Scholar] [CrossRef]
- Mackey, D.A.; Oostra, R.J.; Rosenberg, T.; Nikoskelainen, E.; Bronte-Stewart, J.; Poulton, J.; Harding, A.E.; Govan, G.; Bolhuis, P.A.; Norby, S. Primary pathogenic mtDNA mutations in multigeneration pedigrees with Leber hereditary optic neuropathy. Am. J. Hum. Genet. 1996, 59, 481–485. [Google Scholar]
- Brown, M.D.; Voljavec, A.S.; Lott, M.T.; Torroni, A.; Yang, C.C.; Wallace, D.C. Mitochondrial DNA complex I and III mutations associated with Leber’s hereditary optic neuropathy. Genetics 1992, 130, 163–173. [Google Scholar]
- Johns, D.R.; Neufeld, M.J. Cytochrome b mutations in Leber hereditary optic neuropathy. Biochem. Biophys. Res. Commun. 1991, 181, 1358–1364. [Google Scholar] [CrossRef]
- Haut, S.; Brivet, M.; Touati, G.; Rustin, P.; Lebon, S.; Garcia-Cazorla, A.; Saudubray, J.M.; Boutron, A.; Legrand, A.; Slama, A. A deletion in the human QP-C gene causes a complex III deficiency resulting in hypoglycaemia and lactic acidosis. Hum. Genet. 2003, 113, 118–122. [Google Scholar] [CrossRef] [PubMed]
- Hemrika, W.; De Jong, M.; Berden, J.A.; Grivell, L.A. The C-terminus of the 14-kDa subunit of ubiquinol-cytochrome-c oxidoreductase of the yeast Saccharomyces cerevisiae is involved in the assembly of a functional enzyme. Eur. J. Biochem. 1994, 220, 569–576. [Google Scholar] [CrossRef] [PubMed]
- Barel, O.; Shorer, Z.; Flusser, H.; Ofir, R.; Narkis, G.; Finer, G.; Shalev, H.; Nasasra, A.; Saada, A.; Birk, O.S. Mitochondrial complex III deficiency associated with a homozygous mutation in UQCRQ. Am. J. Hum. Genet. 2008, 82, 1211–1216. [Google Scholar] [CrossRef]
- Miyake, N.; Yano, S.; Sakai, C.; Hatakeyama, H.; Matsushima, Y.; Shiina, M.; Watanabe, Y.; Bartley, J.; Abdenur, J.E.; Wang, R.Y.; et al. Mitochondrial complex III deficiency caused by a homozygous UQCRC2 mutation presenting with neonatal-onset recurrent metabolic decompensation. Hum. Mutat. 2013, 34, 446–452. [Google Scholar] [CrossRef]
- Gaignard, P.; Eyer, D.; Lebigot, E.; Oliveira, C.; Therond, P.; Boutron, A.; Slama, A. UQCRC2 mutation in a patient with mitochondrial complex III deficiency causing recurrent liver failure, lactic acidosis and hypoglycemia. J. Hum. Genet. 2017, 62, 729–731. [Google Scholar] [CrossRef]
- Han, Y.; Wu, P.; Wang, Z.; Zhang, Z.; Sun, S.; Liu, J.; Gong, S.; Gao, P.; Iwakuma, T.; Molina-Vila, M.A.; et al. Ubiquinol-cytochrome C reductase core protein II promotes tumorigenesis by facilitating p53 degradation. EBioMedicine 2019, 40, 92–105. [Google Scholar] [CrossRef]
- Wagener, N.; Ackermann, M.; Funes, S.; Neupert, W. A pathway of protein translocation in mitochondria mediated by the AAA-ATPase Bcs1. Mol. Cell 2011, 44, 191–202. [Google Scholar] [CrossRef]
- Ndi, M.; Marin-Buera, L.; Salvatori, R.; Singh, A.P.; Ott, M. Biogenesis of the bc1 Complex of the Mitochondrial Respiratory Chain. J. Mol. Biol. 2018, 430, 3892–3905. [Google Scholar] [CrossRef]
- Akduman, H.; Eminoglu, T.; Okulu, E.; Erdeve, O.; Atasay, B.; Arsan, S. A Neonate Presenting with Gracile Syndrome and Bjornstad Phenotype Associated with Bcs1l Mutation. Genet. Couns. Geneva Switz. 2016, 27, 509–512. [Google Scholar]
- Serdaroğlu, E.; Takcı, Ş.; Kotarsky, H.; Çil, O.; Utine, E.; Yiğit, Ş.; Fellman, V. A Turkish BCS1L mutation causes GRACILE-like disorder. Turk. J. Pediatr. 2016, 58, 658–661. [Google Scholar] [CrossRef] [PubMed]
- Kasapkara, Ç.S.; Tümer, L.; Ezgü, F.S.; Küçükçongar, A.; Hasanoğlu, A. BCS1L gene mutation causing GRACILE syndrome: Case report. Ren. Fail. 2014, 36, 953–954. [Google Scholar] [CrossRef] [PubMed]
- Lynn, A.M.; King, R.I.; Mackay, R.J.; Florkowski, C.M.; Wilson, C.J. BCS1L gene mutation presenting with GRACILE-like syndrome and complex III deficiency. Ann. Clin. Biochem. 2012, 49, 201–203. [Google Scholar] [CrossRef] [PubMed]
- Kotarsky, H.; Karikoski, R.; Mörgelin, M.; Marjavaara, S.; Bergman, P.; Zhang, D.-L.; Smet, J.; van Coster, R.; Fellman, V. Characterization of complex III deficiency and liver dysfunction in GRACILE syndrome caused by a BCS1L mutation. Mitochondrion 2010, 10, 497–509. [Google Scholar] [CrossRef]
- Fellman, V.; Lemmelä, S.; Sajantila, A.; Pihko, H.; Järvelä, I. Screening of BCS1L mutations in severe neonatal disorders suspicious for mitochondrial cause. J. Hum. Genet. 2008, 53, 554–558. [Google Scholar] [CrossRef]
- Visapää, I.; Fellman, V.; Vesa, J.; Dasvarma, A.; Hutton, J.L.; Kumar, V.; Payne, G.S.; Makarow, M.; Van Coster, R.; Taylor, R.W.; et al. GRACILE syndrome, a lethal metabolic disorder with iron overload, is caused by a point mutation in BCS1L. Am. J. Hum. Genet. 2002, 71, 863–876. [Google Scholar] [CrossRef]
- Falco, M.; Franzè, A.; Iossa, S.; De Falco, L.; Gambale, A.; Marciano, E.; Iolascon, A. Novel compound heterozygous mutations in BCS1L gene causing Bjornstad syndrome in two siblings. Am. J. Med. Genet. Part A 2017, 173, 1348–1352. [Google Scholar] [CrossRef]
- Shigematsu, Y.; Hayashi, R.; Yoshida, K.; Shimizu, A.; Kubota, M.; Komori, M.; Shimomura, Y.; Niizeki, H. Novel heterozygous deletion mutation c.821delC in the AAA domain of BCS1L underlies Björnstad syndrome. J. Dermatol. 2017, 44, e111–e112. [Google Scholar] [CrossRef]
- Zhang, J.; Duo, L.; Lin, Z.; Wang, H.; Yin, J.; Cao, X.; Zhao, J.; Dai, L.; Liu, X.; Zhang, J.; et al. Exome sequencing reveals novel BCS1L mutations in siblings with hearing loss and hypotrichosis. Gene 2015, 566, 84–88. [Google Scholar] [CrossRef]
- Yanagishita, T.; Sugiura, K.; Kawamoto, Y.; Ito, K.; Marubashi, Y.; Taguchi, N.; Akiyama, M.; Watanabe, D. A case of Björnstad syndrome caused by novel compound heterozygous mutations in the BCS1L gene. Br. J. Dermatol. 2014, 170, 970–973. [Google Scholar] [CrossRef]
- Siddiqi, S.; Siddiq, S.; Mansoor, A.; Oostrik, J.; Ahmad, N.; Kazmi, S.A.R.; Kremer, H.; Qamar, R.; Schraders, M. Novel mutation in AAA domain of BCS1L causing Bjornstad syndrome. J. Hum. Genet. 2013, 58, 819–821. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hinson, J.T.; Fantin, V.R.; Schönberger, J.; Breivik, N.; Siem, G.; McDonough, B.; Sharma, P.; Keogh, I.; Godinho, R.; Santos, F.; et al. Missense mutations in the BCS1L gene as a cause of the Björnstad syndrome. N. Engl. J. Med. 2007, 356, 809–819. [Google Scholar] [CrossRef] [PubMed]
- Tegelberg, S.; Tomašić, N.; Kallijärvi, J.; Purhonen, J.; Elmér, E.; Lindberg, E.; Nord, D.G.; Soller, M.; Lesko, N.; Wedell, A.; et al. Respiratory chain complex III deficiency due to mutated BCS1L: A novel phenotype with encephalomyopathy, partially phenocopied in a Bcs1l mutant mouse model. Orphanet J. Rare Dis. 2017, 12, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Blázquez, A.; Gil-Borlado, M.C.; Morán, M.; Verdú, A.; Cazorla-Calleja, M.R.; Martín, M.A.; Arenas, J.; Ugalde, C. Infantile mitochondrial encephalomyopathy with unusual phenotype caused by a novel BCS1L mutation in an isolated complex III-deficient patient. Neuromuscul. Disord. NMD 2009, 19, 143–146. [Google Scholar] [CrossRef]
- Fernandez-Vizarra, E.; Bugiani, M.; Goffrini, P.; Carrara, F.; Farina, L.; Procopio, E.; Donati, A.; Uziel, G.; Ferrero, I.; Zeviani, M. Impaired complex III assembly associated with BCS1L gene mutations in isolated mitochondrial encephalopathy. Hum. Mol. Genet. 2007, 16, 1241–1252. [Google Scholar] [CrossRef]
- de Lonlay, P.; Valnot, I.; Barrientos, A.; Gorbatyuk, M.; Tzagoloff, A.; Taanman, J.W.; Benayoun, E.; Chrétien, D.; Kadhom, N.; Lombès, A.; et al. A mutant mitochondrial respiratory chain assembly protein causes complex III deficiency in patients with tubulopathy, encephalopathy and liver failure. Nat. Genet. 2001, 29, 57–60. [Google Scholar] [CrossRef]
- De Meirleir, L.; Seneca, S.; Damis, E.; Sepulchre, B.; Hoorens, A.; Gerlo, E.; García Silva, M.T.; Hernandez, E.M.; Lissens, W.; Van Coster, R. Clinical and diagnostic characteristics of complex III deficiency due to mutations in the BCS1L gene. Am. J. Med. Genet. Part A 2003, 121, 126–131. [Google Scholar] [CrossRef]
- Gil-Borlado, M.C.; González-Hoyuela, M.; Blázquez, A.; García-Silva, M.T.; Gabaldón, T.; Manzanares, J.; Vara, J.; Martín, M.A.; Seneca, S.; Arenas, J.; et al. Pathogenic mutations in the 5′ untranslated region of BCS1L mRNA in mitochondrial complex III deficiency. Mitochondrion 2009, 9, 299–305. [Google Scholar] [CrossRef]
- Ramos-Arroyo, M.A.; Hualde, J.; Ayechu, A.; De Meirleir, L.; Seneca, S.; Nadal, N.; Briones, P. Clinical and biochemical spectrum of mitochondrial complex III deficiency caused by mutations in the BCS1L gene. Clin. Genet. 2009, 75, 585–587. [Google Scholar] [CrossRef]
- Ezgu, F.; Senaca, S.; Gunduz, M.; Tumer, L.; Hasanoglu, A.; Tiras, U.; Unsal, R.; Bakkaloglu, S.A. Severe renal tubulopathy in a newborn due to BCS1L gene mutation: Effects of different treatment modalities on the clinical course. Gene 2013, 528, 364–366. [Google Scholar] [CrossRef]
- Tuppen, H.A.L.; Fehmi, J.; Czermin, B.; Goffrini, P.; Meloni, F.; Ferrero, I.; He, L.; Blakely, E.L.; McFarland, R.; Horvath, R.; et al. Long-term survival of neonatal mitochondrial complex III deficiency associated with a novel BCS1L gene mutation. Mol. Genet. Metab. 2010, 100, 345–348. [Google Scholar] [CrossRef] [PubMed]
- Oláhová, M.; Ceccatelli Berti, C.; Collier, J.J.; Alston, C.L.; Jameson, E.; Jones, S.A.; Edwards, N.; He, L.; Chinnery, P.F.; Horvath, R.; et al. Molecular genetic investigations identify new clinical phenotypes associated with BCS1L-related mitochondrial disease. Hum. Mol. Genet. 2019, 28, 3766–3776. [Google Scholar] [CrossRef] [PubMed]
- Cui, T.-Z.; Smith, P.M.; Fox, J.L.; Khalimonchuk, O.; Winge, D.R. Late-stage maturation of the Rieske Fe/S protein: Mzm1 stabilizes Rip1 but does not facilitate its translocation by the AAA ATPase Bcs1. Mol. Cell. Biol. 2012, 32, 4400–4409. [Google Scholar] [CrossRef] [PubMed]
- Invernizzi, F.; Tigano, M.; Dallabona, C.; Donnini, C.; Ferrero, I.; Cremonte, M.; Ghezzi, D.; Lamperti, C.; Zeviani, M. A homozygous mutation in LYRM7/MZM1L associated with early onset encephalopathy, lactic acidosis, and severe reduction of mitochondrial complex III activity. Hum. Mutat. 2013, 34, 1619–1622. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Liu, M.; Zhang, Z.; Zhou, L.; Kong, W.; Jiang, Y.; Wang, J.; Xiao, J.; Wu, Y. Genotypic Spectrum and Natural History of Cavitating Leukoencephalopathies in Childhood. Pediatr. Neurol. 2019, 94, 38–47. [Google Scholar] [CrossRef] [PubMed]
- Dallabona, C.; Abbink, T.E.M.; Carrozzo, R.; Torraco, A.; Legati, A.; van Berkel, C.G.M.; Niceta, M.; Langella, T.; Verrigni, D.; Rizza, T.; et al. LYRM7 mutations cause a multifocal cavitating leukoencephalopathy with distinct MRI appearance. Brain J. Neurol. 2016, 139, 782–794. [Google Scholar] [CrossRef] [PubMed]
- Hempel, M.; Kremer, L.S.; Tsiakas, K.; Alhaddad, B.; Haack, T.B.; Löbel, U.; Feichtinger, R.G.; Sperl, W.; Prokisch, H.; Mayr, J.A.; et al. LYRM7—Associated complex III deficiency: A clinical, molecular genetic, MR tomographic, and biochemical study. Mitochondrion 2017, 37, 55–61. [Google Scholar] [CrossRef]
- Kremer, L.S.; L’hermitte-Stead, C.; Lesimple, P.; Gilleron, M.; Filaut, S.; Jardel, C.; Haack, T.B.; Strom, T.M.; Meitinger, T.; Azzouz, H.; et al. Severe respiratory complex III defect prevents liver adaptation to prolonged fasting. J. Hepatol. 2016, 65, 377–385. [Google Scholar] [CrossRef]
- Tucker, E.J.; Wanschers, B.F.J.; Szklarczyk, R.; Mountford, H.S.; Wijeyeratne, X.W.; van den Brand, M.A.M.; Leenders, A.M.; Rodenburg, R.J.; Reljić, B.; Compton, A.G.; et al. Mutations in the UQCC1-interacting protein, UQCC2, cause human complex III deficiency associated with perturbed cytochrome b protein expression. PLoS Genet. 2013, 9, e1004034. [Google Scholar] [CrossRef]
- Feichtinger, R.G.; Brunner-Krainz, M.; Alhaddad, B.; Wortmann, S.B.; Kovacs-Nagy, R.; Stojakovic, T.; Erwa, W.; Resch, B.; Windischhofer, W.; Verheyen, S.; et al. Combined Respiratory Chain Deficiency and UQCC2 Mutations in Neonatal Encephalomyopathy: Defective Supercomplex Assembly in Complex III Deficiencies. Oxid. Med. Cell. Longev. 2017, 2017. [Google Scholar] [CrossRef]
- Bogenhagen, D.F.; Haley, J.D. Pulse-chase SILAC-based analyses reveal selective oversynthesis and rapid turnover of mitochondrial protein components of respiratory complexes. J. Biol. Chem. 2020, 295, 2544–2554. [Google Scholar] [CrossRef] [PubMed]
- Wanschers, B.F.J.; Szklarczyk, R.; van den Brand, M.A.M.; Jonckheere, A.; Suijskens, J.; Smeets, R.; Rodenburg, R.J.; Stephan, K.; Helland, I.B.; Elkamil, A.; et al. A mutation in the human CBP4 ortholog UQCC3 impairs complex III assembly, activity and cytochrome b stability. Hum. Mol. Genet. 2014, 23, 6356–6365. [Google Scholar] [CrossRef] [PubMed]
- Gaignard, P.; Menezes, M.; Schiff, M.; Bayot, A.; Rak, M.; Ogier de Baulny, H.; Su, C.-H.; Gilleron, M.; Lombes, A.; Abida, H.; et al. Mutations in CYC1, encoding cytochrome c1 subunit of respiratory chain complex III, cause insulin-responsive hyperglycemia. Am. J. Hum. Genet. 2013, 93, 384–389. [Google Scholar] [CrossRef] [PubMed]
- Gusic, M.; Schottmann, G.; Feichtinger, R.G.; Du, C.; Scholz, C.; Wagner, M.; Mayr, J.A.; Lee, C.-Y.; Yépez, V.A.; Lorenz, N.; et al. Bi-Allelic UQCRFS1 Variants Are Associated with Mitochondrial Complex III Deficiency, Cardiomyopathy, and Alopecia Totalis. Am. J. Hum. Genet. 2020, 106, 102–111. [Google Scholar] [CrossRef]
- Wikström, M. Identification of the electron transfers in cytochrome oxidase that are coupled to proton-pumping. Nature 1989, 338, 776–778. [Google Scholar] [CrossRef]
- Hell, K.; Tzagoloff, A.; Neupert, W.; Stuart, R.A. Identification of Cox20p, a novel protein involved in the maturation and assembly of cytochrome oxidase subunit 2. J. Biol. Chem. 2000, 275, 4571–4578. [Google Scholar] [CrossRef]
- Sharma, V.; Enkavi, G.; Vattulainen, I.; Róg, T.; Wikström, M. Proton-coupled electron transfer and the role of water molecules in proton pumping by cytochrome c oxidase. Proc. Natl. Acad. Sci. USA 2015, 112, 2040–2045. [Google Scholar] [CrossRef]
- Pitceathly, R.D.S.; Taanman, J.-W. NDUFA4 (Renamed COXFA4) Is a Cytochrome-c Oxidase Subunit. Trends Endocrinol. Metab. TEM 2018, 29, 452–454. [Google Scholar] [CrossRef]
- Hill, B.C. Modeling the sequence of electron transfer reactions in the single turnover of reduced, mammalian cytochrome c oxidase with oxygen. J. Biol. Chem. 1994, 269, 2419–2425. [Google Scholar]
- Brunori, M.; Giuffrè, A.; Sarti, P. Cytochrome c oxidase, ligands and electrons. J. Inorg. Biochem. 2005, 99, 324–336. [Google Scholar] [CrossRef]
- Tzagoloff, A.; Nobrega, M.; Gorman, N.; Sinclair, P. On the functions of the yeast COX10 and COX11 gene products. Biochem. Mol. Biol. Int. 1993, 31, 593–598. [Google Scholar] [PubMed]
- Barros, M.H.; Carlson, C.G.; Glerum, D.M.; Tzagoloff, A. Involvement of mitochondrial ferredoxin and Cox15p in hydroxylation of heme O. FEBS Lett. 2001, 492, 133–138. [Google Scholar] [CrossRef]
- Barros, M.H.; Nobrega, F.G.; Tzagoloff, A. Mitochondrial ferredoxin is required for heme A synthesis in Saccharomyces cerevisiae. J. Biol. Chem. 2002, 277, 9997–10002. [Google Scholar] [CrossRef] [PubMed]
- Bestwick, M.; Jeong, M.-Y.; Khalimonchuk, O.; Kim, H.; Winge, D.R. Analysis of Leigh syndrome mutations in the yeast SURF1 homolog reveals a new member of the cytochrome oxidase assembly factor family. Mol. Cell. Biol. 2010, 30, 4480–4491. [Google Scholar] [CrossRef] [PubMed]
- Taylor, N.G.; Swenson, S.; Harris, N.J.; Germany, E.M.; Fox, J.L.; Khalimonchuk, O. The Assembly Factor Pet117 Couples Heme a Synthase Activity to Cytochrome Oxidase Assembly. J. Biol. Chem. 2017, 292, 1815–1825. [Google Scholar] [CrossRef] [PubMed]
- Barros, M.H.; McStay, G.P. Modular biogenesis of mitochondrial respiratory complexes. Mitochondrion 2020, 50, 94–114. [Google Scholar] [CrossRef]
- Genova, M.L.; Lenaz, G. Functional role of mitochondrial respiratory supercomplexes. Biochim. Biophys. Acta 2014, 1837, 427–443. [Google Scholar] [CrossRef]
- Berndtsson, J.; Aufschnaiter, A.; Rathore, S.; Marin-Buera, L.; Dawitz, H.; Diessl, J.; Kohler, V.; Barrientos, A.; Büttner, S.; Fontanesi, F.; et al. Respiratory supercomplexes enhance electron transport by decreasing cytochrome c diffusion distance. EMBO Rep. 2020, e51015. [Google Scholar] [CrossRef]
- Schägger, H.; Pfeiffer, K. Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J. 2000, 19, 1777–1783. [Google Scholar] [CrossRef]
- Stroh, A.; Anderka, O.; Pfeiffer, K.; Yagi, T.; Finel, M.; Ludwig, B.; Schägger, H. Assembly of respiratory complexes I, III, and IV into NADH oxidase supercomplex stabilizes complex I in Paracoccus denitrificans. J. Biol. Chem. 2004, 279, 5000–5007. [Google Scholar] [CrossRef]
- Rak, M.; Bénit, P.; Chrétien, D.; Bouchereau, J.; Schiff, M.; El-Khoury, R.; Tzagoloff, A.; Rustin, P. Mitochondrial cytochrome c oxidase deficiency. Clin. Sci. Lond. Engl. 1979 2016, 130, 393–407. [Google Scholar] [CrossRef] [PubMed]
- Trueblood, C.E.; Poyton, R.O. Differential effectiveness of yeast cytochrome c oxidase subunit genes results from differences in expression not function. Mol. Cell. Biol. 1987, 7, 3520–3526. [Google Scholar] [CrossRef] [PubMed]
- DiMauro, S.; Tanji, K.; Schon, E.A. The many clinical faces of cytochrome c oxidase deficiency. Adv. Exp. Med. Biol. 2012, 748, 341–357. [Google Scholar] [CrossRef] [PubMed]
- Indrieri, A.; van Rahden, V.A.; Tiranti, V.; Morleo, M.; Iaconis, D.; Tammaro, R.; D’Amato, I.; Conte, I.; Maystadt, I.; Demuth, S.; et al. Mutations in COX7B cause microphthalmia with linear skin lesions, an unconventional mitochondrial disease. Am. J. Hum. Genet. 2012, 91, 942–949. [Google Scholar] [CrossRef]
- van Rahden, V.A.; Fernandez-Vizarra, E.; Alawi, M.; Brand, K.; Fellmann, F.; Horn, D.; Zeviani, M.; Kutsche, K. Mutations in NDUFB11, encoding a complex I component of the mitochondrial respiratory chain, cause microphthalmia with linear skin defects syndrome. Am. J. Hum. Genet. 2015, 96, 640–650. [Google Scholar] [CrossRef]
- Smith, D.; Gray, J.; Mitchell, L.; Antholine, W.E.; Hosler, J.P. Assembly of cytochrome-c oxidase in the absence of assembly protein Surf1p leads to loss of the active site heme. J. Biol. Chem. 2005, 280, 17652–17656. [Google Scholar] [CrossRef]
- Wedatilake, Y.; Brown, R.M.; McFarland, R.; Yaplito-Lee, J.; Morris, A.A.M.; Champion, M.; Jardine, P.E.; Clarke, A.; Thorburn, D.R.; Taylor, R.W.; et al. SURF1 deficiency: A multi-centre natural history study. Orphanet J. Rare Dis. 2013, 8. [Google Scholar] [CrossRef]
- Zhu, Z.; Yao, J.; Johns, T.; Fu, K.; De Bie, I.; Macmillan, C.; Cuthbert, A.P.; Newbold, R.F.; Wang, J.; Chevrette, M.; et al. SURF1, encoding a factor involved in the biogenesis of cytochrome c oxidase, is mutated in Leigh syndrome. Nat. Genet. 1998, 20, 337–343. [Google Scholar] [CrossRef]
- Tiranti, V.; Hoertnagel, K.; Carrozzo, R.; Galimberti, C.; Munaro, M.; Granatiero, M.; Zelante, L.; Gasparini, P.; Marzella, R.; Rocchi, M.; et al. Mutations of SURF-1 in Leigh disease associated with cytochrome c oxidase deficiency. Am. J. Hum. Genet. 1998, 63, 1609–1621. [Google Scholar] [CrossRef]
- Echaniz-Laguna, A.; Ghezzi, D.; Chassagne, M.; Mayençon, M.; Padet, S.; Melchionda, L.; Rouvet, I.; Lannes, B.; Bozon, D.; Latour, P.; et al. SURF1 deficiency causes demyelinating Charcot-Marie-Tooth disease. Neurology 2013, 81, 1523–1530. [Google Scholar] [CrossRef]
- Nobrega, M.P.; Nobrega, F.G.; Tzagoloff, A. COX10 codes for a protein homologous to the ORF1 product of Paracoccus denitrificans and is required for the synthesis of yeast cytochrome oxidase. J. Biol. Chem. 1990, 265, 14220–14226. [Google Scholar] [PubMed]
- Glerum, D.M.; Muroff, I.; Jin, C.; Tzagoloff, A. COX15 codes for a mitochondrial protein essential for the assembly of yeast cytochrome oxidase. J. Biol. Chem. 1997, 272, 19088–19094. [Google Scholar] [CrossRef] [PubMed]
- Glerum, D.M.; Tzagoloff, A. Isolation of a human cDNA for heme A:farnesyltransferase by functional complementation of a yeast cox10 mutant. Proc. Natl. Acad. Sci. USA 1994, 91, 8452–8456. [Google Scholar] [CrossRef] [PubMed]
- Pitceathly, R.D.S.; Taanman, J.-W.; Rahman, S.; Meunier, B.; Sadowski, M.; Cirak, S.; Hargreaves, I.; Land, J.M.; Nanji, T.; Polke, J.M.; et al. COX10 mutations resulting in complex multisystem mitochondrial disease that remains stable into adulthood. JAMA Neurol. 2013, 70, 1556–1561. [Google Scholar] [CrossRef] [PubMed]
- Bugiani, M.; Tiranti, V.; Farina, L.; Uziel, G.; Zeviani, M. Novel mutations in COX15 in a long surviving Leigh syndrome patient with cytochrome c oxidase deficiency. J. Med. Genet. 2005, 42, e28. [Google Scholar] [CrossRef] [PubMed]
- Rigby, K.; Cobine, P.A.; Khalimonchuk, O.; Winge, D.R. Mapping the Functional Interaction of Sco1 and Cox2 in Cytochrome Oxidase Biogenesis. J. Biol. Chem. 2008, 283, 15015–15022. [Google Scholar] [CrossRef]
- Glerum, D.M.; Shtanko, A.; Tzagoloff, A. SCO1 and SCO2 act as high copy suppressors of a mitochondrial copper recruitment defect in Saccharomyces cerevisiae. J. Biol. Chem. 1996, 271, 20531–20535. [Google Scholar] [CrossRef]
- Franco, L.V.R.; Su, C.-H.; McStay, G.P.; Yu, G.J.; Tzagoloff, A. Cox2p of yeast cytochrome oxidase assembles as a stand-alone subunit with the Cox1p and Cox3p modules. J. Biol. Chem. 2018, 293, 16899–16911. [Google Scholar] [CrossRef]
- Gurgel-Giannetti, J.; Oliveira, G.; Brasileiro Filho, G.; Martins, P.; Vainzof, M.; Hirano, M. Mitochondrial cardioencephalomyopathy due to a novel SCO2 mutation in a Brazilian patient: Case report and literature review. JAMA Neurol. 2013, 70, 258–261. [Google Scholar] [CrossRef]
- Rebelo, A.P.; Saade, D.; Pereira, C.V.; Farooq, A.; Huff, T.C.; Abreu, L.; Moraes, C.T.; Mnatsakanova, D.; Mathews, K.; Yang, H.; et al. SCO2 mutations cause early-onset axonal Charcot-Marie-Tooth disease associated with cellular copper deficiency. Brain J. Neurol. 2018, 141, 662–672. [Google Scholar] [CrossRef]
- Wakazono, T.; Miyake, M.; Yamashiro, K.; Yoshikawa, M.; Yoshimura, N. Association between SCO2 mutation and extreme myopia in Japanese patients. Jpn. J. Ophthalmol. 2016, 60, 319–325. [Google Scholar] [CrossRef] [PubMed]
- Tran-Viet, K.-N.; Powell, C.; Barathi, V.A.; Klemm, T.; Maurer-Stroh, S.; Limviphuvadh, V.; Soler, V.; Ho, C.; Yanovitch, T.; Schneider, G.; et al. Mutations in SCO2 are associated with autosomal-dominant high-grade myopia. Am. J. Hum. Genet. 2013, 92, 820–826. [Google Scholar] [CrossRef] [PubMed]
- Shteyer, E.; Saada, A.; Shaag, A.; Al-Hijawi, F.A.; Kidess, R.; Revel-Vilk, S.; Elpeleg, O. Exocrine pancreatic insufficiency, dyserythropoeitic anemia, and calvarial hyperostosis are caused by a mutation in the COX4I2 gene. Am. J. Hum. Genet. 2009, 84, 412–417. [Google Scholar] [CrossRef]
- Abu-Libdeh, B.; Douiev, L.; Amro, S.; Shahrour, M.; Ta-Shma, A.; Miller, C.; Elpeleg, O.; Saada, A. Mutation in the COX4I1 gene is associated with short stature, poor weight gain and increased chromosomal breaks, simulating Fanconi anemia. Eur. J. Hum. Genet. EJHG 2017, 25, 1142–1146. [Google Scholar] [CrossRef] [PubMed]
- Baertling, F.; Al-Murshedi, F.; Sánchez-Caballero, L.; Al-Senaidi, K.; Joshi, N.P.; Venselaar, H.; van den Brand, M.A.; Nijtmans, L.G.; Rodenburg, R.J. Mutation in mitochondrial complex IV subunit COX5A causes pulmonary arterial hypertension, lactic acidemia, and failure to thrive. Hum. Mutat. 2017, 38, 692–703. [Google Scholar] [CrossRef]
- Tamiya, G.; Makino, S.; Hayashi, M.; Abe, A.; Numakura, C.; Ueki, M.; Tanaka, A.; Ito, C.; Toshimori, K.; Ogawa, N.; et al. A mutation of COX6A1 causes a recessive axonal or mixed form of Charcot-Marie-Tooth disease. Am. J. Hum. Genet. 2014, 95, 294–300. [Google Scholar] [CrossRef]
- Laššuthová, P.; Šafka Brožková, D.; Krůtová, M.; Mazanec, R.; Züchner, S.; Gonzalez, M.A.; Seeman, P. Severe axonal Charcot-Marie-Tooth disease with proximal weakness caused by de novo mutation in the MORC2 gene. Brain J. Neurol. 2016, 139, e26. [Google Scholar] [CrossRef]
- Massa, V.; Fernandez-Vizarra, E.; Alshahwan, S.; Bakhsh, E.; Goffrini, P.; Ferrero, I.; Mereghetti, P.; D’Adamo, P.; Gasparini, P.; Zeviani, M. Severe infantile encephalomyopathy caused by a mutation in COX6B1, a nucleus-encoded subunit of cytochrome c oxidase. Am. J. Hum. Genet. 2008, 82, 1281–1289. [Google Scholar] [CrossRef]
- Abdulhag, U.N.; Soiferman, D.; Schueler-Furman, O.; Miller, C.; Shaag, A.; Elpeleg, O.; Edvardson, S.; Saada, A. Mitochondrial complex IV deficiency, caused by mutated COX6B1, is associated with encephalomyopathy, hydrocephalus and cardiomyopathy. Eur. J. Hum. Genet. EJHG 2015, 23, 159–164. [Google Scholar] [CrossRef]
- Calvo, S.E.; Compton, A.G.; Hershman, S.G.; Lim, S.C.; Lieber, D.S.; Tucker, E.J.; Laskowski, A.; Garone, C.; Liu, S.; Jaffe, D.B.; et al. Molecular diagnosis of infantile mitochondrial disease with targeted next-generation sequencing. Sci. Transl. Med. 2012, 4, 118ra10. [Google Scholar] [CrossRef]
- Hallmann, K.; Kudin, A.P.; Zsurka, G.; Kornblum, C.; Reimann, J.; Stüve, B.; Waltz, S.; Hattingen, E.; Thiele, H.; Nürnberg, P.; et al. Loss of the smallest subunit of cytochrome c oxidase, COX8A, causes Leigh-like syndrome and epilepsy. Brain J. Neurol. 2016, 139, 338–345. [Google Scholar] [CrossRef] [PubMed]
- Pitceathly, R.D.S.; Rahman, S.; Wedatilake, Y.; Polke, J.M.; Cirak, S.; Foley, A.R.; Sailer, A.; Hurles, M.E.; Stalker, J.; Hargreaves, I.; et al. NDUFA4 mutations underlie dysfunction of a cytochrome c oxidase subunit linked to human neurological disease. Cell Rep. 2013, 3, 1795–1805. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, R.; Wanschers, B.F.J.; Nijtmans, L.G.; Rodenburg, R.J.; Zschocke, J.; Dikow, N.; van den Brand, M.A.M.; Hendriks-Franssen, M.G.M.; Gilissen, C.; Veltman, J.A.; et al. A mutation in the FAM36A gene, the human ortholog of COX20, impairs cytochrome c oxidase assembly and is associated with ataxia and muscle hypotonia. Hum. Mol. Genet. 2013, 22, 656–667. [Google Scholar] [CrossRef] [PubMed]
- Doss, S.; Lohmann, K.; Seibler, P.; Arns, B.; Klopstock, T.; Zühlke, C.; Freimann, K.; Winkler, S.; Lohnau, T.; Drungowski, M.; et al. Recessive dystonia-ataxia syndrome in a Turkish family caused by a COX20 (FAM36A) mutation. J. Neurol. 2014, 261, 207–212. [Google Scholar] [CrossRef] [PubMed]
- Otero, M.G.; Tiongson, E.; Diaz, F.; Haude, K.; Panzer, K.; Collier, A.; Kim, J.; Adams, D.; Tifft, C.J.; Cui, H.; et al. Novel pathogenic COX20 variants causing dysarthria, ataxia, and sensory neuropathy. Ann. Clin. Transl. Neurol. 2019, 6, 154–160. [Google Scholar] [CrossRef]
- Xu, H.; Ji, T.; Lian, Y.; Wang, S.; Chen, X.; Li, S.; Yin, Y.; Dong, X. Observation of novel COX20 mutations related to autosomal recessive axonal neuropathy and static encephalopathy. Hum. Genet. 2019, 138, 749–756. [Google Scholar] [CrossRef]
- Weraarpachai, W.; Sasarman, F.; Nishimura, T.; Antonicka, H.; Auré, K.; Rötig, A.; Lombès, A.; Shoubridge, E.A. Mutations in C12orf62, a factor that couples COX I synthesis with cytochrome c oxidase assembly, cause fatal neonatal lactic acidosis. Am. J. Hum. Genet. 2012, 90, 142–151. [Google Scholar] [CrossRef]
- Renkema, G.H.; Visser, G.; Baertling, F.; Wintjes, L.T.; Wolters, V.M.; van Montfrans, J.; de Kort, G.A.P.; Nikkels, P.G.J.; van Hasselt, P.M.; van der Crabben, S.N.; et al. Mutated PET117 causes complex IV deficiency and is associated with neurodevelopmental regression and medulla oblongata lesions. Hum. Genet. 2017, 136, 759–769. [Google Scholar] [CrossRef]
- Huigsloot, M.; Nijtmans, L.G.; Szklarczyk, R.; Baars, M.J.H.; van den Brand, M.A.M.; Hendriksfranssen, M.G.M.; van den Heuvel, L.P.; Smeitink, J.A.M.; Huynen, M.A.; Rodenburg, R.J.T. A mutation in C2orf64 causes impaired cytochrome c oxidase assembly and mitochondrial cardiomyopathy. Am. J. Hum. Genet. 2011, 88, 488–493. [Google Scholar] [CrossRef]
- Lim, S.C.; Smith, K.R.; Stroud, D.A.; Compton, A.G.; Tucker, E.J.; Dasvarma, A.; Gandolfo, L.C.; Marum, J.E.; McKenzie, M.; Peters, H.L.; et al. A founder mutation in PET100 causes isolated complex IV deficiency in Lebanese individuals with Leigh syndrome. Am. J. Hum. Genet. 2014, 94, 209–222. [Google Scholar] [CrossRef]
- Oláhová, M.; Haack, T.B.; Alston, C.L.; Houghton, J.A.; He, L.; Morris, A.A.; Brown, G.K.; McFarland, R.; Chrzanowska-Lightowlers, Z.M.; Lightowlers, R.N.; et al. A truncating PET100 variant causing fatal infantile lactic acidosis and isolated cytochrome c oxidase deficiency. Eur. J. Hum. Genet. EJHG 2015, 23, 935–939. [Google Scholar] [CrossRef] [PubMed]
- Baertling, F.; van den Brand, M.A.M.; Hertecant, J.L.; Al-Shamsi, A.; van den Heuvel, L.P.; Distelmaier, F.; Mayatepek, E.; Smeitink, J.A.; Nijtmans, L.G.J.; Rodenburg, R.J.T. Mutations in COA6 cause cytochrome c oxidase deficiency and neonatal hypertrophic cardiomyopathy. Hum. Mutat. 2015, 36, 34–38. [Google Scholar] [CrossRef]
- Ghosh, A.; Trivedi, P.P.; Timbalia, S.A.; Griffin, A.T.; Rahn, J.J.; Chan, S.S.L.; Gohil, V.M. Copper supplementation restores cytochrome c oxidase assembly defect in a mitochondrial disease model of COA6 deficiency. Hum. Mol. Genet. 2014, 23, 3596–3606. [Google Scholar] [CrossRef]
- Ostergaard, E.; Weraarpachai, W.; Ravn, K.; Born, A.P.; Jønson, L.; Duno, M.; Wibrand, F.; Shoubridge, E.A.; Vissing, J. Mutations in COA3 cause isolated complex IV deficiency associated with neuropathy, exercise intolerance, obesity, and short stature. J. Med. Genet. 2015, 52, 203–207. [Google Scholar] [CrossRef] [PubMed]
- Miryounesi, M.; Fardaei, M.; Tabei, S.M.; Ghafouri-Fard, S. Leigh syndrome associated with a novel mutation in the COX15 gene. J. Pediatr. Endocrinol. Metab. JPEM 2016, 29, 741–744. [Google Scholar] [CrossRef] [PubMed]
- Oquendo, C.E.; Antonicka, H.; Shoubridge, E.A.; Reardon, W.; Brown, G.K. Functional and genetic studies demonstrate that mutation in the COX15 gene can cause Leigh syndrome. J. Med. Genet. 2004, 41, 540–544. [Google Scholar] [CrossRef] [PubMed]
- Alfadhel, M.; Lillquist, Y.P.; Waters, P.J.; Sinclair, G.; Struys, E.; McFadden, D.; Hendson, G.; Hyams, L.; Shoffner, J.; Vallance, H.D. Infantile cardioencephalopathy due to a COX15 gene defect: Report and review. Am. J. Med. Genet. Part A 2011, 155, 840–844. [Google Scholar] [CrossRef]
- Antonicka, H.; Mattman, A.; Carlson, C.G.; Glerum, D.M.; Hoffbuhr, K.C.; Leary, S.C.; Kennaway, N.G.; Shoubridge, E.A. Mutations in COX15 produce a defect in the mitochondrial heme biosynthetic pathway, causing early-onset fatal hypertrophic cardiomyopathy. Am. J. Hum. Genet. 2003, 72, 101–114. [Google Scholar] [CrossRef]
- Coenen, M.J.H.; van den Heuvel, L.P.; Ugalde, C.; Ten Brinke, M.; Nijtmans, L.G.J.; Trijbels, F.J.M.; Beblo, S.; Maier, E.M.; Muntau, A.C.; Smeitink, J.A.M. Cytochrome c oxidase biogenesis in a patient with a mutation in COX10 gene. Ann. Neurol. 2004, 56, 560–564. [Google Scholar] [CrossRef]
- Antonicka, H.; Leary, S.C.; Guercin, G.-H.; Agar, J.N.; Horvath, R.; Kennaway, N.G.; Harding, C.O.; Jaksch, M.; Shoubridge, E.A. Mutations in COX10 result in a defect in mitochondrial heme A biosynthesis and account for multiple, early-onset clinical phenotypes associated with isolated COX deficiency. Hum. Mol. Genet. 2003, 12, 2693–2702. [Google Scholar] [CrossRef]
- Pronicka, E.; Piekutowska-Abramczuk, D.; Ciara, E.; Trubicka, J.; Rokicki, D.; Karkucińska-Więckowska, A.; Pajdowska, M.; Jurkiewicz, E.; Halat, P.; Kosińska, J.; et al. New perspective in diagnostics of mitochondrial disorders: Two years’ experience with whole-exome sequencing at a national paediatric centre. J. Transl. Med. 2016, 14. [Google Scholar] [CrossRef] [PubMed]
- Kohda, M.; Tokuzawa, Y.; Kishita, Y.; Nyuzuki, H.; Moriyama, Y.; Mizuno, Y.; Hirata, T.; Yatsuka, Y.; Yamashita-Sugahara, Y.; Nakachi, Y.; et al. A Comprehensive Genomic Analysis Reveals the Genetic Landscape of Mitochondrial Respiratory Chain Complex Deficiencies. PLoS Genet. 2016, 12, e1005679. [Google Scholar] [CrossRef] [PubMed]
- Brix, N.; Jensen, J.M.; Pedersen, I.S.; Ernst, A.; Frost, S.; Bogaard, P.; Petersen, M.B.; Bender, L. Mitochondrial Disease Caused by a Novel Homozygous Mutation (Gly106del) in the SCO1 Gene. Neonatology 2019, 116, 290–294. [Google Scholar] [CrossRef] [PubMed]
- Leary, S.C.; Antonicka, H.; Sasarman, F.; Weraarpachai, W.; Cobine, P.A.; Pan, M.; Brown, G.K.; Brown, R.; Majewski, J.; Ha, K.C.H.; et al. Novel mutations in SCO1 as a cause of fatal infantile encephalopathy and lactic acidosis. Hum. Mutat. 2013, 34, 1366–1370. [Google Scholar] [CrossRef]
- Stiburek, L.; Vesela, K.; Hansikova, H.; Hulkova, H.; Zeman, J. Loss of function of Sco1 and its interaction with cytochrome c oxidase. Am. J. Physiol. Cell Physiol. 2009, 296, C1218–C1226. [Google Scholar] [CrossRef]
- Mitchell, P. Coupling of Phosphorylation to Electron and Hydrogen Transfer by a Chemi-Osmotic type of Mechanism. Nature 1961, 191, 144–148. [Google Scholar] [CrossRef]
- Kagawa, Y.; Racker, E. Partial Resolution of the Enzymes Catalyzing Oxidative Phosphorylation VIII. Properties of a factor conferring oligomycin sensitivity on mitochondrial adenosine triphosphatase. J. Biol. Chem. 1966, 241, 2461–2466. [Google Scholar]
- Boyer, P.D. The Atp synthase—A splendid molecular machine. Annu. Rev. Biochem. 1997, 66, 717–749. [Google Scholar] [CrossRef]
- Velours, J.; Arselin, G. The Saccharomyces cerevisiae ATP Synthase. J. Bioenerg. Biomembr. N. Y. 2000, 32, 383–390. [Google Scholar] [CrossRef]
- Anderson, S.; Bankier, A.T.; Barrell, B.G.; de Bruijn, M.H.L.; Coulson, A.R.; Drouin, J.; Eperon, I.C.; Nierlich, D.P.; Roe, B.A.; Sanger, F.; et al. Sequence and organization of the human mitochondrial genome. Nature 1981, 290, 457–465. [Google Scholar] [CrossRef] [PubMed]
- Dautant, A.; Velours, J.; Giraud, M.-F. Crystal Structure of the Mg·ADP-inhibited State of the Yeast F1c10-ATP Synthase. J. Biol. Chem. 2010, 285, 29502–29510. [Google Scholar] [CrossRef] [PubMed]
- Symersky, J.; Pagadala, V.; Osowski, D.; Krah, A.; Meier, T.; Faraldo-Gómez, J.D.; Mueller, D.M. Structure of the c10 ring of the yeast mitochondrial ATP synthase in the open conformation. Nat. Struct. Mol. Biol. 2012, 19, 485–491. [Google Scholar] [CrossRef] [PubMed]
- Watt, I.N.; Montgomery, M.G.; Runswick, M.J.; Leslie, A.G.W.; Walker, J.E. Bioenergetic cost of making an adenosine triphosphate molecule in animal mitochondria. Proc. Natl. Acad. Sci. USA 2010, 107, 16823–16827. [Google Scholar] [CrossRef]
- Schon, E.A.; Santra, S.; Pallotti, F.; Girvin, M.E. Pathogenesis of primary defects in mitochondrial ATP synthesis. Semin. Cell Dev. Biol. 2001, 12, 441–448. [Google Scholar] [CrossRef] [PubMed]
- Holt, I.J.; Harding, A.E.; Petty, R.K.; Morgan-Hughes, J.A. A new mitochondrial disease associated with mitochondrial DNA heteroplasmy. Am. J. Hum. Genet. 1990, 46, 428–433. [Google Scholar]
- Ganetzky, R.D.; Stendel, C.; McCormick, E.M.; Zolkipli-Cunningham, Z.; Goldstein, A.C.; Klopstock, T.; Falk, M.J. MT-ATP6 mitochondrial disease variants: Phenotypic and biochemical features analysis in 218 published cases and cohort of 14 new cases. Hum. Mutat. 2019, 40, 499–515. [Google Scholar] [CrossRef]
- Rak, M.; Tetaud, E.; Duvezin-Caubet, S.; Ezkurdia, N.; Bietenhader, M.; Rytka, J.; di Rago, J.-P. A yeast model of the neurogenic ataxia retinitis pigmentosa (NARP) T8993G mutation in the mitochondrial ATP synthase-6 gene. J. Biol. Chem. 2007, 282, 34039–34047. [Google Scholar] [CrossRef]
- Franco, L.V.R.; Su, C.-H.; Burnett, J.; Teixeira, L.S.; Tzagoloff, A. Atco, a yeast mitochondrial complex of Atp9 and Cox6, is an assembly intermediate of the ATP synthase. PLoS ONE 2020, 15, e0233177. [Google Scholar] [CrossRef]
- Kabala, A.M.; Lasserre, J.-P.; Ackerman, S.H.; di Rago, J.-P.; Kucharczyk, R. Defining the impact on yeast ATP synthase of two pathogenic human mitochondrial DNA mutations, T9185C and T9191C. Biochimie 2014, 100, 200–206. [Google Scholar] [CrossRef]
- Su, X.; Dautant, A.; Godard, F.; Bouhier, M.; Zoladek, T.; Kucharczyk, R.; di Rago, J.-P.; Tribouillard-Tanvier, D. Molecular Basis of the Pathogenic Mechanism Induced by the m.9191T>C Mutation in Mitochondrial ATP6 Gene. Int. J. Mol. Sci. 2020, 21, 5083. [Google Scholar] [CrossRef]
- Houštěk, J.; Pícková, A.; Vojtíšková, A.; Mráček, T.; Pecina, P.; Ješina, P. Mitochondrial diseases and genetic defects of ATP synthase. Biochim. Biophys. Acta BBA Bioenerg. 2006, 1757, 1400–1405. [Google Scholar] [CrossRef] [PubMed]
- Jonckheere, A.I.; Hogeveen, M.; Nijtmans, L.; van den Brand, M.; Janssen, A.; Diepstra, H.; van den Brandt, F.; van den Heuvel, B.; Hol, F.; Hofste, T.; et al. A novel mitochondrial ATP8 gene mutation in a patient with apical hypertrophic cardiomyopathy and neuropathy. BMJ Case Rep. 2009, 2009. [Google Scholar] [CrossRef] [PubMed]
- Tansel, T.; Paçal, F.; Ustek, D. A novel ATP8 gene mutation in an infant with tetralogy of Fallot. Cardiol. Young Camb. 2014, 24, 531–533. [Google Scholar] [CrossRef]
- Dautant, A.; Meier, T.; Hahn, A.; Tribouillard-Tanvier, D.; di Rago, J.-P.; Kucharczyk, R. ATP Synthase Diseases of Mitochondrial Genetic Origin. Front. Physiol. 2018, 9. [Google Scholar] [CrossRef]
- Kytövuori, L.; Lipponen, J.; Rusanen, H.; Komulainen, T.; Martikainen, M.H.; Majamaa, K. A novel mutation m.8561C>G in MT-ATP6/8 causing a mitochondrial syndrome with ataxia, peripheral neuropathy, diabetes mellitus, and hypergonadotropic hypogonadism. J. Neurol. 2016, 263, 2188–2195. [Google Scholar] [CrossRef]
- Fragaki, K.; Chaussenot, A.; Serre, V.; Acquaviva, C.; Bannwarth, S.; Rouzier, C.; Chabrol, B.; Paquis-Flucklinger, V. A novel variant m.8561C>T in the overlapping region of MT-ATP6 and MT-ATP8 in a child with early-onset severe neurological signs. Mol. Genet. Metab. Rep. 2019, 21. [Google Scholar] [CrossRef]
- Holme, E.; Greter, J.; Jacobson, C.-E.; Larsson, N.-G.; Lindstedt, S.; Nilsson, K.O.; Oldfors, A.; Tulinius, M. Mitochondrial ATP-Synthase Deficiency in a Child with 3-Methylglutaconic Aciduria. Pediatr. Res. 1992, 32, 731–736. [Google Scholar] [CrossRef]
- Houštek, J.; Klement, P.; Floryk, D.; Antonická, H.; Hermanská, J.; Kalous, M.; Hansíková, H.; Houšt’ková, H.; Chowdhury, S.K.R.; Rosipal, Š.; et al. A Novel Deficiency of Mitochondrial ATPase of Nuclear Origin. Hum. Mol. Genet. 1999, 8, 1967–1974. [Google Scholar] [CrossRef][Green Version]
- Sperl, W.; Ješina, P.; Zeman, J.; Mayr, J.A.; DeMeirleir, L.; VanCoster, R.; Pícková, A.; Hansíková, H.; Houšt’ková, H.; Krejčík, Z.; et al. Deficiency of mitochondrial ATP synthase of nuclear genetic origin. Neuromuscul. Disord. 2006, 16, 821–829. [Google Scholar] [CrossRef]
- Sardin, E.; Donadello, S.; di Rago, J.-P.; Tetaud, E. Biochemical investigation of a human pathogenic mutation in the nuclear ATP5E gene using yeast as a model. Front. Genet. 2015, 6. [Google Scholar] [CrossRef][Green Version]
- Oláhová, M.; Yoon, W.H.; Thompson, K.; Jangam, S.; Fernandez, L.; Davidson, J.M.; Kyle, J.E.; Grove, M.E.; Fisk, D.G.; Kohler, J.N.; et al. Biallelic Mutations in ATP5F1D, which Encodes a Subunit of ATP Synthase, Cause a Metabolic Disorder. Am. J. Hum. Genet. 2018, 102, 494–504. [Google Scholar] [CrossRef] [PubMed]
- Feichtinger, R.G.; Schäfer, G.; Seifarth, C.; Mayr, J.A.; Kofler, B.; Klocker, H. Reduced Levels of ATP Synthase Subunit ATP5F1A Correlate with Earlier-Onset Prostate Cancer. Oxid. Med. Cell. Longev. 2018, 2018. [Google Scholar] [CrossRef] [PubMed]
- Franco, L.V.R.; Su, C.H.; Tzagoloff, A. Modular assembly of yeast mitochondrial ATP synthase and cytochrome oxidase. Biol. Chem. 2020, 401. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.-G.; Sheluho, D.; Gatti, D.L.; Ackerman, S.H. The α-subunit of the mitochondrial F1 ATPase interacts directly with the assembly factor Atp12p. EMBO J. 2000, 19, 1486–1493. [Google Scholar] [CrossRef]
- Kratochvílová, H.; Hejzlarová, K.; Vrbacký, M.; Mráček, T.; Karbanová, V.; Tesařová, M.; Gombitová, A.; Cmarko, D.; Wittig, I.; Zeman, J.; et al. Mitochondrial membrane assembly of TMEM70 protein. Mitochondrion 2014, 15, 1–9. [Google Scholar] [CrossRef]
- Sánchez-Caballero, L.; Elurbe, D.M.; Baertling, F.; Guerrero-Castillo, S.; van den Brand, M.; van Strien, J.; van Dam, T.J.P.; Rodenburg, R.; Brandt, U.; Huynen, M.A.; et al. TMEM70 functions in the assembly of complexes I and V. Biochim. Biophys. Acta BBA Bioenerg. 2020, 1861. [Google Scholar] [CrossRef]
- De Meirleir, L. Respiratory chain complex V deficiency due to a mutation in the assembly gene ATP12. J. Med. Genet. 2004, 41, 120–124. [Google Scholar] [CrossRef]
- Lefebvre-Legendre, L.; Vaillier, J.; Benabdelhak, H.; Velours, J.; Slonimski, P.P.; Rago, J.-P. di Identification of a Nuclear Gene (FMC1) Required for the Assembly/Stability of Yeast Mitochondrial F1-ATPase in Heat Stress Conditions. J. Biol. Chem. 2001, 276, 6789–6796. [Google Scholar] [CrossRef]
- Turunen, M.; Olsson, J.; Dallner, G. Metabolism and function of coenzyme Q. Biochim. Biophys. Acta BBA Biomembr. 2004, 1660, 171–199. [Google Scholar] [CrossRef]
- Tran, U.C.; Clarke, C.F. Endogenous synthesis of coenzyme Q in eukaryotes. Mitochondrion 2007, 7, S62–S71. [Google Scholar] [CrossRef]
- Kawamukai, M. Biosynthesis and bioproduction of coenzyme Q10 by yeasts and other organisms. Biotechnol. Appl. Biochem. 2009, 53, 217–226. [Google Scholar] [CrossRef] [PubMed]
- Tzagoloff, A.; Akai, A.; Needleman, R.B. Assembly of the mitochondrial membrane system. Characterization of nuclear mutants of Saccharomyces cerevisiae with defects in mitochondrial ATPase and respiratory enzymes. J. Biol. Chem. 1975, 250, 8228–8235. [Google Scholar] [PubMed]
- Marbois, B.; Gin, P.; Gulmezian, M.; Clarke, C.F. The yeast Coq4 polypeptide organizes a mitochondrial protein complex essential for coenzyme Q biosynthesis. Biochim. Biophys. Acta BBA Mol. Cell Biol. Lipids 2009, 1791, 69–75. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, K.; Jochem, A.; Le Vasseur, M.; Lewis, S.; Paulson, B.R.; Reddy, T.R.; Russell, J.D.; Coon, J.J.; Pagliarini, D.J.; Nunnari, J. Coenzyme Q biosynthetic proteins assemble in a substrate-dependent manner into domains at ER–mitochondria contacts. J. Cell Biol. 2019, 218, 1353–1369. [Google Scholar] [CrossRef] [PubMed]
- Awad, A.M.; Bradley, M.C.; Fernández-del-Río, L.; Nag, A.; Tsui, H.S.; Clarke, C.F. Coenzyme Q10 deficiencies: Pathways in yeast and humans. Essays Biochem. 2018, 62, 361–376. [Google Scholar] [CrossRef]
- Gloor, U.; Wiss, O. On the biosynthesis of ubiquinone (50). Arch. Biochem. Biophys. 1959, 83, 216–222. [Google Scholar] [CrossRef]
- Pierrel, F.; Hamelin, O.; Douki, T.; Kieffer-Jaquinod, S.; Mühlenhoff, U.; Ozeir, M.; Lill, R.; Fontecave, M. Involvement of Mitochondrial Ferredoxin and Para-Aminobenzoic Acid in Yeast Coenzyme Q Biosynthesis. Chem. Biol. 2010, 17, 449–459. [Google Scholar] [CrossRef]
- Payet, L.-A.; Leroux, M.; Willison, J.C.; Kihara, A.; Pelosi, L.; Pierrel, F. Mechanistic Details of Early Steps in Coenzyme Q Biosynthesis Pathway in Yeast. Cell Chem. Biol. 2016, 23, 1241–1250. [Google Scholar] [CrossRef]
- Stefely, J.A.; Pagliarini, D.J. Biochemistry of Mitochondrial Coenzyme Q Biosynthesis. Trends Biochem. Sci. 2017, 42, 824–843. [Google Scholar] [CrossRef]
- Ashby, M.N.; Edwards, P.A. Elucidation of the deficiency in two yeast coenzyme Q mutants. Characterization of the structural gene encoding hexaprenyl pyrophosphate synthetase. J. Biol. Chem. 1990, 265, 13157–13164. [Google Scholar]
- Ashby, M.N.; Kutsunai, S.Y.; Ackerman, S.; Tzagoloff, A.; Edwards, P.A. COQ2 is a candidate for the structural gene encoding para-hydroxybenzoate: Polyprenyltransferase. J. Biol. Chem. 1992, 267, 4128–4136. [Google Scholar]
- Gin, P.; Hsu, A.Y.; Rothman, S.C.; Jonassen, T.; Lee, P.T.; Tzagoloff, A.; Clarke, C.F. The Saccharomyces cerevisiae COQ6 Gene Encodes a Mitochondrial Flavin-dependent Monooxygenase Required for Coenzyme Q Biosynthesis. J. Biol. Chem. 2003, 278, 25308–25316. [Google Scholar] [CrossRef]
- Ozeir, M.; Mühlenhoff, U.; Webert, H.; Lill, R.; Fontecave, M.; Pierrel, F. Coenzyme Q Biosynthesis: Coq6 Is Required for the C5-Hydroxylation Reaction and Substrate Analogs Rescue Coq6 Deficiency. Chem. Biol. 2011, 18, 1134–1142. [Google Scholar] [CrossRef]
- Marbois, B.N.; Clarke, C.F. The COQ7 Gene Encodes a Protein in Saccharomyces cerevisiae Necessary for Ubiquinone Biosynthesis. J. Biol. Chem. 1996, 271, 2995–3004. [Google Scholar] [CrossRef]
- Poon, W.W.; Barkovich, R.J.; Hsu, A.Y.; Frankel, A.; Lee, P.T.; Shepherd, J.N.; Myles, D.C.; Clarke, C.F. Yeast and Rat Coq3 and Escherichia coli UbiG Polypeptides Catalyze Both O-Methyltransferase Steps in Coenzyme Q Biosynthesis. J. Biol. Chem. 1999, 274, 21665–21672. [Google Scholar] [CrossRef]
- Barkovich, R.J.; Shtanko, A.; Shepherd, J.A.; Lee, P.T.; Myles, D.C.; Tzagoloff, A.; Clarke, C.F. Characterization of the COQ5 Gene from Saccharomyces cerevisiae evidence for a C-methyltransferase in ubiquinone biosynthesis. J. Biol. Chem. 1997, 272, 9182–9188. [Google Scholar] [CrossRef] [PubMed]
- Xie, L.X.; Hsieh, E.J.; Watanabe, S.; Allan, C.M.; Chen, J.Y.; Tran, U.C.; Clarke, C.F. Expression of the human atypical kinase ADCK3 rescues coenzyme Q biosynthesis and phosphorylation of Coq polypeptides in yeast coq8 mutants. Biochim. Biophys. Acta BBA Mol. Cell Biol. Lipids 2011, 1811, 348–360. [Google Scholar] [CrossRef] [PubMed]
- Tauche, A.; Krause-Buchholz, U.; Rödel, G. Ubiquinone biosynthesis in Saccharomyces cerevisiae: The molecular organization of O-methylase Coq3p depends on Abc1p/Coq8p. FEMS Yeast Res. 2008, 8, 1263–1275. [Google Scholar] [CrossRef] [PubMed]
- Lagier-Tourenne, C.; Tazir, M.; López, L.C.; Quinzii, C.M.; Assoum, M.; Drouot, N.; Busso, C.; Makri, S.; Ali-Pacha, L.; Benhassine, T.; et al. ADCK3, an Ancestral Kinase, Is Mutated in a Form of Recessive Ataxia Associated with Coenzyme Q10 Deficiency. Am. J. Hum. Genet. 2008, 82, 661–672. [Google Scholar] [CrossRef]
- Hsieh, E.J.; Gin, P.; Gulmezian, M.; Tran, U.C.; Saiki, R.; Marbois, B.N.; Clarke, C.F. Saccharomyces cerevisiae Coq9 polypeptide is a subunit of the mitochondrial coenzyme Q biosynthetic complex. Arch. Biochem. Biophys. 2007, 463, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Awad, A.M.; Nag, A.; Pham, N.V.B.; Bradley, M.C.; Jabassini, N.; Nathaniel, J.; Clarke, C.F. Intragenic suppressor mutations of the COQ8 protein kinase homolog restore coenzyme Q biosynthesis and function in Saccharomyces cerevisiae. PLoS ONE 2020, 15, e0234192. [Google Scholar] [CrossRef] [PubMed]
- Bradley, M.C.; Yang, K.; Fernández-del-Río, L.; Ngo, J.; Ayer, A.; Tsui, H.S.; Novales, N.A.; Stocker, R.; Shirihai, O.S.; Barros, M.H.; et al. COQ11 deletion mitigates respiratory deficiency caused by mutations in the gene encoding the coenzyme Q chaperone protein Coq10. J. Biol. Chem. 2020, 295, 6023–6042. [Google Scholar] [CrossRef] [PubMed]
- Allan, C.M.; Awad, A.M.; Johnson, J.S.; Shirasaki, D.I.; Wang, C.; Blaby-Haas, C.E.; Merchant, S.S.; Loo, J.A.; Clarke, C.F. Identification of Coq11, a New Coenzyme Q Biosynthetic Protein in the CoQ-Synthome in Saccharomyces cerevisiae. J. Biol. Chem. 2015, 290, 7517–7534. [Google Scholar] [CrossRef]
- Ogasahara, S.; Engel, A.G.; Frens, D.; Mack, D. Muscle coenzyme Q deficiency in familial mitochondrial encephalomyopathy. Proc. Natl. Acad. Sci. USA 1989, 86, 2379–2382. [Google Scholar] [CrossRef] [PubMed]
- Emmanuele, V.; López, L.C.; Berardo, A.; Naini, A.; Tadesse, S.; Wen, B.; D’Agostino, E.; Solomon, M.; DiMauro, S.; Quinzii, C.; et al. Heterogeneity of Coenzyme Q10 Deficiency: Patient Study and Literature Review. Arch. Neurol. 2012, 69, 978–983. [Google Scholar] [CrossRef] [PubMed]
- Desbats, M.A.; Lunardi, G.; Doimo, M.; Trevisson, E.; Salviati, L. Genetic bases and clinical manifestations of coenzyme Q10 (CoQ10) deficiency. J. Inherit. Metab. Dis. 2015, 38, 145–156. [Google Scholar] [CrossRef] [PubMed]
- Acosta, M.J.; Vazquez Fonseca, L.; Desbats, M.A.; Cerqua, C.; Zordan, R.; Trevisson, E.; Salviati, L. Coenzyme Q biosynthesis in health and disease. Biochim. Biophys. Acta BBA Bioenerg. 2016, 1857, 1079–1085. [Google Scholar] [CrossRef]
- Malicdan, M.C.V.; Vilboux, T.; Ben-Zeev, B.; Guo, J.; Eliyahu, A.; Pode-Shakked, B.; Dori, A.; Kakani, S.; Chandrasekharappa, S.C.; Ferreira, C.R.; et al. A novel inborn error of the coenzyme Q10 biosynthesis pathway: Cerebellar ataxia and static encephalomyopathy due to COQ5 C-methyltransferase deficiency. Hum. Mutat. 2018, 39, 69–79. [Google Scholar] [CrossRef]
- Montini, G.; Malaventura, C.; Salviati, L. Early Coenzyme Q10 Supplementation in Primary Coenzyme Q10 Deficiency. N. Engl. J. Med. 2008, 26, 2849–2850. [Google Scholar] [CrossRef]
- Quinzii, C.M.; Hirano, M. Coenzyme Q and mitochondrial disease. Dev. Disabil. Res. Rev. 2010, 16, 183–188. [Google Scholar] [CrossRef]
- López, L.C.; Quinzii, C.M.; Area, E.; Naini, A.; Rahman, S.; Schuelke, M.; Salviati, L.; DiMauro, S.; Hirano, M. Treatment of CoQ10 Deficient Fibroblasts with Ubiquinone, CoQ Analogs, and Vitamin C: Time- and Compound-Dependent Effects. PLoS ONE 2010, 5, e11897. [Google Scholar] [CrossRef] [PubMed]
- Do, T.Q.; Hsu, A.Y.; Jonassen, T.; Lee, P.T.; Clarke, C.F. A Defect in Coenzyme Q Biosynthesis Is Responsible for the Respiratory Deficiency in Saccharomyces cerevisiae abc1Mutants. J. Biol. Chem. 2001, 276, 18161–18168. [Google Scholar] [CrossRef] [PubMed]
- Zampol, M.A.; Busso, C.; Gomes, F.; Ferreira-Junior, J.R.; Tzagoloff, A.; Barros, M.H. Over-expression of COQ10 in Saccharomyces cerevisiae inhibits mitochondrial respiration. Biochem. Biophys. Res. Commun. 2010, 402, 82–87. [Google Scholar] [CrossRef]
- Busso, C.; Ferreira-Júnior, J.R.; Paulela, J.A.; Bleicher, L.; Demasi, M.; Barros, M.H. Coq7p relevant residues for protein activity and stability. Biochimie 2015, 119, 92–102. [Google Scholar] [CrossRef]
- Jonassen, T.; Clarke, C.F. Isolation and Functional Expression of Human COQ3, a Gene Encoding a Methyltransferase Required for Ubiquinone Biosynthesis. J. Biol. Chem. 2000, 275, 12381–12387. [Google Scholar] [CrossRef]
- Barros, M.H.; Johnson, A.; Gin, P.; Marbois, B.N.; Clarke, C.F.; Tzagoloff, A. The Saccharomyces cerevisiae COQ10 Gene Encodes a START Domain Protein Required for Function of Coenzyme Q in Respiration. J. Biol. Chem. 2005, 280, 42627–42635. [Google Scholar] [CrossRef] [PubMed]
- He, C.H.; Black, D.S.; Allan, C.M.; Meunier, B.; Rahman, S.; Clarke, C.F. Human COQ9 Rescues a coq9 Yeast Mutant by Enhancing Coenzyme Q Biosynthesis from 4-Hydroxybenzoic Acid and Stabilizing the CoQ-Synthome. Front. Physiol. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.P.T.; Casarin, A.; Desbats, M.A.; Doimo, M.; Trevisson, E.; Santos-Ocaña, C.; Navas, P.; Clarke, C.F.; Salviati, L. Molecular characterization of the human COQ5 C-methyltransferase in coenzyme Q10 biosynthesis. Biochim. Biophys. Acta BBA Mol. Cell Biol. Lipids 2014, 1841, 1628–1638. [Google Scholar] [CrossRef] [PubMed]
- Mollet, J.; Giurgea, I.; Schlemmer, D.; Dallner, G.; Chretien, D.; Delahodde, A.; Bacq, D.; de Lonlay, P.; Munnich, A.; Rötig, A. Prenyldiphosphate synthase, subunit 1 (PDSS1) and OH-benzoate polyprenyltransferase (COQ2) mutations in ubiquinone deficiency and oxidative phosphorylation disorders. J. Clin. Investig. 2007, 117, 765–772. [Google Scholar] [CrossRef]
- López-Martín, J.M.; Salviati, L.; Trevisson, E.; Montini, G.; DiMauro, S.; Quinzii, C.; Hirano, M.; Rodriguez-Hernandez, A.; Cordero, M.D.; Sánchez-Alcázar, J.A.; et al. Missense mutation of the COQ2 gene causes defects of bioenergetics and de novo pyrimidine synthesis. Hum. Mol. Genet. 2007, 16, 1091–1097. [Google Scholar] [CrossRef]
- Salviati, L.; Trevisson, E.; Hernandez, M.A.R.; Casarin, A.; Pertegato, V.; Doimo, M.; Cassina, M.; Agosto, C.; Desbats, M.A.; Sartori, G.; et al. Haploinsufficiency of COQ4 causes coenzyme Q10 deficiency. J. Med. Genet. 2012, 49, 187–191. [Google Scholar] [CrossRef] [PubMed]
- Heeringa, S.F.; Chernin, G.; Chaki, M.; Zhou, W.; Sloan, A.J.; Ji, Z.; Xie, L.X.; Salviati, L.; Hurd, T.W.; Vega-Warner, V.; et al. COQ6 mutations in human patients produce nephrotic syndrome with sensorineural deafness. J. Clin. Investig. 2011, 121, 2013–2024. [Google Scholar] [CrossRef] [PubMed]
- Duncan, A.J.; Bitner-Glindzicz, M.; Meunier, B.; Costello, H.; Hargreaves, I.P.; López, L.C.; Hirano, M.; Quinzii, C.M.; Sadowski, M.I.; Hardy, J.; et al. A Nonsense Mutation in COQ9 Causes Autosomal-Recessive Neonatal-Onset Primary Coenzyme Q10 Deficiency: A Potentially Treatable Form of Mitochondrial Disease. Am. J. Hum. Genet. 2009, 84, 558–566. [Google Scholar] [CrossRef] [PubMed]
- Lake, N.J.; Compton, A.G.; Rahman, S.; Thorburn, D.R. Leigh syndrome: One disorder, more than 75 monogenic causes. Ann. Neurol. 2016, 79, 190–203. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Lin, S.; Li, L.; Tang, Z.; Hu, Y.; Ban, X.; Zeng, T.; Zhou, Y.; Zhu, Y.; Gao, S.; et al. PDSS2 Deficiency Induces Hepatocarcinogenesis by Decreasing Mitochondrial Respiration and Reprogramming Glucose Metabolism. Cancer Res. 2018, 78, 4471–4481. [Google Scholar] [CrossRef]
- Quinzii, C.; Naini, A.; Salviati, L.; Trevisson, E.; Navas, P.; DiMauro, S.; Hirano, M. A Mutation in Para-Hydroxybenzoate-Polyprenyl Transferase (COQ2) Causes Primary Coenzyme Q10 Deficiency. Am. J. Hum. Genet. 2006, 78, 345–349. [Google Scholar] [CrossRef]
- Caglayan, A.O.; Gumus, H.; Sandford, E.; Kubisiak, T.L.; Ma, Q.; Ozel, A.B.; Per, H.; Li, J.Z.; Shakkottai, V.G.; Burmeister, M. COQ4 Mutation Leads to Childhood-Onset Ataxia Improved by CoQ10 Administration. Cerebellum Lond. Engl. 2019, 18, 665–669. [Google Scholar] [CrossRef]
- Lu, M.; Zhou, Y.; Wang, Z.; Xia, Z.; Ren, J.; Guo, Q. Clinical phenotype, in silico and biomedical analyses, and intervention for an East Asian population-specific c.370G>A (p.G124S) COQ4 mutation in a Chinese family with CoQ10 deficiency-associated Leigh syndrome. J. Hum. Genet. 2019, 64, 297–304. [Google Scholar] [CrossRef]
- Song, C.-C.; Hong, Q.; Geng, X.-D.; Wang, X.; Wang, S.-Q.; Cui, S.-Y.; Guo, M.-D.; Li, O.; Cai, G.-Y.; Chen, X.-M.; et al. New Mutation of Coenzyme Q10 Monooxygenase 6 Causing Podocyte Injury in a Focal Segmental Glomerulosclerosis Patient. Chin. Med. J. 2018, 131, 2666–2675. [Google Scholar] [CrossRef]
- Freyer, C.; Stranneheim, H.; Naess, K.; Mourier, A.; Felser, A.; Maffezzini, C.; Lesko, N.; Bruhn, H.; Engvall, M.; Wibom, R.; et al. Rescue of primary ubiquinone deficiency due to a novel COQ7 defect using 2,4–dihydroxybensoic acid. J. Med. Genet. 2015, 52, 779–783. [Google Scholar] [CrossRef]
- Wang, Y.; Smith, C.; Parboosingh, J.S.; Khan, A.; Innes, M.; Hekimi, S. Pathogenicity of two COQ7 mutations and responses to 2,4-dihydroxybenzoate bypass treatment. J. Cell. Mol. Med. 2017, 21, 2329–2343. [Google Scholar] [CrossRef] [PubMed]
- Kwong, A.K.-Y.; Chiu, A.T.-G.; Tsang, M.H.-Y.; Lun, K.-S.; Rodenburg, R.J.T.; Smeitink, J.; Chung, B.H.-Y.; Fung, C.-W. A fatal case of COQ7-associated primary coenzyme Q10 deficiency. JIMD Rep. 2019, 47, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Cullen, J.K.; Abdul Murad, N.; Yeo, A.; McKenzie, M.; Ward, M.; Chong, K.L.; Schieber, N.L.; Parton, R.G.; Lim, Y.C.; Wolvetang, E.; et al. AarF Domain Containing Kinase 3 (ADCK3) Mutant Cells Display Signs of Oxidative Stress, Defects in Mitochondrial Homeostasis and Lysosomal Accumulation. PLoS ONE 2016, 11, e0148213. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Song, X.; Fang, X.; Tang, X.; Cao, Q.; Zhai, Y.; Chen, J.; Liu, J.; Zhang, Z.; Xiang, T.; Qian, Y.; et al. COQ8B nephropathy: Early detection and optimal treatment. Mol. Genet. Genomic Med. 2020, 8, e1360. [Google Scholar] [CrossRef] [PubMed]
- Lohman, D.C.; Aydin, D.; Von Bank, H.C.; Smith, R.W.; Linke, V.; Weisenhorn, E.; McDevitt, M.T.; Hutchins, P.; Wilkerson, E.M.; Wancewicz, B.; et al. An Isoprene Lipid-Binding Protein Promotes Eukaryotic Coenzyme Q Biosynthesis. Mol. Cell 2019, 73, 763–774.e10. [Google Scholar] [CrossRef]
- Danhauser, K.; Herebian, D.; Haack, T.B.; Rodenburg, R.J.; Strom, T.M.; Meitinger, T.; Klee, D.; Mayatepek, E.; Prokisch, H.; Distelmaier, F. Fatal neonatal encephalopathy and lactic acidosis caused by a homozygous loss-of-function variant in COQ9. Eur. J. Hum. Genet. EJHG 2016, 24, 450–454. [Google Scholar] [CrossRef]
- Smith, A.C.; Ito, Y.; Ahmed, A.; Schwartzentruber, J.A.; Beaulieu, C.L.; Aberg, E.; Majewski, J.; Bulman, D.E.; Horsting-Wethly, K.; Koning, D.V.; et al. A family segregating lethal neonatal coenzyme Q10 deficiency caused by mutations in COQ9. J. Inherit. Metab. Dis. 2018, 41, 719–729. [Google Scholar] [CrossRef]
- Olgac, A.; Öztoprak, Ü.; Kasapkara, Ç.S.; Kılıç, M.; Yüksel, D.; Derinkuyu, E.B.; Taşçı Yıldız, Y.; Ceylaner, S.; Ezgu, F.S. A rare case of primary coenzyme Q10 deficiency due to COQ9 mutation. J. Pediatr. Endocrinol. Metab. JPEM 2020, 33, 165–170. [Google Scholar] [CrossRef]
- Luna-Sánchez, M.; Díaz-Casado, E.; Barca, E.; Tejada, M.Á.; Montilla-García, Á.; Cobos, E.J.; Escames, G.; Acuña-Castroviejo, D.; Quinzii, C.M.; López, L.C. The clinical heterogeneity of coenzyme Q10 deficiency results from genotypic differences in the Coq9 gene. EMBO Mol. Med. 2015, 7, 670–687. [Google Scholar] [CrossRef]
- Quinzii, C.M.; Luna-Sanchez, M.; Ziosi, M.; Hidalgo-Gutierrez, A.; Kleiner, G.; Lopez, L.C. The Role of Sulfide Oxidation Impairment in the Pathogenesis of Primary CoQ Deficiency. Front. Physiol. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Morison, I.M.; Cramer Bordé, E.M.; Cheesman, E.J.; Cheong, P.L.; Holyoake, A.J.; Fichelson, S.; Weeks, R.J.; Lo, A.; Davies, S.M.K.; Wilbanks, S.M.; et al. A mutation of human cytochrome c enhances the intrinsic apoptotic pathway but causes only thrombocytopenia. Nat. Genet. 2008, 40, 387–389. [Google Scholar] [CrossRef] [PubMed]
- De Rocco, D.; Cerqua, C.; Goffrini, P.; Russo, G.; Pastore, A.; Meloni, F.; Nicchia, E.; Moraes, C.T.; Pecci, A.; Salviati, L.; et al. Mutations of cytochrome c identified in patients with thrombocytopenia THC4 affect both apoptosis and cellular bioenergetics. Biochim. Biophys. Acta BBA Mol. Basis Dis. 2014, 1842, 269–274. [Google Scholar] [CrossRef] [PubMed]
- Johnson, B.; Lowe, G.C.; Futterer, J.; Lordkipanidzé, M.; MacDonald, D.; Simpson, M.A.; Sanchez-Guiú, I.; Drake, S.; Bem, D.; Leo, V.; et al. Whole exome sequencing identifies genetic variants in inherited thrombocytopenia with secondary qualitative function defects. Haematologica 2016, 101, 1170–1179. [Google Scholar] [CrossRef] [PubMed]
- Uchiyama, Y.; Yanagisawa, K.; Kunishima, S.; Shiina, M.; Ogawa, Y.; Nakashima, M.; Hirato, J.; Imagawa, E.; Fujita, A.; Hamanaka, K.; et al. A novel CYCS mutation in the α-helix of the CYCS C-terminal domain causes non-syndromic thrombocytopenia. Clin. Genet. 2018, 94, 548–553. [Google Scholar] [CrossRef] [PubMed]
- Ong, L.; Morison, I.M.; Ledgerwood, E.C. Megakaryocytes from CYCS mutation-associated thrombocytopenia release platelets by both proplatelet-dependent and -independent processes. Br. J. Haematol. 2017, 176, 268–279. [Google Scholar] [CrossRef]
- Schwarz, Q.P.; Cox, T.C. Complementation of a yeast CYC3 deficiency identifies an X-linked mammalian activator of apocytochrome c. Genomics 2002, 79, 51–57. [Google Scholar] [CrossRef]
- van Rahden, V.A.; Rau, I.; Fuchs, S.; Kosyna, F.K.; de Almeida, H.L.; Fryssira, H.; Isidor, B.; Jauch, A.; Joubert, M.; Lachmeijer, A.M.A.; et al. Clinical spectrum of females with HCCS mutation: From no clinical signs to a neonatal lethal form of the microphthalmia with linear skin defects (MLS) syndrome. Orphanet J. Rare Dis. 2014, 9. [Google Scholar] [CrossRef]
- Indrieri, A.; Conte, I.; Chesi, G.; Romano, A.; Quartararo, J.; Tatè, R.; Ghezzi, D.; Zeviani, M.; Goffrini, P.; Ferrero, I.; et al. The impairment of HCCS leads to MLS syndrome by activating a non-canonical cell death pathway in the brain and eyes. EMBO Mol. Med. 2013, 5, 280–293. [Google Scholar] [CrossRef]
- Luttik, M.A.; Overkamp, K.M.; Kötter, P.; de Vries, S.; van Dijken, J.P.; Pronk, J.T. The Saccharomyces cerevisiae NDE1 and NDE2 genes encode separate mitochondrial NADH dehydrogenases catalyzing the oxidation of cytosolic NADH. J. Biol. Chem. 1998, 273, 24529–24534. [Google Scholar] [CrossRef]
- Fassone, E.; Rahman, S. Complex I deficiency: Clinical features, biochemistry and molecular genetics. J. Med. Genet. 2012, 49, 578–590. [Google Scholar] [CrossRef]
- Forte, M.; Palmerio, S.; Bianchi, F.; Volpe, M.; Rubattu, S. Mitochondrial complex I deficiency and cardiovascular diseases: Current evidence and future directions. J. Mol. Med. Berl. Ger. 2019, 97, 579–591. [Google Scholar] [CrossRef] [PubMed]
- Perry, C.N.; Huang, C.; Liu, W.; Magee, N.; Carreira, R.S.; Gottlieb, R.A. Xenotransplantation of Mitochondrial Electron Transfer Enzyme, Ndi1, in Myocardial Reperfusion Injury. PLoS ONE 2011, 6, e16288. [Google Scholar] [CrossRef] [PubMed]
- Kerscher, S.; Dröse, S.; Zwicker, K.; Zickermann, V.; Brandt, U. Yarrowia lipolytica, a yeast genetic system to study mitochondrial complex I. Biochim. Biophys. Acta BBA Bioenerg. 2002, 1555, 83–91. [Google Scholar] [CrossRef]
- Kerscher, S.; Grgic, L.; Garofano, A.; Brandt, U. Application of the yeast Yarrowia lipolytica as a model to analyse human pathogenic mutations in mitochondrial complex I (NADH:ubiquinone oxidoreductase). Biochim. Biophys. Acta 2004, 1659, 197–205. [Google Scholar] [CrossRef] [PubMed]
- Gerber, S.; Ding, M.G.; Gérard, X.; Zwicker, K.; Zanlonghi, X.; Rio, M.; Serre, V.; Hanein, S.; Munnich, A.; Rotig, A.; et al. Compound heterozygosity for severe and hypomorphic NDUFS2 mutations cause non-syndromic LHON-like optic neuropathy. J. Med. Genet. 2017, 54, 346–356. [Google Scholar] [CrossRef]
- Varghese, F.; Atcheson, E.; Bridges, H.R.; Hirst, J. Characterization of clinically identified mutations in NDUFV1, the flavin-binding subunit of respiratory complex I, using a yeast model system. Hum. Mol. Genet. 2015, 24, 6350–6360. [Google Scholar] [CrossRef]
- Maclean, A.E.; Kimonis, V.E.; Balk, J. Pathogenic mutations in NUBPL affect complex I activity and cold tolerance in the yeast model Yarrowia lipolytica. Hum. Mol. Genet. 2018, 27, 3697–3709. [Google Scholar] [CrossRef]
- Parey, K.; Haapanen, O.; Sharma, V.; Köfeler, H.; Züllig, T.; Prinz, S.; Siegmund, K.; Wittig, I.; Mills, D.J.; Vonck, J.; et al. High-resolution cryo-EM structures of respiratory complex I: Mechanism, assembly, and disease. Sci. Adv. 2019, 5. [Google Scholar] [CrossRef]
- Parey, K.; Brandt, U.; Xie, H.; Mills, D.J.; Siegmund, K.; Vonck, J.; Kühlbrandt, W.; Zickermann, V. Cryo-EM structure of respiratory complex I at work. eLife 2018, 7. [Google Scholar] [CrossRef]
- Wood, V.; Lock, A.; Harris, M.A.; Rutherford, K.; Bähler, J.; Oliver, S.G. Hidden in plain sight: What remains to be discovered in the eukaryotic proteome? Open Biol. 2019, 9. [Google Scholar] [CrossRef]
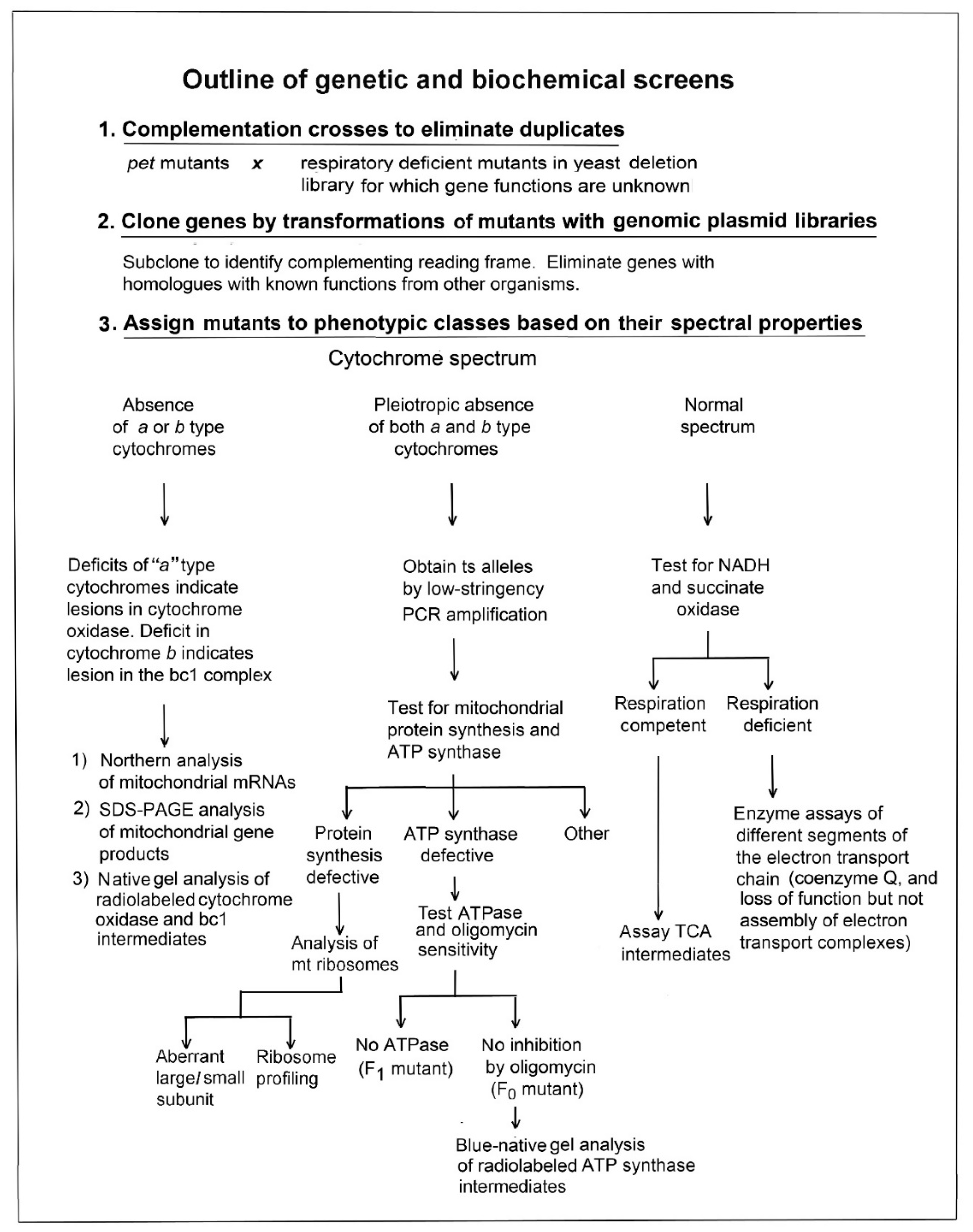
| Human Gene | Yeast Gene | Clinical Phenotype 1 | Mutation | Confirmation in Yeast | Reference |
|---|---|---|---|---|---|
| SDHA | SDH1 | Leigh syndrome | homozygous R554W | yes | [25] |
| compound heterozygous A524V, M1L | no | [45] | |||
| compound heterozygous W119*, A83V | no | [46] | |||
| homozygous G555E | no | [27] | |||
| late-onset optic atrophy, ataxia, myopathy | heterozygous R408C | no | [47] | ||
| neonatal isolated cardiomyopathy | homozygous G555E | no | [28] | ||
| undefined 2 | homozygous G555E | no | [29] | ||
| encephalopathy | compound heterozygous—stop codons at residues 56 and 81 | no | [48] | ||
| cardiomyopathy, leukodystrophy | compound heterozygous T508I, S509L | no | [49] | ||
| optic atrophy, progressive, polyneuropathy, cardiomyopathy | heterozygous R451C | no | [50] | ||
| SDHB | SDH2 | hypotonia, leukodystrophy | homozygous D48V | yes | [49] |
| leukoencephalopathy | compound heterozygous D48V, R230H; homozygous L257V 3 | no | [51] | ||
| SDHD | SDH4 | early progressive encephalomyopathy | compound heterozygous E69K, *164Lext*3 | no | [52] |
| SDHAF1 | SDH6 | leukoencephalopathy | homozygous G57R; homozygous R55P 3 | yes | [34] |
| homozygous R55P; homozygous Q8*; homozygous G57E 3 | no | [37] |
| Yeast Gene | Genome | pet Mutant | Human Gene | Function | |
|---|---|---|---|---|---|
| Enzyme Subunits | COB | mitochondrial | N/A | MT-CYB | catalysis |
| CYT1 | nuclear | yes | CYC1 | catalysis | |
| RIP1 | nuclear | yes | UQCRFS1 | catalysis | |
| COR1 | nuclear | yes | UQCRC1 | structure | |
| COR2 | nuclear | yes | UQCRC2 | structure | |
| QCR6 | nuclear | no | UQCRH | structure | |
| QCR8 | nuclear | yes | UQCRQ | structure | |
| QCR7 | nuclear | yes | UQCRB | structure | |
| QCR9 | nuclear | no | UQCR10 | structure | |
| QCR10 | nuclear | no | UQCR11 | structure | |
| Assembly Factors | CBP3 | nuclear | yes | UQCC1 | translation/assembly of cyt. b |
| CBP6 | nuclear | yes | UQCC2 | translation/assembly of cyt. b | |
| CBP4 | nuclear | yes | UQCC3 | assembly of cyt. b | |
| MZM1 | nuclear | no | LYRM7 | assembly of Rieske protein | |
| BCS1 | nuclear | yes | BCS1L | assembly of Rieske protein |
| Human Gene | Yeast Gene | Clinical Phenotype | Mutation | Confirmation in Yeast | Reference |
|---|---|---|---|---|---|
| MT-CYB | COB | MELAS, LHON, hearing loss, exercise intolerance | many | [70] 1 | |
| UQCR4 | CYT1 | ketoacidosis and insulin-responsive hyperglycemia | homozygous L215F; homozygous T96C 2 | yes | [115] |
| UQCRFS1 | RIP1 | cardiomyopathy and alopecia totalis | homozygous V72_T81del10; heterozygous V14D; heterozygous R204∗ 2 | no | [116] |
| UQCRC2 | COR2 | neonatal onset of hypoglycemia | homozygous R183W | no | [77,78] |
| UQCRB | QCR7 | hypoglycemia | homozygous 4 bp deletion at nucleotides 338–341 | yes [75] | [74] |
| UQCRQ | QCR8 | severe psychomotor retardation, dystonia, athetosis, ataxia, dementia | homozygous S45F | no | [76] |
| UQCC2 | CBP6 | intrauterine growth retardation, renal tubular dysfunction | homozygous c.214-3C>G 3 | no | [111] |
| neonatal encephalomyopathy | homozygous R8P and L10F 4 | no | [112] | ||
| UQCC3 | CBP4 | hypoglycemia, hypotonia, delayed development | homozygous V20E | no | [114] |
| BCS1L | BCS1 | GRACILE syndrome, Björstand syndrome, encephalopathy, muscle weakness | many | see main text | |
| LYRM7 | MZM1 | early onset severe encephalopathy | homozygous D25N | yes | [106] |
| leukoencephalopathy | homozygous c.243_244 + 2del 3 | no | [107] | ||
| several | yes | [108] | |||
| homozygous 4bp deletion c.[243_244 + 2delGAGT] 3 | no | [109] | |||
| liver dysfunction | homozygous R18Dfs*12 | no | [110] |
| Yeast Gene | Genome | pet Mutant | Human Gene | Function | |
|---|---|---|---|---|---|
| Enzyme Subunits | COX1 | mitochondrial | N/A | MTCO1 | catalysis |
| COX2 | mitochondrial | N/A | MTCO2 | catalysis | |
| COX3 | mitochondrial | N/A | MTCO3 | structure/catalysis | |
| COX5a | nuclear | yes | COX4 | structure | |
| COX5b1 | nuclear | no | - | structure | |
| COX6 | nuclear | yes | COX5A | structure | |
| COX4 | nuclear | yes | COX5B | structure | |
| COX13 | nuclear | no | COX6A | structure | |
| COX12 | nuclear | yes | COX6B | structure | |
| COX9 | nuclear | yes | COX6C | structure | |
| COX7 | nuclear | yes | COX7A | structure | |
| - | nuclear | - | COX7B | structure | |
| COX8 | nuclear | no | COX7C | structure | |
| - | nuclear | - | COX8 | structure | |
| - | nuclear | - | NDUFA4/COXFA4 | structure | |
| Assembly Factors | COX20 | nuclear | yes | COX20/FAM36A | membrane insertion of Cox2 |
| COX14 | nuclear | yes | COX14 | regulation of COX1 expression, maintenance of monomeric Cox1 | |
| PET117 | nuclear | yes | PET117 | couples synthesis of heme a to COX assembly | |
| PET191 | nuclear | yes | COA5 | required for COX assembly | |
| PET100 | nuclear | yes | PET100 | required for COX assembly | |
| COA6 | nuclear | no | COA6 | maturation of the CuA site | |
| COX25/COA3 | nuclear | yes | COA3 | translational regulation of COX1 mRNA | |
| COX15 | nuclear | yes | COX15 | conversion of heme o to heme a | |
| COX10 | nuclear | yes | COX10 | farnesylation of heme b | |
| SCO1 | nuclear | yes | SCO1 | maturation of the CuA site | |
| SCO2 | nuclear | no | SCO2 | maturation of the CuA site | |
| SHY1 | nuclear | yes | SURF1 | hemylation of Cox1 |
| Human Gene | Yeast Gene | Clinical Phenotype | Mutation | Confirmation in Yeast | Reference |
|---|---|---|---|---|---|
| MTCO1 | COX1 | prostate cancer, LHON, SNHL, DEAF | many | See main text | |
| MTCO2 | COX2 | progressive encephalomyopathy, HCM, SNHL, DEAF, LHON | many | See main text | |
| MTCO3 | COX3 | LHON, Alzheimer’s disease | many | See main text | |
| COX4 | COX5a | pancreatic insufficiency, dyserythropoeitic anemia, calvarial hyperostosis | homozygous E138K | no | [155] |
| short stature, poor weight gain, mild dysmorphic features with highly suspected Fanconi anemia | homozygous K101N | no | [156] | ||
| COX5A | COX6 | early-onset pulmonary arterial hypertension, failure to thrive | homozygous R107C | no | [157] |
| COX6a | COX13 | axonal or mixed form of Charcot-Marie-Tooth disease | homozygous c.247−10_247−6delCACTC 3 | no | [158,159] |
| COX6b | COX12 | infantile encephalomyopathy, myopathy, growth retardation | homozygous R19H | yes | [160] |
| encephalomyopathy, hydrocephalus, HCM | homozygous R20C | no | [161] | ||
| cystic leukodystrophy | homozygous c.241A>C | no | [162] | ||
| COX7b | microphthalmia with linear skin lesions | heterozygous c.196delC; heterozygous c.41-2A>G 3; heterozygous Q19∗ 2 | no | [136] | |
| COX8 | Leigh-like syndrome, epilepsy | homozygous c.115-1G>C 3 | no | [163] | |
| NDUFA4 | Leigh syndrome | homozygous c.42+1G>C 3 | no | [164] | |
| FAM36A | COX20 | dystonia and ataxia | homozygous T52P | no | [165,166] |
| dysarthria, ataxia, sensory neuropathy | compound heterozygous K14R, G114S, c.157+3G>C 3 | no | [167] | ||
| axonal neuropathy, static encephalopathy | compound heterozygous L14R, W74C | no | [168] | ||
| COX14 | COX14 | fatal neonatal lactic acidosis, dysmorphic features | homozygous M19I | no | [169] |
| PET117 | PET117 | neurodevelopmental regression, medulla oblongata lesions | homozygous c.172C>T (stop codon at residue 58) | no | [170] |
| COA5 | PET191 | fatal neonatal cardiomyopathy | homozygous A53P | no | [171] |
| PET100 | PET100 | Leigh syndrome | homozygous c.3G>C (p.Met1?) | no | [172] |
| fatal infantile lactic acidosis | homozygous Q48* | no | [173] | ||
| COA6 | COA6 | HCM | homozygous W66R | no | [174] |
| compound heterozygous W59C, E87* | yes [175] | [162] | |||
| COA3 | COX25 | neuropathy, exercise intolerance, obesity, short stature | compound heterozygous L67Pfs*21, Y72C | no | [176] |
| COX15 | COX15 | Leigh syndrome | homozygous L139V | no | [177] |
| homozygous R217W | no | [178] | |||
| compound heterozygous S152*, S344P | no | [147] | |||
| infantile cardioencephalopathy | compound heterozygous S151*, R217W | no | [179] | ||
| early-onset fatal HCM | compound heterozygous R217W, c.C447-3G 3 | no | [180] | ||
| COX10 | COX10 | severe muscle weakness, hypotonia, ataxia, ptosis, pyramidal syndrome, status epilepticus | homozygous N204K | yes | [8] |
| Leigh-like disease | homozygous T>C in the ATG start codon | no | [181] | ||
| anemia, sensorineural deafness, fatal infantile HCM | compound heterozygous T196K, P225L | no | [182] | ||
| Leigh syndrome, anemia | compound heterozygous D336V, D336G | no | [182] | ||
| myopathy, demyelinating neuropathy, premature ovarian failure, short stature, hearing loss | compound heterozygous D336V, R339W | yes | [146] | ||
| Leigh syndrome, anemia | homozygous P225L | no | [183] | ||
| hypotony, sideroblastic anemia, progressive encephalopathy | compound heterozygous M344V, L424Pfs | no | [183] | ||
| developmental delay, short stature | compound heterozygous R228H, deletion disrupting the last 2 exons | no | [184] | ||
| SCO1 | SCO1 | encephalopathy, hepatopathy, hypotonia, or cardiac involvement | homozygous G106del | no | [185] |
| fatal infantile encephalopathy | compound heterozygous M294V, V93* | no | [186] | ||
| early onset HCM, encephalopathy, hypotonia, hepatopathy | homozygous G132S | no | [187] | ||
| neonatal-onset hepatic failure, encephalopathy | compound heterozygous P174L, ΔGA nt 363–364 | no | [8] | ||
| SCO2 | SCO2 | cardioencephalomyopathy, Leigh syndrome, high myopia, Charcot-Marie-Tooth disease 2 | many | [151,152,154] 1 | |
| SURF1 | SHY1 | Leigh syndrome, Charcot-Marie-Tooth disease 2 | many | [139,142] 1; |
| ATP Synthase Sector | Human Subunit | Human Gene | Genome in Human | Yeast Subunit | Yeast Gene | Genome in Yeast | pet Mutant |
|---|---|---|---|---|---|---|---|
| F1 catalytic barrel | α | ATP5F1A | nuclear | α | ATP1 | nuclear | yes |
| β | ATP5F1B | nuclear | β | ATP2 | nuclear | yes | |
| F1 central stalk | γ | ATP5F1C | nuclear | γ | ATP3 | nuclear | yes |
| ε | ATP5F1E | nuclear | ε | ATP15 | nuclear | yes | |
| δ | ATP5F1D | nuclear | δ | ATP16 | nuclear | yes | |
| Peripheral stalk | b | ATP5PB | nuclear | b | ATP4 | nuclear | yes |
| d | ATP5PD | nuclear | d | ATP7 | nuclear | yes | |
| OSCP | ATP5PO | nuclear | OSCP | ATP5 | nuclear | yes | |
| F6 | ATP5PF | nuclear | h | ATP14 | nuclear | yes | |
| Fo rotor | Atp6 | MT-ATP6 | mitochondrial | Atp6 | ATP6 | mitochondrial | N/A |
| Atp8 | MT-ATP8 | mitochondrial | Atp8 | ATP8 | mitochondrial | N/A | |
| c1 | ATP5MC1 | nuclear | Atp9 | ATP9 | mitochondrial | N/A | |
| c2 | ATP5MC2 | nuclear | |||||
| c3 | ATP5MC3 | nuclear | |||||
| Fo supernumerary | f | ATP5MF | nuclear | f | ATP17 | nuclear | yes |
| 6.8 PL | ATP5MPL | nuclear | i/j | ATP18 | nuclear | no | |
| DAPIT | ATP5MD | nuclear | k | ATP19 | nuclear | no | |
| g | ATP5MG | nuclear | g | ATP20 | nuclear | no | |
| e | ATP5ME | nuclear | e | ATP21 | nuclear | no |
| Human Gene | Yeast Gene | Clinical Phenotype | Mutation | Confirmation in Yeast | Reference |
|---|---|---|---|---|---|
| MT-ATP6 | ATP6 | NARP, FBSN, Leigh syndrome, Charcot-Marie-Tooth disorder, HCM, MLASA | many | [198] 1 | |
| MT-ATP8 | ATP8 | apical HCM, neuropathy | homoplasmic W55* | no | [204] |
| tetralogy of Fallot | homoplasmic G9804A, C8481T, heteroplamic T7501C 3 | no | [205] | ||
| ATP5F1D | ATP16 | metabolic disorders, encephalopathy | homozygous P82L; homozygous V106G 2 | no | [213] |
| ATP5F1E | ATP15 | mild mental retardation, developed peripheral neuropathy | homozygous Y12C | no | [6] |
| ATP5F1A | ATP1 | fatal neonatal encephalopathy | heterozygous R329C | no | [5] |
| ATPAF2 | ATP12 | microcephaly etc | homozygous W94R | yes | [7,211] |
| Yeast Gene | Human Gene |
|---|---|
| COQ1 | PDSS1 |
| COQ1 | PDSS2 |
| COQ2 | COQ2 |
| COQ3 | COQ3 |
| COQ4 | COQ4 |
| COQ5 | COQ5 |
| COQ6 | COQ6 |
| COQ7/CAT5 | COQ7 |
| COQ8/ABC1 | COQ8A |
| COQ8/ABC2 | COQ8B |
| COQ9 | COQ9 |
| COQ10 | COQ10A, COQ10B |
| COQ11 | NDUFA9 |
| Human Gene | Yeast Gene | Clinical Phenotype | Mutation | Confirmation in Yeast | Reference |
|---|---|---|---|---|---|
| PDSS1 | COQ1 | infantile multisystemic disease | homozygous D308E | yes | [261] |
| PDSS2 | COQ1 | infantile multisystemic disease, SRNS, LS, hepatocellular carcinoma | compound heterozygous Q322*, S382L | yes | [262] |
| COQ2 | COQ2 | SRNS, SRNS with encephalomyopathy resembling MELAS, fatal infantile multisystemic disease | homozygous Y297C | yes | [268] |
| COQ4 | COQ4 | LS, cerebellar atrophy, lactic acidosis, etc. | many | [263,269,270] 1 | |
| COQ5 | COQ5 | cerebellar ataxia, dysarthria, myoclonic jerks | biallelic 9590 bp duplication | no | [250] |
| COQ6 | COQ6 | SRNS, infantile multisystemic disease | many | [264] 1 | |
| COQ7 | COQ7 | spasticity, sensorineural hearing loss etc. | homozygous V141E; homozygous L111P; compound heterozygous R107W, K200Ifs*56 2 | no | [272,273,274] |
| COQ8A | COQ8 | ARCA, seizures, dystonia, spasticity | many | [248] 1 | |
| COQ8B | COQ8 | SRNS | many | [248,276] 1 | |
| COQ9 | COQ9 | infantile multisystemic disease, Leigh-like Syndrome, microcephaly | many | See main text |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Franco, L.V.R.; Bremner, L.; Barros, M.H. Human Mitochondrial Pathologies of the Respiratory Chain and ATP Synthase: Contributions from Studies of Saccharomyces cerevisiae. Life 2020, 10, 304. https://doi.org/10.3390/life10110304
Franco LVR, Bremner L, Barros MH. Human Mitochondrial Pathologies of the Respiratory Chain and ATP Synthase: Contributions from Studies of Saccharomyces cerevisiae. Life. 2020; 10(11):304. https://doi.org/10.3390/life10110304
Chicago/Turabian StyleFranco, Leticia V. R., Luca Bremner, and Mario H. Barros. 2020. "Human Mitochondrial Pathologies of the Respiratory Chain and ATP Synthase: Contributions from Studies of Saccharomyces cerevisiae" Life 10, no. 11: 304. https://doi.org/10.3390/life10110304
APA StyleFranco, L. V. R., Bremner, L., & Barros, M. H. (2020). Human Mitochondrial Pathologies of the Respiratory Chain and ATP Synthase: Contributions from Studies of Saccharomyces cerevisiae. Life, 10(11), 304. https://doi.org/10.3390/life10110304





