Understanding the Mechanism of Recognition of Gab2 by the N-SH2 Domain of SHP2
Abstract
:1. Introduction
2. Materials and Methods
2.1. Expression and Purification of the N-SH2 Site-Directed Variants of SHP2
2.2. Stopped-Flow Kinetic Binding and Displacement Experiments
3. Results and Discussion
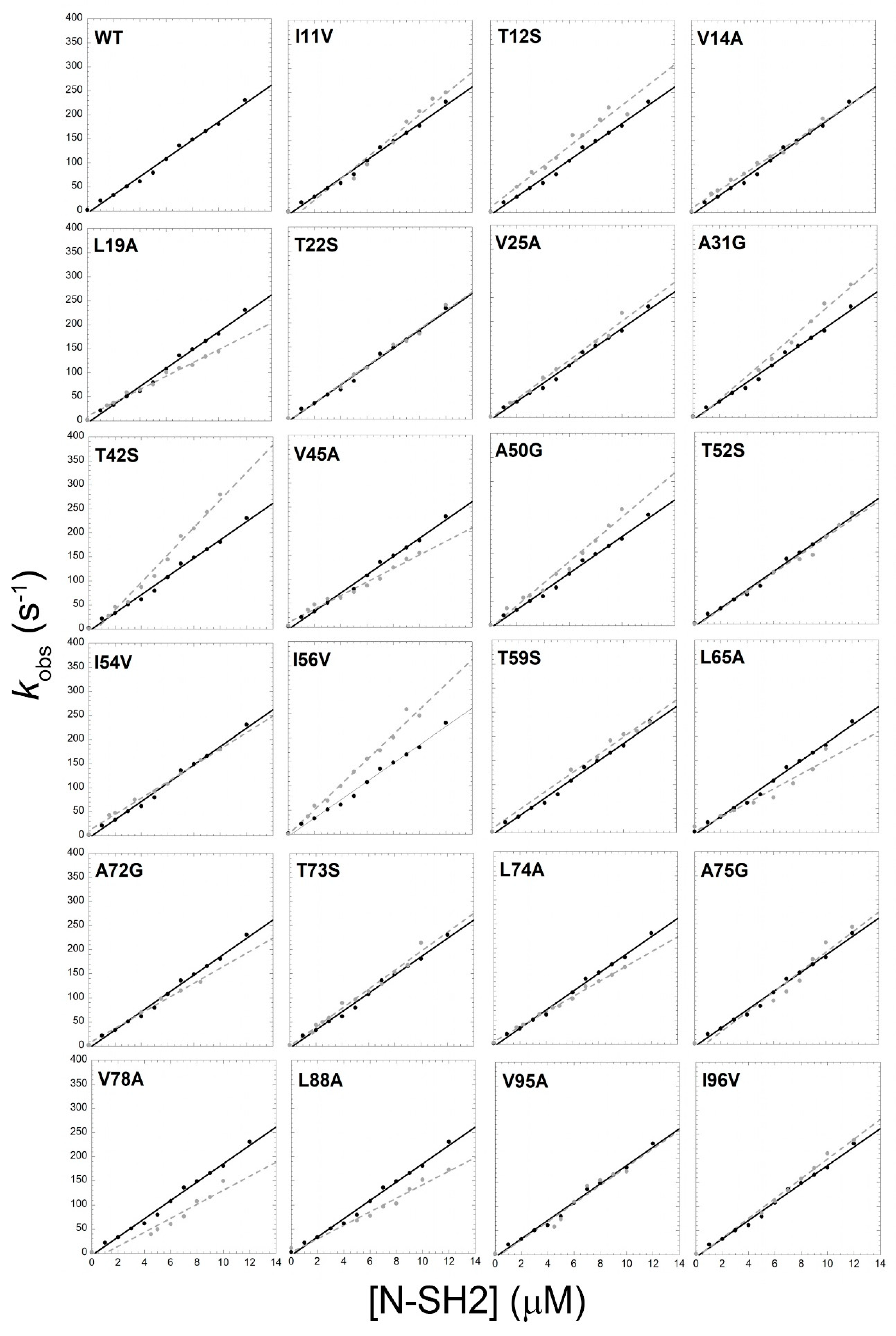
3.1. Mutational Analysis of the Kinetics of Binding between the N-SH2 of SHP2 and Gab2
3.2. N-SH2 SHP2: Gab2 Complex Is Stabilized by Weak Interactions
3.3. Does a Conserved Mechanism of Binding Characterize the SH2 Domain Family?
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Bhattacharyya, R.P.; Reményi, A.; Yeh, B.J.; Lim, W.A. Domains, motifs, and scaffolds: The role of modular interactions in the evolution and wiring of cell signaling circuits. Annu. Rev. Biochem. 2006, 75, 655–680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teyra, J.; Sidhu, S.S.; Kim, P.M. Elucidation of the binding preferences of peptide recognition modules: SH3 and PDZ domains. FEBS Lett. 2012, 586, 2631–2637. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kurochkina, N.; Guha, U. SH3 domains: Modules of protein-protein interactions. Biophys. Rev. 2013, 5, 29–39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pawson, T.; Scott, J.D. Signaling through scaffold, anchoring, and adaptor proteins. Science 1997, 278, 2075–2080. [Google Scholar] [CrossRef] [Green Version]
- Macias, M.J.; Hyvönen, M.; Baraldi, E.; Schultz, J.; Sudol, M.; Saraste, M.; Oschkinat, H. Structure of the WW domain of a kinase-associated protein complexed with a proline-rich peptide. Nature 1996, 382, 646–649. [Google Scholar] [CrossRef]
- Ivarsson, Y.; Jemth, P. Affinity and specificity of motif-based protein-protein interactions. Curr. Opin. Struct. Biol. 2019, 54, 26–33. [Google Scholar] [CrossRef] [Green Version]
- Malagrinò, F.; Troilo, F.; Bonetti, D.; Toto, A.; Gianni, S. Mapping the allosteric network within a SH3 domain. Sci. Rep. 2019, 9, 8279. [Google Scholar] [CrossRef]
- Gianni, S.; Haq, S.R.; Montemiglio, L.C.; Jürgens, M.C.; Engström, Å.; Chi, C.N.; Brunori, M.; Jemth, P. Sequence-specific long range networks in PSD-95/discs large/ZO-1 (PDZ) domains tune their binding selectivity. J. Biol. Chem. 2011, 286, 27167–27175. [Google Scholar] [CrossRef] [Green Version]
- Lockless, S.W.; Ranganathan, R. Evolutionarily conserved pathways of energetic connectivity in protein families. Science 1999, 286, 295–299. [Google Scholar] [CrossRef] [Green Version]
- Visconti, L.; Toto, A.; Jarvis, J.A.; Troilo, F.; Malagrinò, F.; De Simone, A.; Gianni, S. Demonstration of Binding Induced Structural Plasticity in a SH2 Domain. Front. Mol. Biosci. 2020, 7, 89. [Google Scholar] [CrossRef]
- Motlagh, H.N.; Wrabl, J.O.; Li, J.; Hilser, V.J. The ensemble nature of allostery. Nature 2014, 508, 331–339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kern, D.; Zuiderweg, E.R.P. The role of dynamics in allosteric regulation. Curr. Opin. Struct. Biol. 2003, 13, 748–757. [Google Scholar] [CrossRef]
- Good, M.C.; Zalatan, J.G.; Lim, W.A. Scaffold proteins: Hubs for controlling the flow of cellular information. Science 2011, 332, 680–686. [Google Scholar] [CrossRef] [Green Version]
- Adams, S.J.; Aydin, I.T.; Celebi, J.T. GAB2—A scaffolding protein in cancer. Mol. Cancer Res. 2012, 10, 1265–1270. [Google Scholar] [CrossRef] [Green Version]
- Ding, C.-B.; Yu, W.-N.; Feng, J.-H.; Luo, J.-M. Structure and function of Gab2 and its role in cancer (Review). Mol. Med. Rep. 2015, 12, 4007–4014. [Google Scholar] [CrossRef] [Green Version]
- Kong, M.; Mounier, C.; Dumas, V.; Posner, B.I. Epidermal Growth Factor-induced DNA Synthesis: KEY ROLE FOR Src PHOSPHORYLATION OF THE DOCKING PROTEIN Gab2. J. Biol. Chem. 2003, 278, 5837–5844. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Lavoie, G.; Fort, L.; Huttlin, E.L.; Tcherkezian, J.; Galan, J.A.; Gu, H.; Gygi, S.P.; Carreno, S.; Roux, P.P. Gab2 Phosphorylation by RSK Inhibits Shp2 Recruitment and Cell Motility. Mol. Cell. Biol. 2013, 33, 1657–1670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Xue, J.; Zadorozny, E.V.; Robinson, L.J. G-CSF stimulates Jak2-dependent Gab2 phosphorylation leading to Erk1/2 activation and cell proliferation. Cell. Signal. 2008, 20, 1890–1899. [Google Scholar] [CrossRef] [Green Version]
- Neel, B.G.; Gu, H.; Pao, L. The ’Shp’ing news: SH2 domain-containing tyrosine phosphatases in cell signaling. Trends Biochem. Sci. 2003, 28, 284–293. [Google Scholar] [CrossRef]
- Hof, P.; Pluskey, S.; Dhe-Paganon, S.; Eck, M.J.; Shoelson, S.E. Crystal structure of the tyrosine phosphatase SHP-2. Cell 1998, 92, 441–450. [Google Scholar] [CrossRef] [Green Version]
- Waksman, G.; Kominos, D.; Robertson, S.C.; Pant, N.; Baltimore, D.; Birge, R.B.; Cowburn, D.; Hanafusa, H.; Mayer, B.J.; Overduin, M.; et al. Crystal structure of the phosphotyrosine recognition domain SH2 of v-src complexed with tyrosine-phosphorylated peptides. Nature 1992, 358, 646–653. [Google Scholar] [CrossRef]
- Waksman, G.; Shoelson, S.E.; Pant, N.; Cowburn, D.; Kuriyan, J. Binding of a high affinity phosphotyrosyl peptide to the Src SH2 domain: Crystal structures of the complexed and peptide-free forms. Cell 1993, 72, 779–790. [Google Scholar] [CrossRef]
- Payne, G.; Stolz, L.A.; Pei, D.; Band, H.; Shoelson, S.E.; Walsh, C.T. The phosphopeptide-binding specificity of Src family SH2 domains. Chem. Biol. 1994, 1, 99–105. [Google Scholar] [CrossRef]
- Schaffhausen, B. SH2 domain structure and function. Biochim. Biophys. Acta 1995, 1242, 61–75. [Google Scholar] [CrossRef]
- McCallister, E.L.; Alm, E.; Baker, D. Critical role of beta-hairpin formation in protein G folding. Nat. Struct. Biol. 2000, 7, 669–673. [Google Scholar] [CrossRef]
- Bonetti, D.; Troilo, F.; Toto, A.; Travaglini-Allocatelli, C.; Brunori, M.; Gianni, S. Mechanism of Folding and Binding of the N-Terminal SH2 Domain from SHP2. J. Phys. Chem. B 2018, 122, 11108–11114. [Google Scholar] [CrossRef]
- Visconti, L.; Malagrinò, F.; Gianni, S.; Toto, A. Structural characterization of an on-pathway intermediate and transition state in the folding of the N-terminal SH2 domain from SHP2. FEBS J. 2019, 286, 4769–4777. [Google Scholar] [CrossRef]
- Antonini, E.; Brunori, M. Hemoglobin and Myoglobin in Their Reactions with Ligands; North-Holland: Amsterdam, The Netherlands, 1971. [Google Scholar]
- Toto, A.; Malagrinò, F.; Visconti, L.; Troilo, F.; Gianni, S. Unveiling the Molecular Basis of the Noonan Syndrome-Causing Mutation T42A of SHP2. IJMS 2020, 21, 461. [Google Scholar] [CrossRef] [Green Version]
- Fersht, A.R. Relationship of Leffler (Bronsted) alpha values and protein folding Phi values to position of transition-state structures on reaction coordinates. Proc. Natl. Acad. Sci. USA 2004, 101, 14338–14342. [Google Scholar] [CrossRef] [Green Version]
- Mallis, R.J.; Brazin, K.N.; Fulton, D.B.; Andreotti, A.H. Structural characterization of a proline-driven conformational switch within the Itk SH2 domain. Nat. Struct. Biol. 2002, 9, 900–905. [Google Scholar] [CrossRef]
- Breheny, P.J.; Laederach, A.; Fulton, D.B.; Andreotti, A.H. Ligand specificity modulated by prolyl imide bond Cis/Trans isomerization in the Itk SH2 domain: A quantitative NMR study. J. Am. Chem. Soc. 2003, 125, 15706–15707. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pletneva, E.V.; Sundd, M.; Fulton, D.B.; Andreotti, A.H. Molecular details of Itk activation by prolyl isomerization and phospholigand binding: The NMR structure of the Itk SH2 domain bound to a phosphopeptide. J. Mol. Biol. 2006, 357, 550–561. [Google Scholar] [CrossRef] [PubMed]
- Ladbury, J.E.; Arold, S. Searching for specificity in SH domains. Chem. Biol. 2000, 7, R3–R8. [Google Scholar] [CrossRef] [Green Version]


| kon (μM−1 s−1) | koff (s−1) | KD (nM) | Activation Free Energy ∆∆G# (kcal mol−1) | Equilibrium Free Energy ∆∆Geq (kcal mol−1) | |
|---|---|---|---|---|---|
| WT | 19.0 ± 0.5 | 1.85 ± 0.01 | 100 ± 5 | ||
| I11V | 22.2 ± 1.5 | 1.81 ± 0.01 | 80 ± 6 | −0.09 ± 0.02 | −0.10 ± 0.01 |
| T12S | 21.2 ± 1.4 | 1.73 ± 0.01 | 80 ± 6 | −0.06 ± 0.01 | −0.10 ± 0.02 |
| V14A | 17.9 ± 0.6 | 2.81 ± 0.01 | 160 ± 5 | 0.03 ± 0.01 | 0.27 ± 0.03 |
| L19A | 13.8 ± 0.5 | 2.39 ± 0.01 | 170 ± 6 | 0.18 ± 0.02 | 0.32 ± 0.03 |
| T22S | 19.2 ± 0.8 | 1.93 ± 0.01 | 100 ± 4 | −0.01 ± 0.01 | 0.02 ± 0.01 |
| V25A | 20.2 ± 0.9 | 2.04 ± 0.07 | 100 ± 6 | −0.04 ± 0.01 | 0.02 ± 0.01 |
| A31G | 23.5 ± 1.1 | 1.62 ± 0.01 | 70 ± 3 | −0.12 ± 0.02 | −0.19 ± 0.02 |
| T42S | 28.6 ± 1.1 | 0.14 ± 0.01 | 5.0 ± 0.5 | −0.23 ± 0.02 | −1.69 ± 0.10 |
| V45A | 14.1 ± 0.7 | 3.23 ± 0.01 | 230 ± 11 | 0.17 ± 0.10 | 0.48 ± 0.05 |
| A50G | 23.0 ± 1.0 | 1.63 ± 0.07 | 70 ± 4 | −0.11 ± 0.01 | −0.18 ± 0.02 |
| T52S | 18.7 ± 1.0 | 0.20 ± 0.01 | 10 ± 1 | 0.01 ± 0.01 | −1.24 ± 0.10 |
| I54V | 17.0 ± 0.5 | 2.53 ± 0.20 | 150 ± 13 | 0.06 ± 0.03 | 0.24 ± 0.02 |
| I56V | 26.0 ± 1.1 | 0.68 ± 0.01 | 30 ± 1 | −0.18 ± 0.02 | −0.74 ± 0.07 |
| T59S | 19.1 ± 0.9 | 2.30 ± 0.01 | 120 ± 6 | 0.00 ± 0.01 | 0.12 ± 0.01 |
| L65A | 14.8 ± 1.5 | 12.70 ± 0.10 | 860 ± 90 | 0.14 ± 0.05 | 1.22 ± 0.10 |
| A72G | 14.7 ± 0.9 | 2.39 ± 0.01 | 170 ± 10 | 0.14 ± 0.02 | 0.29 ± 0.03 |
| T73S | 19.7 ± 0.8 | 2.09 ± 0.02 | 110 ± 4 | −0.02 ± 0.01 | 0.05 ± 0.01 |
| L74A | 15.6 ± 1.9 | 3.23 ± 0.01 | 210 ± 25 | 0.11 ± 0.05 | 0.43 ± 0.04 |
| A75G | 20.7 ± 2.1 | 2.35 ± 0.02 | 110 ± 10 | −0.05 ± 0.02 | 0.09 ± 0.01 |
| V78A | 14.5 ± 1.7 | 2.73 ± 0.40 | 190 ± 35 | 0.15 ± 0.04 | 0.37 ± 0.03 |
| L88A | 14.0 ± 0.9 | 10.30 ± 0.05 | 730 ± 50 | 0.17 ± 0.05 | 1.13 ± 0.10 |
| V95A | 18.9 ± 1.7 | 2.44 ± 0.01 | 130 ± 12 | 0.00 ± 0.02 | 0.16 ± 0.01 |
| I96V | 20.5 ± 0.8 | 2.06 ± 0.01 | 100 ± 4 | −0.04 ± 0.02 | 0.02 ± 0.01 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Visconti, L.; Malagrinò, F.; Pagano, L.; Toto, A. Understanding the Mechanism of Recognition of Gab2 by the N-SH2 Domain of SHP2. Life 2020, 10, 85. https://doi.org/10.3390/life10060085
Visconti L, Malagrinò F, Pagano L, Toto A. Understanding the Mechanism of Recognition of Gab2 by the N-SH2 Domain of SHP2. Life. 2020; 10(6):85. https://doi.org/10.3390/life10060085
Chicago/Turabian StyleVisconti, Lorenzo, Francesca Malagrinò, Livia Pagano, and Angelo Toto. 2020. "Understanding the Mechanism of Recognition of Gab2 by the N-SH2 Domain of SHP2" Life 10, no. 6: 85. https://doi.org/10.3390/life10060085
APA StyleVisconti, L., Malagrinò, F., Pagano, L., & Toto, A. (2020). Understanding the Mechanism of Recognition of Gab2 by the N-SH2 Domain of SHP2. Life, 10(6), 85. https://doi.org/10.3390/life10060085






