Abstract
Rodent models have been widely used as analogs for estimating spaceflight-relevant molecular mechanisms in human tissues. NASA GeneLab provides access to numerous spaceflight omics datasets that can potentially generate novel insights and hypotheses about fundamental space biology when analyzed in new and integrated fashions. Here, we performed a pilot study to elucidate space biological mechanisms across tissues by reanalyzing mouse RNA-sequencing spaceflight data archived on NASA GeneLab. Our results showed that clock gene expressions in spaceflight mice were altered compared with those in ground control mice. Furthermore, the results suggested that spaceflight promotes asynchrony of clock gene expressions between peripheral tissues. Abnormal circadian rhythms are associated not only with jet lag and sleep disorders but also with cancer, lifestyle-related diseases, and mental disorders. Overall, our findings highlight the importance of elucidating the causes of circadian rhythm disruptions using the unique approach of space biology research to one day potentially develop countermeasures that benefit humans on Earth and in space.
Keywords:
circadian rhythm; RNA-seq; bioinformatics; genomics; gene expression; microgravity; spaceflight; space biology 1. Introduction
The unique environment during spaceflight affects various physiological functions in astronauts [1,2]. With increasing opportunities for human spaceflight, a better understanding of the effects of spaceflight on the human body at the molecular level is essential. Unfortunately, solid tissue biopsies cannot be performed in the full human body in most cases. Human biopsies can usually only be obtained by using less invasive approaches, rendering it all the more difficult to investigate human tissue-level mechanisms during spaceflight. The impact of spaceflight itself is challenging to understand owing to its inherent complexity of environmental stressors, such as confinement in a closed environment, microgravity, radiation, and noise. Since the dawn of human spaceflight, basic life science experiments have been actively conducted in space to learn more about disease processes here on Earth from a new and unique perspective. Rodent models are often used as analogs for estimating biological mechanisms inside human tissues [3,4]. Mice studies under spaceflight conditions have been successfully conducted to elicit molecular mechanisms at the tissue and cellular levels [3,5,6,7,8].
Omics analysis has led to many novel discoveries for researchers through advances in genome-wide analysis techniques [9]. Since the NASA GeneLab database was launched in 2015, the open-access database and its multi-omics format have provided researchers with countless new insights [10,11,12,13,14,15]. The NASA GeneLab database stores a variety of open-access, spaceflight-related omics datasets for numerous organisms, including microbes, plants, animals, and humans [16,17]. Datasets are stored as microarrays, RNA-seq, bisulfite sequencing, proteomics, and metabolomics [16,17]. Omics data are rich with information on the different molecules that make up living organisms. Furthermore, GeneLab Analysis Working Groups (AWGs) regularly update data standardization workflows in the database to meet the latest recommendations from the bioinformatics discipline. As such, multi-omics data from a platform like NASA GeneLab can be continuously re-investigated with newer and deeper analyses to potentially extract novel insights and hypotheses that assess the biological risks of space missions.
This study aimed to reanalyze archived RNA-seq data from NASA GeneLab to unravel molecular responses to spaceflight. We conducted enrichment analysis on data from multiple tissues to investigate what molecular changes were revealed under spaceflight conditions. Enrichment analysis indicated that common ontologies and critical regulators between peripheral tissues were associated with circadian rhythms. Our results suggested that clock genes across tissues were affected by expression changes under spaceflight conditions. Furthermore, we observed asynchrony in the expression of the clock genes between certain peripheral tissues. Circadian rhythm disruption is associated not only with astronaut health and performance but also with cancer, lifestyle-related diseases, and mental disorders here on Earth [18,19]. Therefore, further studies on how spaceflight affects tissue functions should be an essential agenda item in order to better understand mammalian physiology and potentially create countermeasures that benefit humans on Earth and in space. Our study did not determine whether clock genes were primarily affected by spaceflight factors (such as microgravity and radiation) and/or extraneous factors (such as rearing environment and sample processing) [20]. Our findings not only generate new insights into space biological mechanisms, but also underline the critical need for space biology researchers to maintain and share detailed experimental metadata so that meta-analyses may one day disentangle exactly which factors cause circadian rhythm disruption during spaceflight.
2. Materials and Methods
2.1. Data Preparation and Processing
The transcriptome datasets used for this study were from openly available data housed on NASA’s GeneLab platform and are updated to meet the latest recommendations from the analysis workflow by NASA GeneLab AWGs (genelab.nasa.gov). Specifically, 8 RNA-seq datasets from NASA GeneLab (GLDS-98, -99, -101, -102, -103, -104, -105, and -168) were used for downstream analysis [21,22,23,24,25,26,27,28]. These data were all derived from the same mission. Metadata on NASA GeneLab indicated that 16-week-old C57BL/6 J female mice were used and that the mice were maintained under a 12 h light/dark cycle throughout the 37-day spaceflight mission. Each dataset corresponded to a different mouse tissue: adrenal glands, extensor digitorum longus muscle, gastrocnemius muscle, kidneys, quadriceps muscle, soleus muscle, tibialis anterior muscle, and liver, respectively. In addition, each dataset contained about 5–6 spaceflight (FLT) and 5–6 ground control (GC). We used the External RNA Control Consortium (ERCC) results for the quantitative values of GLDS-168. We performed 2 main forms of differentially expressed gene (DEG) analyses: 1 tissue-wide analysis and 8 individual tissue analyses. For the tissue-wide analysis, we compared all the FLT samples regardless of the tissue (n = 47) with all GC samples regardless of the tissue (n = 46). Quantile normalization was performed on each “rna_seq_Unnormalized counts.csv” file stored in each “GeneLab Processed RNA-Seq Files” directory on NASA GeneLab using CLC Genomics Workbench (version 11.0.2, QIAGEN, Hilden, Germany). The t test was used for the statistical analysis, and the statistical threshold was defined with a false discovery rate (FDR) < 0.1. For each of the 8 individual tissue analyses, we downloaded the “rna_seq_differential_expression.csv” files stored in the “GeneLab Processed RNA-Sseq Files” directory on NASA GeneLab, in which statistical analysis had already been conducted to determine DEGs. The statistical threshold was defined as FDR < 0.05.
2.2. Data Visualization
Principal component analysis (PCA) was performed using the scikit-learn package 0.17.1 in Python v3.7 (https://scikit-learn.org). Heatmaps were generated using the Python seaborn.clustermap function (https://seaborn.pydata.org).
2.3. Enrichment Analysis
Enrichment analysis was performed on DEGs using Metascape with default settings [29].
3. Results
3.1. Spaceflight Could Cause Significant Changes in the Expression of Clock Genes in Multiple Tissues
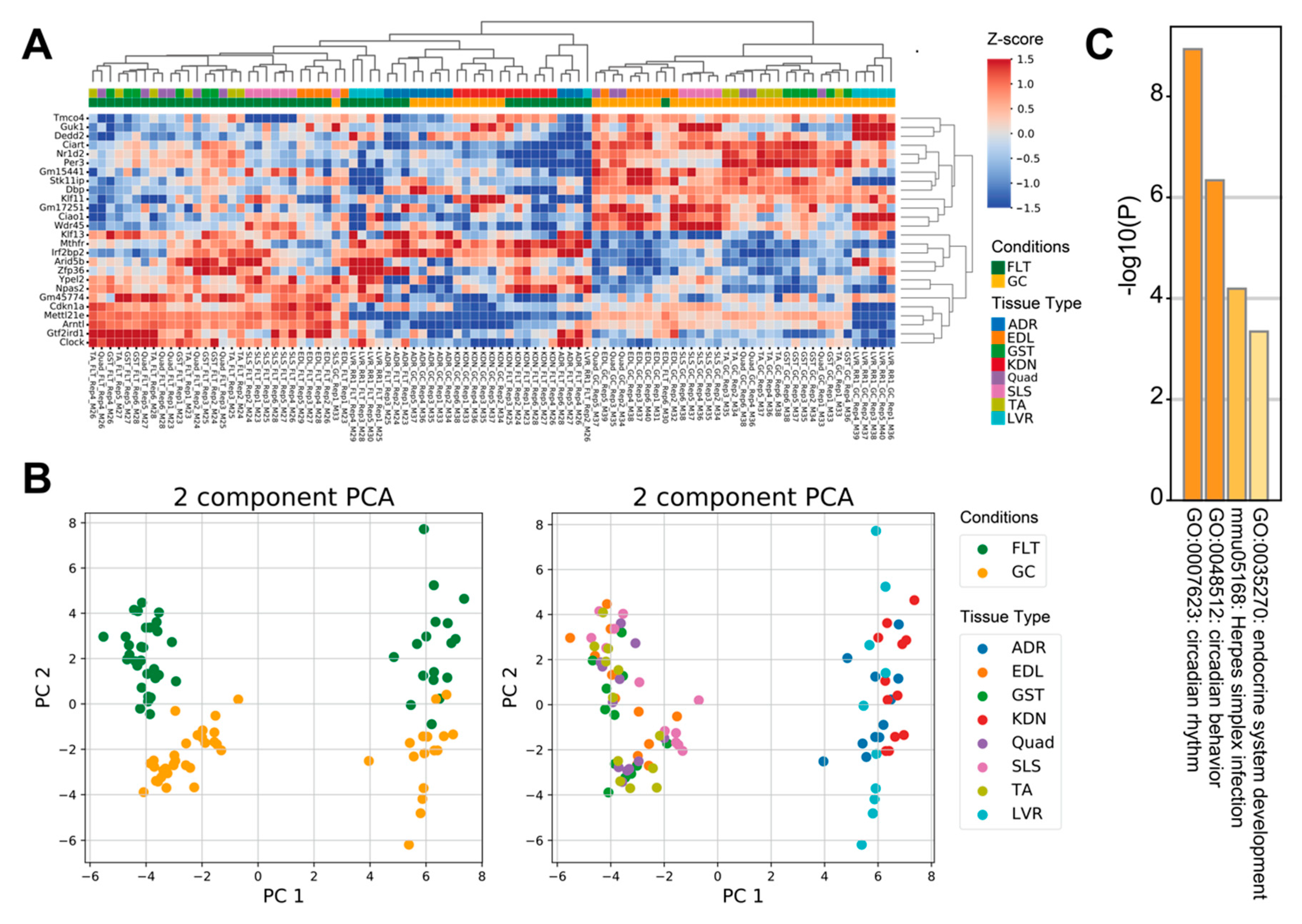
To examine how spaceflight affects prevalent molecular mechanisms across tissues, we reanalyzed 8 RNA-seq datasets archived on the NASA GeneLab database using tissue-wide analysis. Comparison between FLT and GC identified 13 up-regulated genes and 13 down-regulated genes (FDR < 0.1) (Figure 1A). PCA plots of the 26 tissue-wide DEGs showed that the first principal component separated the 5 muscle tissues from the adrenal glands, kidneys, and liver tissues, whereas the second principal component separated the FLT group from the GC group (Figure 1B). Enrichment analysis showed that terms related to circadian rhythms, herpes simplex infection, and endocrine system development were significantly enriched in the 26 DEGs of the tissue-wide analysis (Figure 1C).
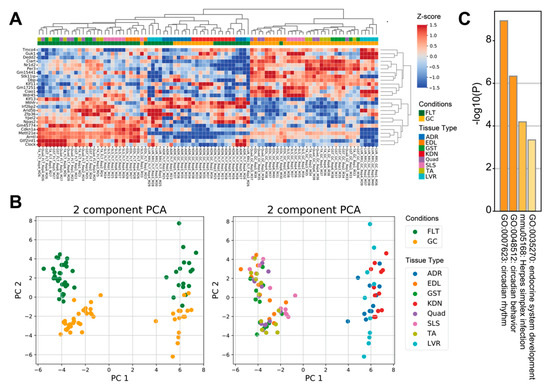
Figure 1.
Differential gene expression between spaceflight and ground control across mice tissues. (A) is the heatmap of the 26 tissue-wide DEGs between FLT and GC. (B) are PCA plots of the 26 tissue-wide DEGs between FLT and GC conditions. PCA plots are colored by conditions (left) and tissue types (right). (C) is the bar chart of enrichment ontology categories in the tissue-wide DEGs between FLT and GC. Abbreviations: FLT, spaceflight mice; GC, ground control mice; ADR, adrenal glands; EDL, extensor digitorum longus; GST, gastrocnemius; KDN, kidneys; Quad, quadriceps; SLS, soleus; TA, tibialis anterior; LVR, liver.
3.2. Spaceflight Could Enrich Key Regulators Related to Circadian Rhythms in Peripheral Tissues
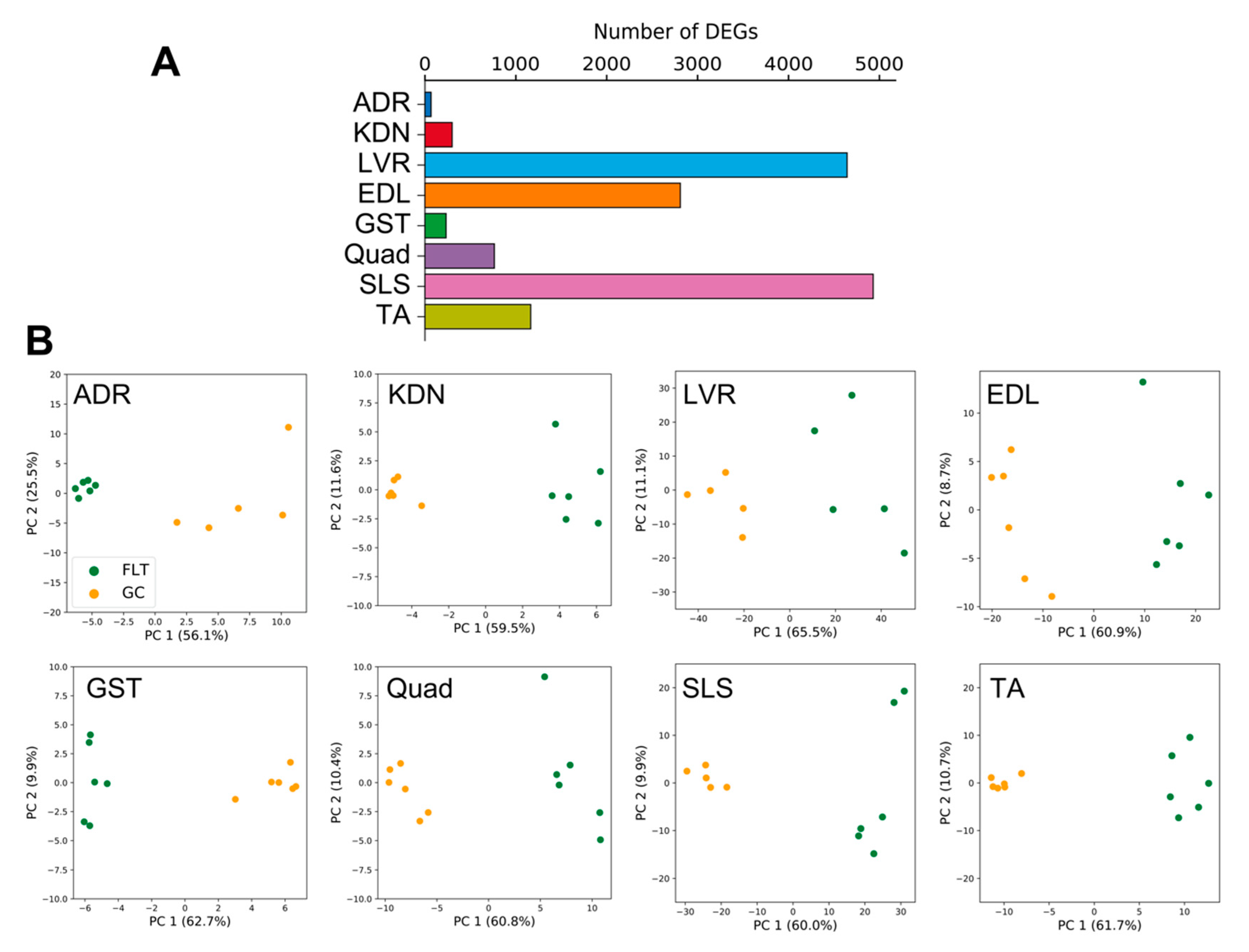
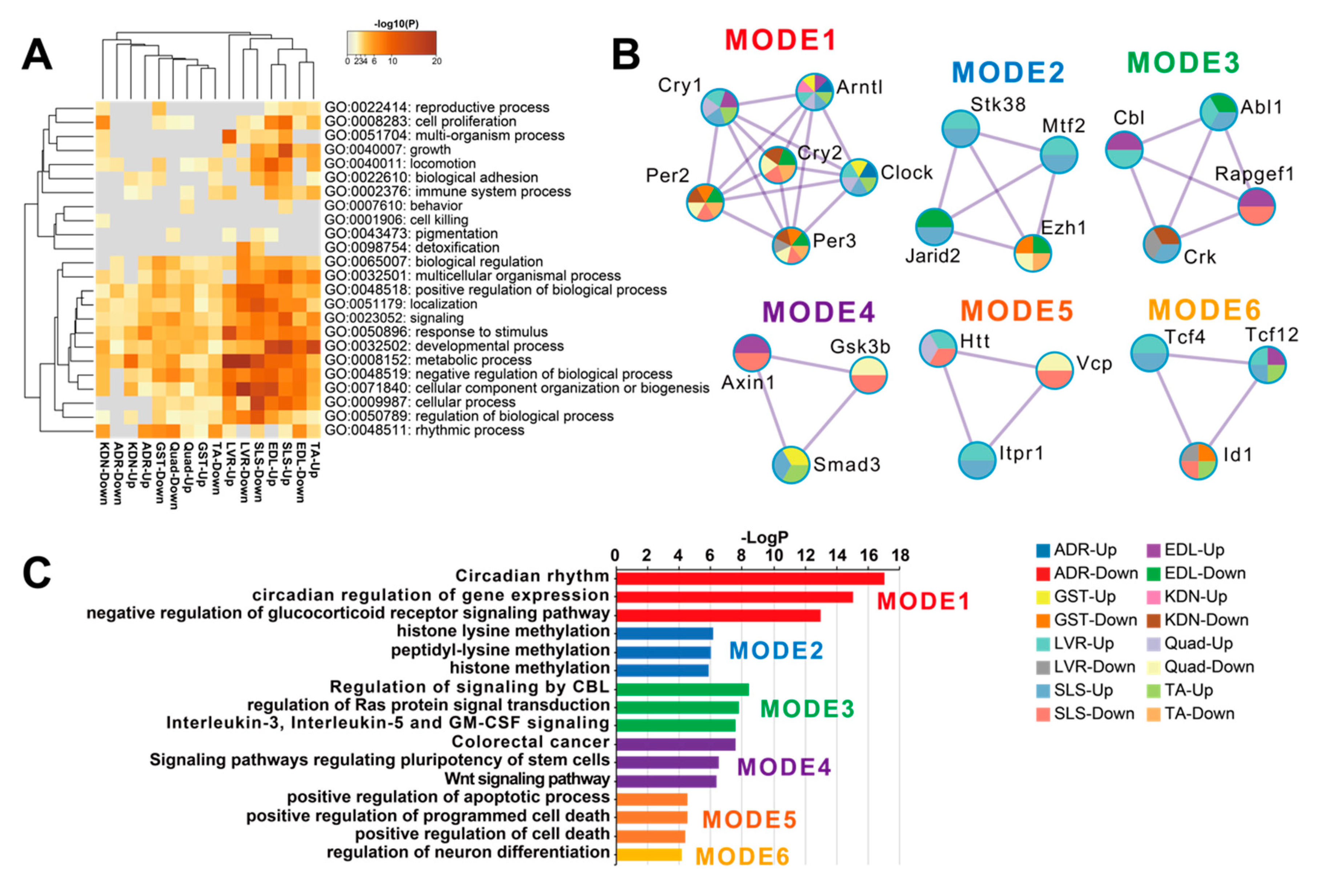
DEGs between FLT and GC were identified at the FDR < 0.05 level separately for each of the 8 individual tissues: adrenal glands (n = 67), kidneys (n = 299), liver (n = 4644), extensor digitorum longus (n = 2809), gastrocnemius (n = 232), quadriceps (n = 763), soleus (n = 4931), and tibialis anterior (n = 1163) (Figure 2A). A previous report that analyzed these data [13] found that spaceflight induced the largest number of DEGs in the soleus (similar to our current results). However, despite this consistent ranking, our results were not consistent with the previous study in terms of the actual DEG counts [13]. We note that Beheshti et al. used the same dataset as we did but analyzed it using a different RNA-seq workflow that included a t-test with a p-value ≤ 0.05 [13]. This indicates that RNA-seq results highly depend on the standardized processing workflows. To allow sufficient reproducibility, the NASA Genelab team has established standardized processing workflows for omics datasets (https://genelab-data.ndc.nasa.gov/genelab/projects). Our study used DEGs that were defined by analysis pipelines developed by GeneLab AWGs, which regularly update and share data standardization workflows in the database to meet the latest recommendations from the bioinformatics discipline. Therefore, the reanalysis of archived datasets might be worthwhile for finding novel insights. Our findings showed that the liver was the tissue with the second largest number of DEGs. This discrepancy may be due to the liver datasets being re-sequenced between the previous report and our study as described in GLDS-48 (https://genelab-data.ndc.nasa.gov/genelab/accession/GLDS-48). PCA plots demonstrated remarkable differences between FLT and GC conditions in all tissues (Figure 2B). These results suggested that gene expression changes occurred in each peripheral tissue during spaceflight. To examine the underlying molecular mechanisms across tissues caused by spaceflight, enrichment analysis was performed on the DEGs of the 8 tissues and showed shared terms between tissues (Figure 3A). Clustering analysis of protein–protein interaction (PPI) networks using MODE showed that the terms related to circadian rhythms were most significantly enriched in MODE1 (Figure 3B,C).
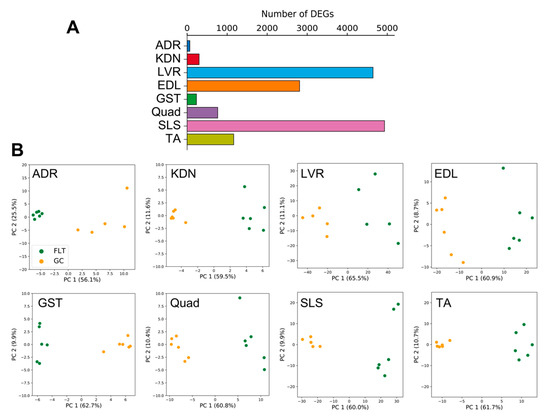
Figure 2.
DEG numbers and PCA plots for each peripheral tissue. (A) is the number of DEGs detected between FLT and GC conditions. (B) are PCA plots for each of the 8 different tissues.
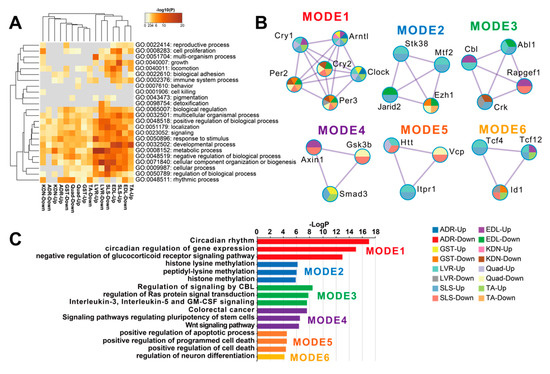
Figure 3.
Functional enrichment analysis of DEGs in each tissue. (A) is the heatmap of the top-level Gene Ontology (GO) biological process categories for DEGs in each tissue. Gray color indicates a lack of significant term. (B) are the clustered PPI networks by MCODE identified from the combined list of all DEGs in each tissue. (C) is the bar plot of clusters of enrichment categories detected by the MCODE algorithm. Abbreviations: Up, up-regulated genes; Down, down-regulated genes.
3.3. Spaceflight Could Induce Asynchrony in Clock Genes between Peripheral Tissues
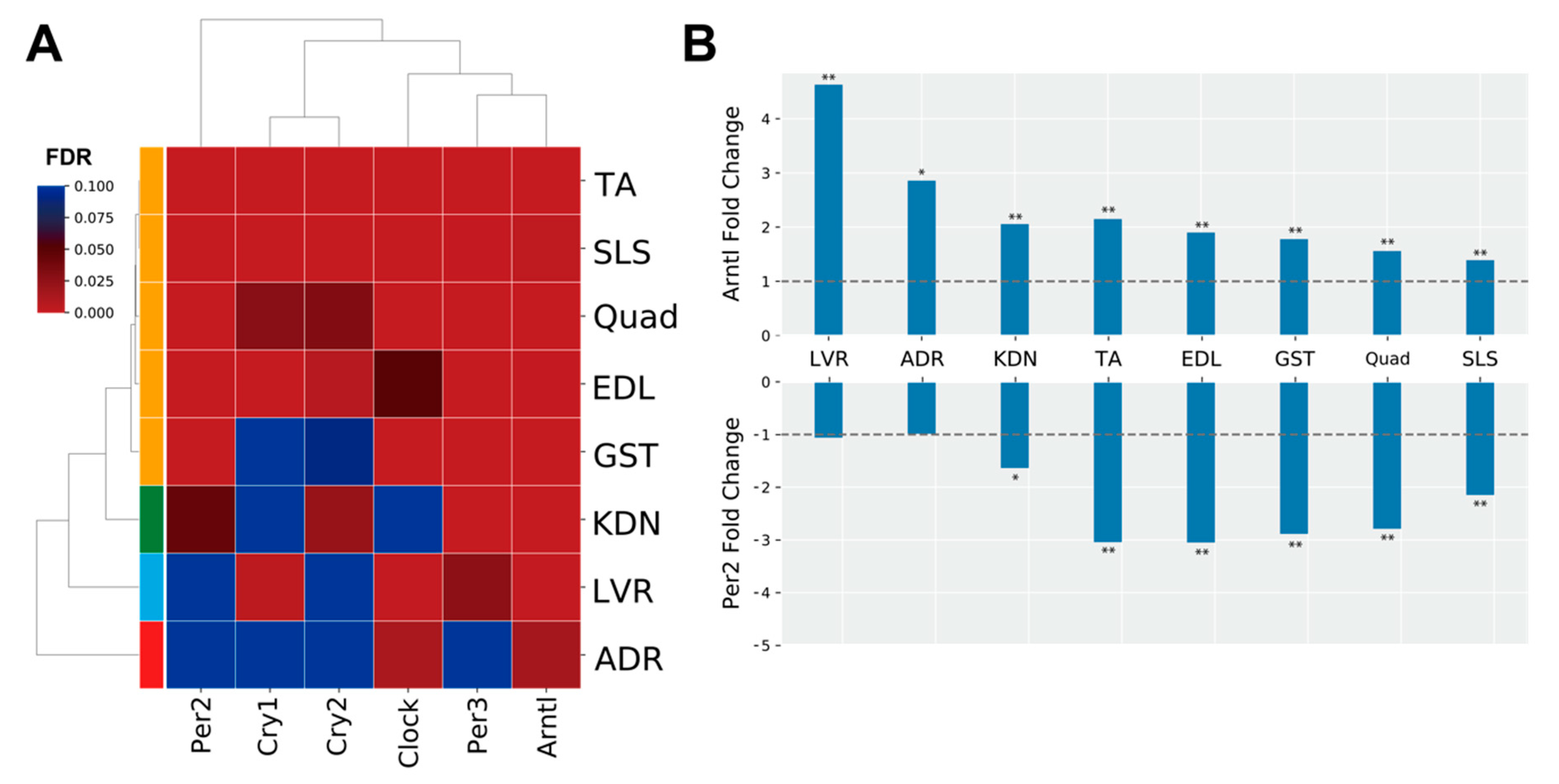
The genes in MODE1 are known to play a role in a core loop that regulates circadian rhythm cycles via a feedback mechanism, altogether synchronizing circadian rhythms between tissues from signals of the central nervous system via light input to the eye (Figure 3B) [30]. Therefore, we predicted that clock genes under spaceflight would show gene expression patterns that were consistent between peripheral tissues. To investigate this hypothesis, we focused on the genes in the MODE1 cluster and confirmed their FDR values (Figure 4A). We found that some clock genes did not significantly change expression during spaceflight in the adrenal glands, kidneys, and liver (Figure 4A). For example, Cry and Per gene expressions were not significantly altered under spaceflight in the adrenal glands (Figure 4A). On the other hand, most muscle tissues showed a significantly changed expression for most of these same clock genes under spaceflight (Figure 4A). Furthermore, we plotted the fold changes of Arntl (also known as Bmal1) and Per2 as representative clock genes because they are known to be in antiphase with each other (Figure 4B) [31]. Arntl was consistently up-regulated and Per2 was consistently down-regulated across all muscle tissues in the spaceflight condition (Figure 4B). Arntl was also consistently up-regulated in the adrenal glands, kidneys, and liver. However, Per2 did not show significant changes in the adrenal glands and liver, and only showed significant changes at less conservative thresholds in the kidneys compared to the muscle tissues (Figure 4B). In summary, our results suggested that the clock genes became asynchronous between certain peripheral tissues by some undetermined stimuli under spaceflight conditions.
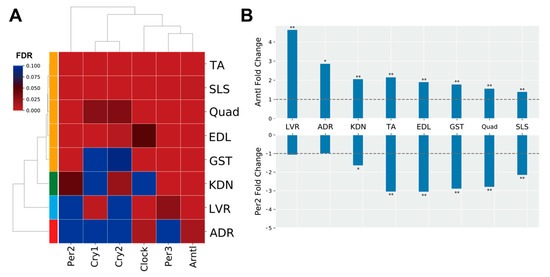
Figure 4.
Clock gene expression patterns. (A) is the heatmap of the FDR values of clock genes for each tissue. (B) is the fold change (FLT/GC) of representative clock genes (Arntl and Per2), which are known to be in antiphase oscillation with each other. The dashed horizontal lines indicate fold change magnitudes with values of one as visual references. * FDR < 0.05, ** FDR < 0.01.
4. Discussion
Our purpose was to elucidate the biological mechanisms across tissues by reanalyzing integrated RNA-seq data of mice during spaceflight. We found that the FLT group showed significantly altered clock gene expressions across tissues compared to the GC group (Figure 1A–C). Astronauts experience sleep and circadian rhythm shifts that impair health, alertness, and performance during spaceflight [18,19]. Therefore, understanding the effects of the spaceflight environment on the circadian rhythm system is important for long-term crewed space missions. It would also be important as the commercial spaceflight field expands for passengers and pilots who regularly fly orbital or suborbital flights between, say, New York City and Tokyo. This is because the circadian rhythms of the cells in most peripheral tissues are uncoupled into their own circadian rhythms by crossing time zones in a jet plane or spaceflight [32]. Moreover, enrichment analysis in the tissue-wide analysis enriched herpes simplex infection and endocrine system development as prevalent molecular mechanisms across tissues (Figure 1C). Previous studies have reported immune alterations during spaceflight, consisting of reductions in T cell and Natural Killer (NK) cell function, and the reactivation of latent herpes viruses [33,34,35]. Spaceflight could induce increased levels of stress hormones including cortisol, dehydroepiandrosterone, epinephrine, and norepinephrine through hypothalamic-pituitary-adrenal (HPA) and sympathetic-adrenal-medullary (SAM) axes activation [33,34,35]. These stress hormones are reported to affect the immune system such as causing the reactivation of viruses under spaceflight [33,34,35]. Circulating stress hormones, such as glucocorticoids, play a role in the mediation of the circadian clock and the synchronization of peripheral clocks [36,37]. Therefore, the interaction of the endocrine axis and the immune system with regard to the circadian clock could be associated with the underlying molecular mechanisms across tissues under spaceflight.
The central clock localized in the suprachiasmatic nucleus (SCN) unifies circadian rhythms between tissues through neurotransmission and hormonal signals [30]. After the SCN receives light input from the eye, the resulting signal from the SCN synchronizes the peripheral clocks of peripheral tissues. At the same time, this light-induced mechanism can be altered by external stimuli such as food intake, which can cause peripheral tissues to perform their own phase of circadian rhythm [31]. In other words, other events may be driving the observed circadian rhythm changes other than the light–dark cycle in peripheral tissues. Indeed, previous mice studies during spaceflight have implemented carefully controlled 12 h light–dark cycles but still observed circadian rhythm-related disruptions at both the behavioral and molecular biological levels. For instance, studies have reported that the FLT group showed significant increases in behavior activity during the dark cycle and in food intake throughout the mission compared to the GC group [3,38]. Likewise, other studies reported that clock-related gene expressions were changed in the liver and eyes of mice that underwent spaceflight for about one month [10,30,39]. A previous study suggested that the circadian rhythm could contribute to signs of non-alcoholic fatty liver disease (NAFLD) at the molecular level in mice under spaceflight [10]. It is essential for future studies to investigate the molecular details of how circadian rhythms are disrupted during spaceflight.
Our results using DEGs from the 8 tissues showed that circadian rhythms were commonly enriched (Figure 3A). Clustering analysis of PPI networks showed that MODE1 was most significantly enriched in terms and genes related to the circadian rhythm (Figure 3B,C). Genes in MODE1 positively regulate clock gene expressions, such as those of Per and Cry, by forming heterodimers of the transcription factors (TFs) Arntl and Clock [31]. On the other hand, the proteins produced from these same clock genes negatively regulate the transcriptional activities of Arntl and Clock to form the core loop of the diurnal fluctuation rhythm [31]. We found that most of the clock genes in the MODE1 cluster showed significantly altered expression across the muscle tissues under spaceflight (Figure 4A). In contrast, most clock genes (except Arntl) in the MODE1 cluster did not significantly change in the adrenal glands, kidneys, and liver (Figure 4A). While the fold change of Arntl was consistently up-regulated across all tissues, Per2 did not show significant changes in the adrenal glands and liver, and only showed significant changes at less conservative thresholds in the kidneys compared to the muscle tissues (Figure 4B). Given that the data we used came from studies that reported treating GC mice as similarly as possible to FLT mice by implementing controlled light–dark cycles and food resources within the same housing devices [3,38], we predicted that clock genes under spaceflight would show gene expression patterns that were in synchrony between peripheral tissues. However, our results suggested that other factors besides the environmental light–dark cycle (such as gravity, radiation, and rearing environment) may be driving asynchrony of the circadian rhythm between certain peripheral tissues during spaceflight.
We observed that the adrenal glands, kidneys, and liver showed similar expression distribution in the PCA plot (Figure 1B). Enrichment analysis for the tissue-wide analysis showed that endocrine system development terms were enriched as prevalent molecular mechanisms across tissues. In addition, these 3 tissues showed inconsistent clock gene expression changes compared to most muscle tissues (Figure 4A,B). These 3 tissues relate to functions known to be affected by the circadian rhythm, such as the endocrine systems (renin-angiotensin-aldosterone system, erythropoietin, vasopressin, and glucocorticoids) and water–mineral balance regulation [40,41,42,43]. Indeed, spaceflight conditions in humans are believed to induce activation of the endocrine systems, including renin-aldosterone, glucocorticoid, and catecholamines, and changes in water–mineral balance [18,44,45,46]. In addition, during a Mars simulation study in humans, the hormones aldosterone and cortisol fluctuated for longer-than-usual periods despite constant salt intake, suggesting that clock genes may be involved in water–mineral balance [47]. Few studies have examined the responses of adrenal glands and kidneys in spaceflight [45,47,48,49,50,51]. Therefore, our observed asynchrony of clock genes in these three tissues may provide new molecular-level insights about the endocrine systems and water–mineral balance regulation during spaceflight.
Arntl, Clock, Per2, Per3, Cry1, and Cry2 were enriched across some tissues in MODE1 of the PPI network analysis (Figure 3B). In addition, Arntl, Clock, and Per3 were significantly changed in the tissue-wide analysis (Figure 1A). Since TFs of these genes form core clock components in circadian rhythm mechanisms [31], these clock genes could play a role as key regulators associated with underlying mechanisms of physiology and metabolism under spaceflight [18,52]. For example, previous studies showed that clock genes have demonstrated a role of circadian rhythm in muscle atrophy and bone remodeling [53,54,55]. In addition, these clock genes have been implicated in NAFLD [56], the symptoms of which have been observed in mice during spaceflight [10]. Furthermore, Arntl plays a role in the circadian regulation of acute glucocorticoid secretion in the adrenal glands in response to stress [57]. Mice models carrying a conditional allele for Arntl showed several disorders, such as increased urine volume, changes in the circadian rhythm of urinary sodium excretion, increased glomerular filtration rate, and significantly reduced plasma aldosterone levels [58]. Clock null mice showed abnormal circadian rhythmicity of plasma aldosterone levels and changes in circadian gene expression patterns in the kidney [59]. Glucocorticoids could shift the phase of circadian oscillations of Per1 and Per2 expressions in peripheral tissues [60]. Cry genes are associated with changes in the transcriptional response to glucocorticoids in mouse embryonic fibroblasts [61]. In addition, Cry1 and Cry2 null mice indicated salt-sensitive hypertension via abnormally high synthesis of the mineralocorticoid aldosterone by the adrenal gland [62]. Given that circulating stress hormones are increased and circadian rhythms are altered in astronauts and mice [34,35,63], spaceflight could contribute to circadian rhythm disruption and asynchrony between peripheral tissues. Therefore, these results suggest that these clock genes could modulate physiology and metabolism mechanisms as key regulators during spaceflight environmental changes. However, the role of clock genes is unknown in peripheral tissues under spaceflight. In a simulated microgravity study, for example, Arntl disrupted diurnal oscillation in rat cerebrovascular contractility by changing circadian regulation of the miR-103/CaV 1.2 signal pathway [64]. Oscillations of Arntl were amplified under simulated microgravity in human keratinocytes [65]. Future studies could determine clock gene roles in peripheral tissues under spaceflight, such as by using clock genes null mice under spaceflight [66].
This study has several limitations. First, the RNA-seq data in this study only reflect the effects of spaceflight after 1-month missions. We note that major clock gene expressions of Drosophila melanogaster were unaffected after short-term spaceflight for 13 days, despite being exposed to the same 12 h light/dark cycles that the mice were exposed to in the data we analyzed [20]. Hence, our pilot study provides new motivation to elucidate both shorter and longer time series of molecular circadian mechanisms under spaceflight in mammals. Second, it is unclear whether our mice results generalize to human levels; future spaceflight experiments can explore clock-related gene expression changes in human tissue samples derived from liquid biopsies. Third, although the data we used came from studies that reported treating GC mice as similarly as possible to FLT mice, clock gene expressions may be changed by factors that may not be routinely controlled within and between spaceflight datasets, such as sample collection and dissection schedules [20,31]. It is crucial for principal investigators to provide detailed metadata when publishing their data; this includes sample handling information, such as dissection and freezing schedules. In addition, the current study provided preliminary findings related to circadian rhythm disruptions during spaceflight using limited types of tissue datasets that were all from the same mission. However, the circadian rhythm is known to depend on other systems that we did not explore (such as cardiovascular and nervous systems) [18]; these types of data are currently not available on NASA GeneLab collectively from the same mission and sample manipulation. Future studies may expand upon our pilot study and investigate all systems related to circadian rhythms if they become available in NASA GeneLab.
In summary, since the safety and performance of astronauts and commercial spaceflight pilots can be affected by disruption of circadian clocks, our study highlights the necessity of further investigation into the impact of spaceflight on mammalian tissue functions. Our results shed novel insights into possible health consequences under spaceflight conditions, including the overall disruption of clock gene synchronization between peripheral tissues. As of now, our pilot study remains inconclusive as to whether disruptions in the clock gene expressions in mice during spaceflight are due to the space environment itself or to sample operation artifacts or to both. The current cross-data study was possible due to open-source space omics databases that implement state-of-the-art metadata normalization. As metadata standardization and multi-omics approaches to space biology continue to improve, future studies may more conclusively determine causes and countermeasures to circadian rhythm disruptions in space.
Author Contributions
The following contributions were made by the authors of this manuscript: conceptualization, S.-i.F., L.R., Q.O. and M.M.; methodology, S.-i.F.; formal analysis, S.-i.F.; investigation, S.-i.F.; gathering and analyzing the data, S.-i.F.; writing—original draft preparation, S.-i.F.; writing—review and editing, L.R., Q.O. and M.M.; visualization, S.-i.F.; supervision, M.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by JSPS (15K21745, 20H03234).
Acknowledgments
We thank NASA GeneLab for making space omics datasets available. We thank Haruka Ozaki (University of Tsukuba) for helpful advice and Flaminia Miyamasu (University of Tsukuba) for editing a draft of this manuscript. This work was supported by JSPS KAKENHI Grant Numbers JP15K21745, JP 20H03234.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Kandarpa, K.; Schneider, V.; Ganapathy, K. Human health during space travel: An overview. Neurol. India 2019, 67, 176. [Google Scholar] [CrossRef]
- Barratt, M.R.; Baker, E.S.; Pool, S.L. (Eds.) Principles of Clinical Medicine for Space Flight; Springer: New York, NY, USA, 2019; ISBN 978-1-4939-9887-6. [Google Scholar]
- Choi, S.Y.; Saravia-Butler, A.; Shirazi-Fard, Y.; Leveson-Gower, D.; Stodieck, L.S.; Cadena, S.M.; Beegle, J.; Solis, S.; Ronca, A.; Globus, R.K. Validation of a New Rodent Experimental System to Investigate Consequences of Long Duration Space Habitation. Sci. Rep. 2020, 10, 2336. [Google Scholar] [CrossRef] [PubMed]
- Furukawa, S.; Nagamatsu, A.; Nenoi, M.; Fujimori, A.; Kakinuma, S.; Katsube, T.; Wang, B.; Tsuruoka, C.; Shirai, T.; Nakamura, A.J.; et al. Space Radiation Biology for “Living in Space”. Available online: https://www.hindawi.com/journals/bmri/2020/4703286/ (accessed on 11 April 2020).
- Shiba, D.; Mizuno, H.; Yumoto, A.; Shimomura, M.; Kobayashi, H.; Morita, H.; Shimbo, M.; Hamada, M.; Kudo, T.; Shinohara, M.; et al. Development of new experimental platform ’MARS’-Multiple Artificial-gravity Research System-to elucidate the impacts of micro/partial gravity on mice. Sci. Rep. 2017, 7, 10837. [Google Scholar] [CrossRef] [PubMed]
- Bradbury, P.; Wu, H.; Choi, J.U.; Rowan, A.E.; Zhang, H.; Poole, K.; Lauko, J.; Chou, J. Modeling the Impact of Microgravity at the Cellular Level: Implications for Human Disease. Front. Cell Dev. Biol. 2020, 8, 96. [Google Scholar] [CrossRef] [PubMed]
- Chatziravdeli, V.; Katsaras, G.N.; Lambrou, G.I. Gene Expression in Osteoblasts and Osteoclasts under Microgravity Conditions: A Systematic Review. Curr. Genom. 2019, 20, 184–198. [Google Scholar] [CrossRef] [PubMed]
- Matsuda, C.; Kato, T.; Inoue-Suzuki, S.; Kikuchi, J.; Ohta, T.; Kagawa, M.; Hattori, M.; Kobayashi, H.; Shiba, D.; Shirakawa, M.; et al. Dietary intervention of mice using an improved Multiple Artificial-gravity Research System (MARS) under artificial 1 g. Npj Microgravity 2019, 5, 16. [Google Scholar] [CrossRef]
- Shendure, J.; Balasubramanian, S.; Church, G.M.; Gilbert, W.; Rogers, J.; Schloss, J.A.; Waterston, R.H. DNA sequencing at 40: Past, present and future. Nature 2017, 550, 345–353. [Google Scholar] [CrossRef]
- Beheshti, A.; Chakravarty, K.; Fogle, H.; Fazelinia, H.; da Silveira, W.A.; Boyko, V.; Polo, S.-H.L.; Saravia-Butler, A.M.; Hardiman, G.; Taylor, D.; et al. Multi-omics analysis of multiple missions to space reveal a theme of lipid dysregulation in mouse liver. Sci. Rep. 2019, 9, 19195. [Google Scholar] [CrossRef]
- Beheshti, A.; McDonald, J.T.; Miller, J.; Grabham, P.; Costes, S.V. GeneLab Database Analyses Suggest Long-Term Impact of Space Radiation on the Cardiovascular System by the Activation of FYN through Reactive Oxygen Species. Int. J. Mol. Sci. 2019, 20, 661. [Google Scholar] [CrossRef]
- Beheshti, A.; Cekanaviciute, E.; Smith, D.J.; Costes, S.V. Global transcriptomic analysis suggests carbon dioxide as an environmental stressor in spaceflight: A systems biology GeneLab case study. Sci. Rep. 2018, 8, 4191. [Google Scholar] [CrossRef]
- Beheshti, A.; Ray, S.; Fogle, H.; Berrios, D.; Costes, S.V. A microRNA signature and TGF-β1 response were identified as the key master regulators for spaceflight response. PLoS ONE 2018, 13, e0199621. [Google Scholar] [CrossRef] [PubMed]
- McDonald, J.T.; Stainforth, R.; Miller, J.; Cahill, T.; da Silveira, W.A.; Rathi, K.S.; Hardiman, G.; Taylor, D.; Costes, S.V.; Chauhan, V.; et al. NASA GeneLab Platform Utilized for Biological Response to Space Radiation in Animal Models. Cancers 2020, 12, 381. [Google Scholar] [CrossRef] [PubMed]
- Karouia, F.; Peyvan, K.; Pohorille, A. Toward biotechnology in space: High-throughput instruments for in situ biological research beyond Earth. Biotechnol. Adv. 2017, 35, 905–932. [Google Scholar] [CrossRef]
- Berrios, D.; Weitz, E.; Grigorev, K.; Costes, S.; Gebre, S.; Beheshti, A. Visualizing Omics Data from Spaceflight Samples using the NASA GeneLab Platform. In Proceedings of the 12th International Conference, San Francisco, CA, USA, 23–25 March 2020; pp. 89–98. [Google Scholar]
- Ray, S.; Gebre, S.; Fogle, H.; Berrios, D.C.; Tran, P.B.; Galazka, J.M.; Costes, S.V. GeneLab: Omics database for spaceflight experiments. Bioinformatics 2019, 35, 1753–1759. [Google Scholar] [CrossRef] [PubMed]
- Astaburuaga, R.; Basti, A.; Li, Y.; Herms, D.; Relógio, A. Circadian regulation of physiology: Relevance for space medicine. REACH 2019, 14–15, 100029. [Google Scholar] [CrossRef]
- Guo, J.-H.; Qu, W.-M.; Chen, S.-G.; Chen, X.-P.; Lv, K.; Huang, Z.-L.; Wu, Y.-L. Keeping the right time in space: Importance of circadian clock and sleep for physiology and performance of astronauts. Mil. Med. Res. 2014, 1, 1–7. [Google Scholar] [CrossRef]
- Ma, L.; Ma, J.; Xu, K. Effect of Spaceflight on the Circadian Rhythm, Lifespan and Gene Expression of Drosophila melanogaster. PLoS ONE 2015, 10, e0121600. [Google Scholar] [CrossRef]
- Galazka, J.; Globus, R. Rodent Research-1 (RR1) NASA Validation Flight: Mouse Adrenal Gland Transcriptomic, Proteomic, and Epigenomic Data; Version 7; GeneLab: Mountain View, CA, USA. [CrossRef]
- Galazka, J.; Globus, R. Rodent Research-1 (RR1) NASA Validation Flight: Mouse Extensor Digitorum Longus Muscle Transcriptomic and Epigenomic Data; Version 4; GeneLab: Mountain View, CA, USA. [CrossRef]
- Galazka, J.; Globus, R. Rodent Research-1 (RR1) NASA Validation Flight: Mouse Gastrocnemius Muscle Transcriptomic, Proteomic, and Epigenomic Data; Version 4; GeneLab: Mountain View, CA, USA. [CrossRef]
- Galazka, J.; Globus, R. Rodent Research-1 (RR1) NASA Validation Flight: Mouse Kidney Transcriptomic, Proteomic, and Epigenomic Data; Version 4; GeneLab: Mountain View, CA, USA. [CrossRef]
- Galazka, J.; Globus, R. Rodent Research-1 (RR1) NASA Validation Flight: Mouse Quadriceps Muscle Transcriptomic, Proteomic, and Epigenomic Data; Version 4; GeneLab: Mountain View, CA, USA. [CrossRef]
- Galazka, J.; Globus, R. Rodent Research-1 (RR1) NASA Validation Flight: Mouse Soleus Muscle Transcriptomic and Epigenomic Data; Version 4; GeneLab: Mountain View, CA, USA. [CrossRef]
- Galazka, J.; Globus, R. Rodent Research-1 (RR1) NASA Validation Flight: Mouse Tibialis Anterior Muscle Transcriptomic, Proteomic, and Epigenomic Data; Version 4; GeneLab: Mountain View, CA, USA. [CrossRef]
- Galazka, J. RR-1 and RR-3 Mouse Liver Transcriptomics with and without ERCC Control RNA Spike-Ins; Version 8; GeneLab: Mountain View, CA, USA. [CrossRef]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun 2019, 10, 1523. [Google Scholar] [CrossRef]
- Mao, X.W.; Nishiyama, N.C.; Byrum, S.D.; Stanbouly, S.; Jones, T.; Drew, A.; Sridharan, V.; Boerma, M.; Tackett, A.J.; Zawieja, D.; et al. Characterization of mouse ocular response to a 35-day spaceflight mission: Evidence of blood-retinal barrier disruption and ocular adaptations. Sci. Rep. 2019, 9, 8215. [Google Scholar] [CrossRef]
- Preußner, M.; Heyd, F. Post-transcriptional control of the mammalian circadian clock: Implications for health and disease. Pflug. Arch. 2016, 468, 983–991. [Google Scholar] [CrossRef]
- Foster, R.G.; Kreitzman, L. The rhythms of life: What your body clock means to you! Exp. Physiol. 2014, 99, 599–606. [Google Scholar] [CrossRef]
- Crucian, B.E.; Makedonas, G.; Sams, C.F.; Pierson, D.L.; Simpson, R.; Stowe, R.P.; Smith, S.M.; Zwart, S.R.; Krieger, S.S.; Rooney, B.; et al. Countermeasures-based Improvements in Stress, Immune System Dysregulation and Latent Herpesvirus Reactivation onboard the International Space Station—Relevance for Deep Space Missions and Terrestrial Medicine. Neurosci. Biobehav. Rev. 2020, 115, 68–76. [Google Scholar] [CrossRef] [PubMed]
- Mann, V.; Sundaresan, A.; Mehta, S.K.; Crucian, B.; Doursout, M.F.; Devakottai, S. Effects of microgravity and other space stressors in immunosuppression and viral reactivation with potential nervous system involvement. Neurol. India 2019, 67, 198. [Google Scholar] [CrossRef]
- Akiyama, T.; Horie, K.; Hinoi, E.; Hiraiwa, M.; Kato, A.; Maekawa, Y.; Takahashi, A.; Furukawa, S. How does spaceflight affect the acquired immune system? NPJ Micrograv. 2020, 6, 14. [Google Scholar] [CrossRef] [PubMed]
- Dumbell, R.; Matveeva, O.; Oster, H. Circadian Clocks, Stress, and Immunity. Front. Endocrinol. 2016, 7, 37. [Google Scholar] [CrossRef]
- Edgar, R.S.; Stangherlin, A.; Nagy, A.D.; Nicoll, M.P.; Efstathiou, S.; O’Neill, J.S.; Reddy, A.B. Cell autonomous regulation of herpes and influenza virus infection by the circadian clock. Proc. Natl. Acad. Sci. USA 2016, 113, 10085–10090. [Google Scholar] [CrossRef]
- Ronca, A.E.; Moyer, E.L.; Talyansky, Y.; Lowe, M.; Padmanabhan, S.; Choi, S.; Gong, C.; Cadena, S.M.; Stodieck, L.; Globus, R.K. Behavior of mice aboard the International Space Station. Sci. Rep. 2019, 9, 4717. [Google Scholar] [CrossRef]
- Overbey, E.G.; da Silveira, W.A.; Stanbouly, S.; Nishiyama, N.C.; Roque-Torres, G.D.; Pecaut, M.J.; Zawieja, D.C.; Wang, C.; Willey, J.S.; Delp, M.D.; et al. Spaceflight influences gene expression, photoreceptor integrity, and oxidative stress-related damage in the murine retina. Sci. Rep. 2019, 9, 13304. [Google Scholar] [CrossRef]
- Gizowski, C.; Trudel, E.; Bourque, C.W. Central and peripheral roles of vasopressin in the circadian defense of body hydration. Best Pract. Res. Clin. Endocrinol. Metab. 2017, 31, 535–546. [Google Scholar] [CrossRef]
- Kalsbeek, A.; van der Spek, R.; Lei, J.; Endert, E.; Buijs, R.M.; Fliers, E. Circadian rhythms in the hypothalamo–pituitary–adrenal (HPA) axis. Mol. Cell. Endocrinol. 2012, 349, 20–29. [Google Scholar] [CrossRef]
- Pasqualetti, P.; Casale, R. Circadian rhythm of serum erythropoietin in healthy subjects. Riv. Eur. Sci. Med. Farm. 1996, 18, 91–93. [Google Scholar]
- Xie, Y.; Tang, Q.; Chen, G.; Xie, M.; Yu, S.; Zhao, J.; Chen, L. New Insights into the Circadian Rhythm and Its Related Diseases. Front. Physiol. 2019, 10, 682. [Google Scholar] [CrossRef]
- Afonin, B.V.; Grigor’ev, A.I.; Pavlova, E.A. Effect of short-term space flights on the activity of the renin-angiotensin-aldosterone system and on the blood concentration of cyclic nucleotides and prostaglandins. Kosm. Biol. Aviakosm. Med. 1986, 20, 27–30. [Google Scholar] [PubMed]
- Christensen, N.J.; Drummer, C.; Norsk, P. Renal and sympathoadrenal responses in space. Am. J. Kidney Dis. 2001, 38, 679–683. [Google Scholar] [CrossRef] [PubMed]
- Drummer, C.; Norsk, P.; Heer, M. Water and sodium balance in space. Am. J. Kidney Dis. 2001, 38, 684–690. [Google Scholar] [CrossRef] [PubMed]
- Rakova, N.; Jüttner, K.; Dahlmann, A.; Schröder, A.; Linz, P.; Kopp, C.; Rauh, M.; Goller, U.; Beck, L.; Agureev, A.; et al. Long-Term Space Flight Simulation Reveals Infradian Rhythmicity in Human Na+ Balance. Cell Metab. 2013, 17, 125–131. [Google Scholar] [CrossRef]
- Miyake, M.; Yamasaki, M.; Waki, H.; Katahira, K.; O.-ishi, H.; Katsuda, S.; Nagayama, T.; Ijiri, K.; Hazama, A.; Shimizu, T. Morphological characteristics of the kidney and lung in the neonatal rats observed after 16 days spaceflight. Biol. Sci. Space 2003, 17, 173–174. [Google Scholar]
- Liakopoulos, V.; Leivaditis, K.; Eleftheriadis, T.; Dombros, N. The kidney in space. Int. Urol. Nephrol. 2012, 44, 1893–1901. [Google Scholar] [CrossRef]
- Hammond, T.G.; Allen, P.L.; Birdsall, H.H. Effects of Space Flight on Mouse Liver versus Kidney: Gene Pathway Analyses. Int. J. Mol. Sci. 2018, 19, 4106. [Google Scholar] [CrossRef]
- Santucci, D.; Kawano, F.; Ohira, T.; Terada, M.; Nakai, N.; Francia, N.; Alleva, E.; Aloe, L.; Ochiai, T.; Cancedda, R.; et al. Evaluation of gene, protein and neurotrophin expression in the brain of mice exposed to space environment for 91 days. PLoS ONE 2012, 7, e40112. [Google Scholar] [CrossRef]
- Strollo, F.; Gentile, S.; Strollo, G.; Mambro, A.; Vernikos, J. Recent Progress in Space Physiology and Aging. Front. Physiol. 2018, 9, 1551. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.-Y.; Wang, J.; Tong, X.; Chen, J.; Wang, B.; Miao, Z.-N.; Li, X.; Ye, J.-X.; Yuan, F.-L. Emerging role of circadian rhythm in bone remodeling. J. Mol. Med. 2019, 97, 19–24. [Google Scholar] [CrossRef] [PubMed]
- Aoyama, S.; Kojima, S.; Sasaki, K.; Ishikawa, R.; Tanaka, M.; Shimoda, T.; Hattori, Y.; Aoki, N.; Takahashi, K.; Hirooka, R.; et al. Day-Night Oscillation of Atrogin1 and Timing-Dependent Preventive Effect of Weight-Bearing on Muscle Atrophy. EBioMedicine 2018, 37, 499–508. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, S.; Ma, K. Circadian clock regulation of skeletal muscle growth and repair. F1000Research 2016, 5, 1549. [Google Scholar] [CrossRef] [PubMed]
- Shi, D.; Chen, J.; Wang, J.; Yao, J.; Huang, Y.; Zhang, G.; Bao, Z. Circadian Clock Genes in the Metabolism of Non-alcoholic Fatty Liver Disease. Front. Physiol. 2019, 10, 423. [Google Scholar] [CrossRef] [PubMed]
- Leliavski, A.; Shostak, A.; Husse, J.; Oster, H. Impaired glucocorticoid production and response to stress in Arntl-deficient male mice. Endocrinology 2014, 155, 133–142. [Google Scholar] [CrossRef] [PubMed]
- Tokonami, N.; Mordasini, D.; Pradervand, S.; Centeno, G.; Jouffe, C.; Maillard, M.; Bonny, O.; Gachon, F.; Gomez, R.A.; Sequeira-Lopez, M.L.S.; et al. Local Renal Circadian Clocks Control Fluid–Electrolyte Homeostasis and BP. JASN 2014, 25, 1430–1439. [Google Scholar] [CrossRef] [PubMed]
- Nikolaeva, S.; Pradervand, S.; Centeno, G.; Zavadova, V.; Tokonami, N.; Maillard, M.; Bonny, O.; Firsov, D. The Circadian Clock Modulates Renal Sodium Handling. J. Am. Soc. Nephrol. 2012, 23, 1019–1026. [Google Scholar] [CrossRef]
- Nicolaides, N.C.; Charmandari, E.; Kino, T.; Chrousos, G.P. Stress-Related and Circadian Secretion and Target Tissue Actions of Glucocorticoids: Impact on Health. Front. Endocrinol. (Lausanne) 2017, 8, 70. [Google Scholar] [CrossRef]
- Lamia, K.A.; Papp, S.J.; Yu, R.T.; Barish, G.D.; Uhlenhaut, N.H.; Jonker, J.W.; Downes, M.; Evans, R.M. Cryptochromes mediate rhythmic repression of the glucocorticoid receptor. Nature 2011, 480, 552–556. [Google Scholar] [CrossRef]
- Doi, M.; Takahashi, Y.; Komatsu, R.; Yamazaki, F.; Yamada, H.; Haraguchi, S.; Emoto, N.; Okuno, Y.; Tsujimoto, G.; Kanematsu, A.; et al. Salt-sensitive hypertension in circadian clock-deficient Cry-null mice involves dysregulated adrenal Hsd3b6. Nat. Med. 2010, 16, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Larina, I.M.; Witson, P.; Smirnova, T.M.; Chen, Y.-M. Circadian rhythms of salivary cortisol content during long-term space flight. Hum. Physiol. 2000, 26, 462–467. [Google Scholar] [CrossRef]
- Chen, L.; Zhang, B.; Yang, L.; Bai, Y.-G.; Song, J.-B.; Ge, Y.-L.; Ma, H.-Z.; Cheng, J.-H.; Ma, J.; Xie, M.-J. BMAL1 Disrupted Intrinsic Diurnal Oscillation in Rat Cerebrovascular Contractility of Simulated Microgravity Rats by Altering Circadian Regulation of miR-103/CaV1.2 Signal Pathway. Int. J. Mol. Sci. 2019, 20, 3947. [Google Scholar] [CrossRef]
- Ranieri, D.; Cucina, A.; Bizzarri, M.; Alimandi, M.; Torrisi, M.R. Microgravity influences circadian clock oscillation in human keratinocytes. FEBS Open Bio 2015, 5, 717–723. [Google Scholar] [CrossRef] [PubMed]
- Cadena, S.M.; Zhang, Y.; Fang, J.; Brachat, S.; Kuss, P.; Giorgetti, E.; Stodieck, L.S.; Kneissel, M.; Glass, D.J. Skeletal muscle in MuRF1 null mice is not spared in low-gravity conditions, indicating atrophy proceeds by unique mechanisms in space. Sci. Rep. 2019, 9, 9397. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).