Proteomic Profiling of Plasma-Derived Biomarkers in Patients with Bladder Cancer: A Step towards Clinical Translation
Abstract
:1. Introduction
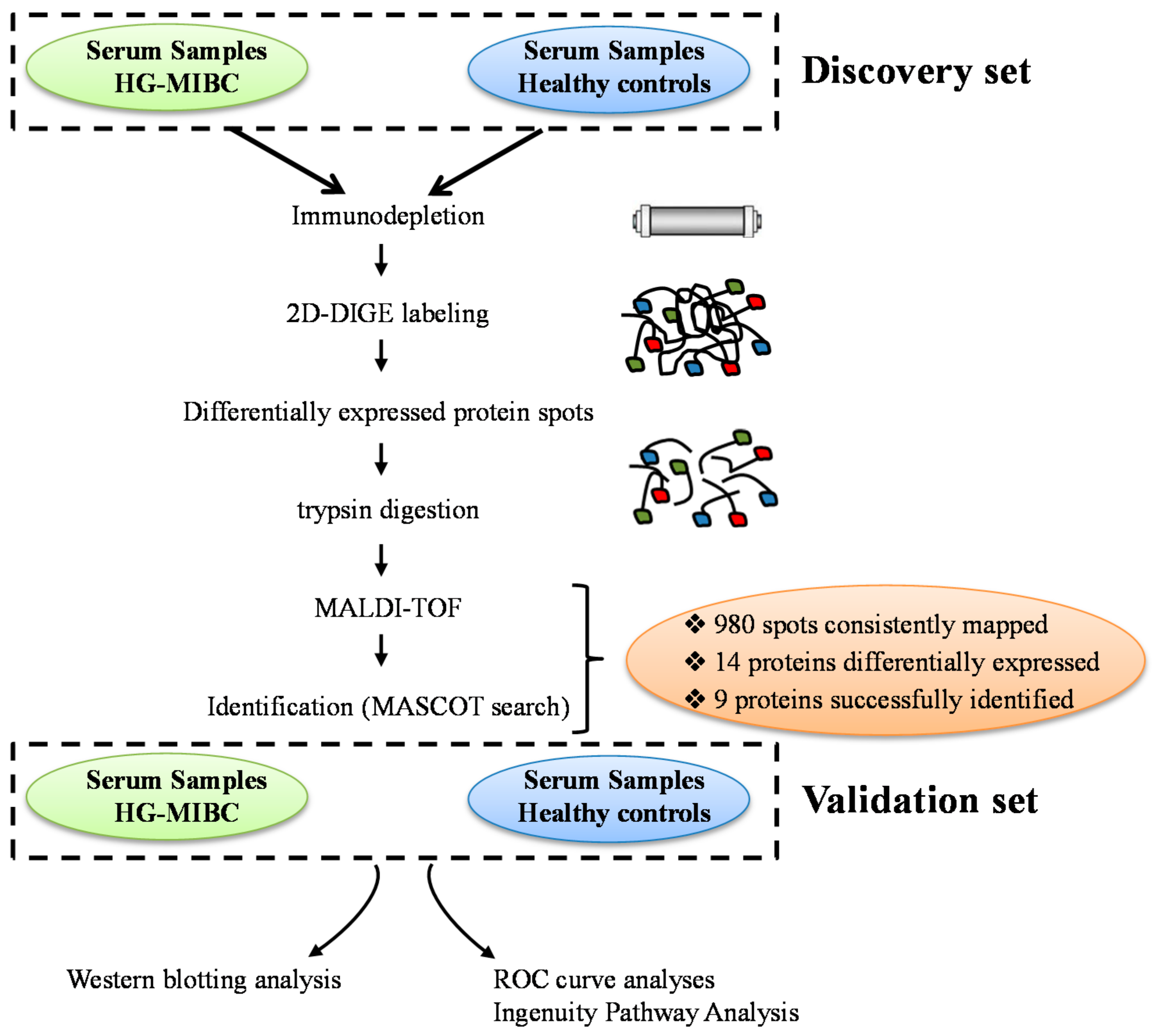
2. Materials and Methods
2.1. Patient and Samples Collection
2.2. Depletion of Abundant Proteins
2.3. Two-D DIGE Labeling
2.4. Two-Dimensional Gel Electrophoresis and Mass Spectrometry Protein Identification
2.5. Protein Interaction and Network Analysis
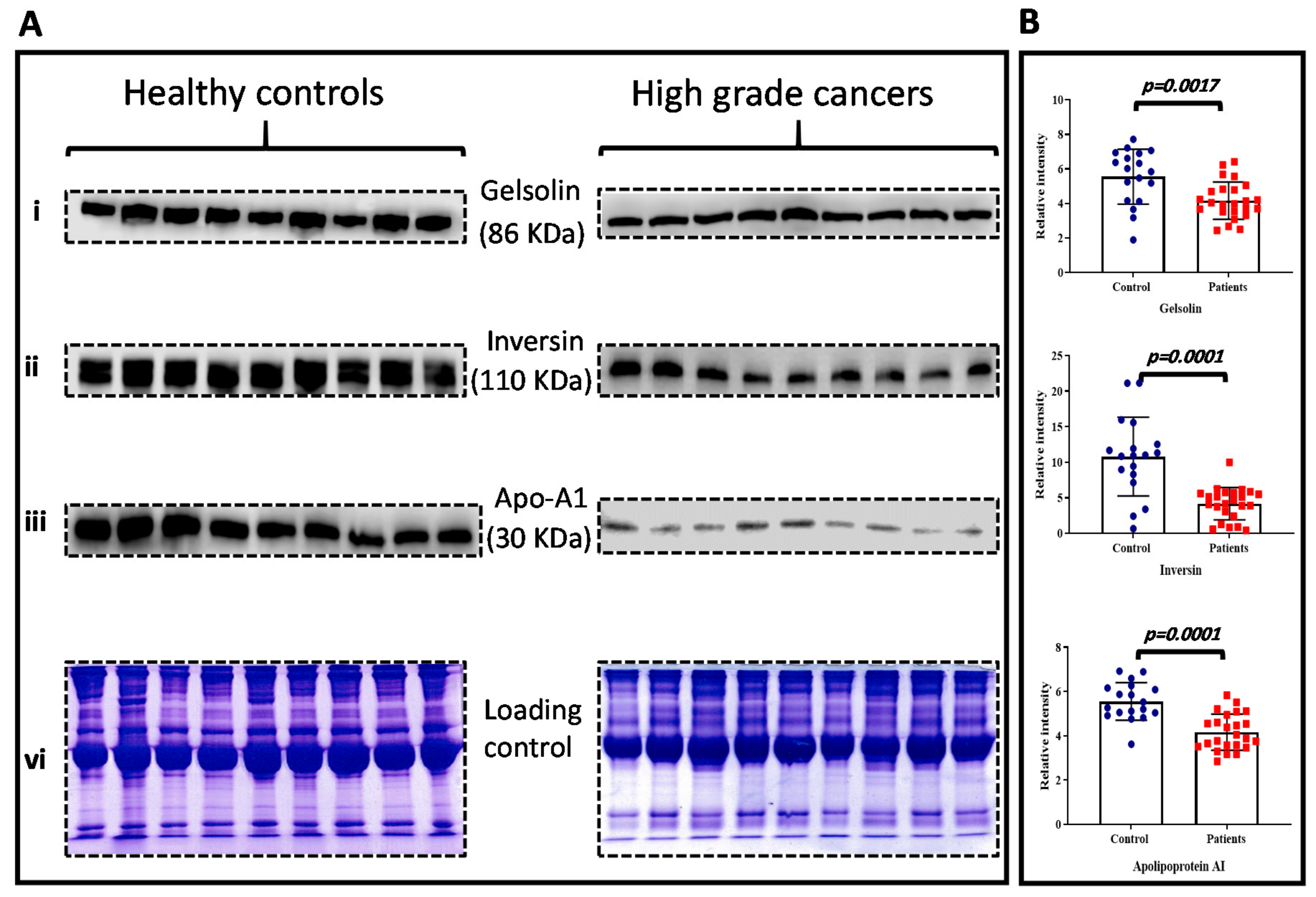
2.6. Data Validation: Western Blot Analysis
2.7. Statistical Analysis
3. Results
3.1. Patients’ Samples
3.2. Proteomic Analysis and Identification of Candidate Biomarkers
3.3. Pattern of Variation of Differentially Expressed Proteins
3.4. Functional Characteristics and Pathway Analysis
3.5. Data Validation
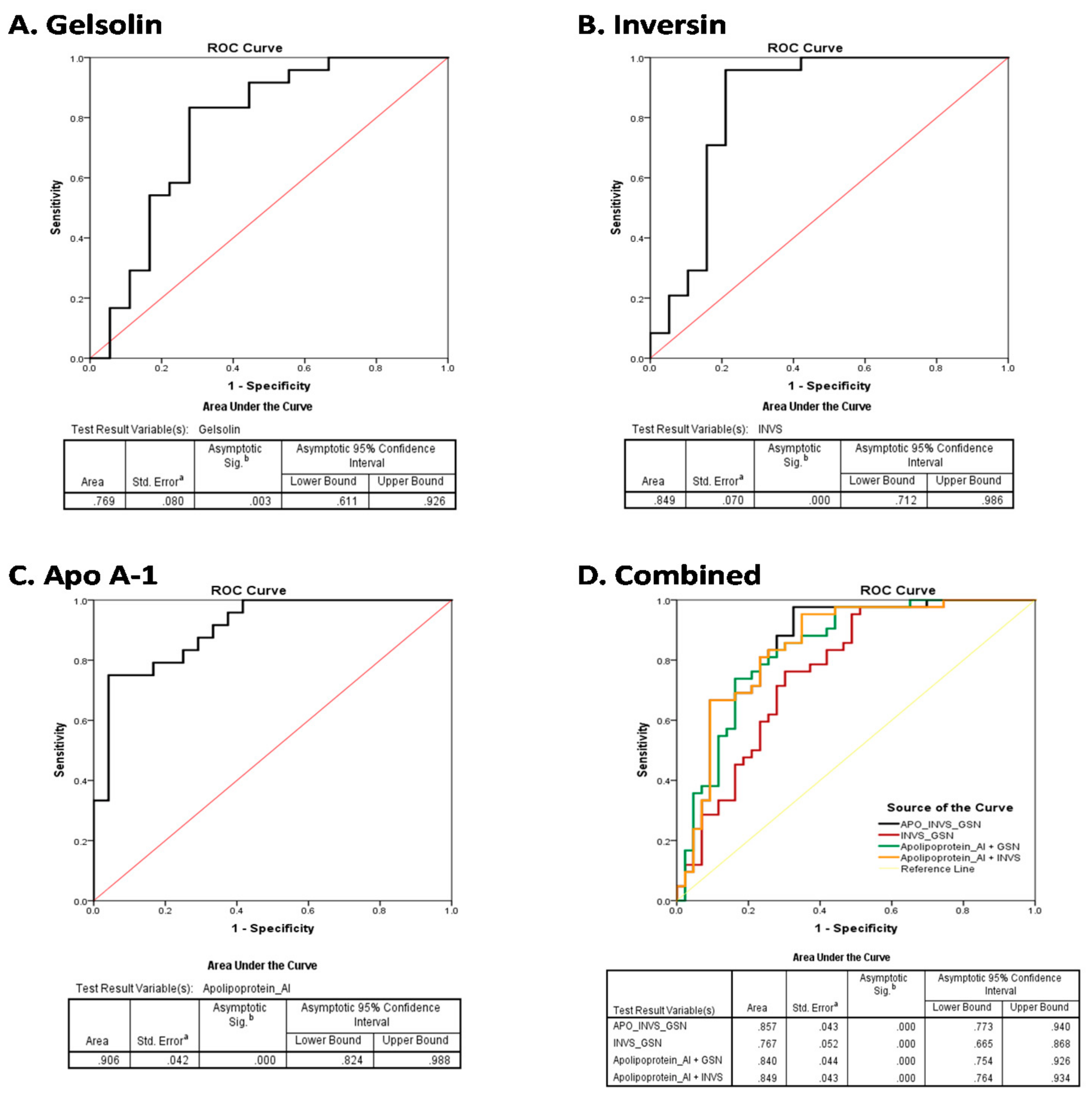
3.6. Diagnostic Efficiency of the Plasma Biomarkers
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [Green Version]
- Sun, M.; Trinh, Q.D. Diagnosis and staging of bladder cancer. Hematol. Oncol. Clin. N. Am. 2015, 29, 205–218. [Google Scholar] [CrossRef]
- Kaufman, D.S.; Shipley, W.U.; Feldman, A.S. Bladder cancer. Lancet 2009, 374, 239–249. [Google Scholar] [CrossRef]
- González, D.A.A.; De Velasco, G.; Lainez, N.; Maroto, P.; Morales-Barrera, R.; Muñoz-Langa, J.; Perez-Valderrama, B.; Basterretxea, L.; Caballero, C.; Vazquez, S. SEOM clinical guideline for treatment of muscle-invasive and metastatic urothelial bladder cancer. Clin. Transl. Oncol. 2019, 21, 64–74. [Google Scholar] [CrossRef] [Green Version]
- Witjes, J.A.; Compérat, E.; Cowan, N.C.; De Santis, M.; Gakis, G.; Lebret, T.; Ribal, M.J.; Van der Heijden, A.G.; Sherif, A. EAU Guidelines on Muscle-invasive and Metastatic Bladder Cancer: Summary of the 2013 Guidelines. Eur. Urol. 2014, 65, 778–792. [Google Scholar] [CrossRef]
- Del Bene, G.; Calabrò, F.; Giannarelli, D.; Plimack, E.R.; Harshman, L.C.; Yu, E.Y.; Crabb, S.J.; Pal, S.K.; Alva, A.S.; Powles, T.; et al. Neoadjuvant vs. Adjuvant Chemotherapy in Muscle Invasive Bladder Cancer (MIBC): Analysis from the RISC Database. Front. Oncol. 2018, 8, 463–471. [Google Scholar] [CrossRef]
- Gogalic, S.; Sauer, U.; Doppler, S.; Preininger, C. Bladder cancer biomarker array to detect aberrant levels of proteins in urine. Analyst 2015, 140, 724–735. [Google Scholar] [CrossRef] [PubMed]
- Xylinas, E.; Kent, M.; Kluth, L.; Pycha, A.; Comploj, E.; Svatek, R.S.; Lotan, Y.; Trinh, Q.-D.; Karakiewicz, P.I.; Holmang, S.; et al. Accuracy of the EORTC risk tables and of the CUETO scoring model to predict outcomes in non-muscle-invasive urothelial carcinoma of the bladder. Br. J. Cancer 2013, 109, 1460–1466. [Google Scholar] [CrossRef]
- Knowles, M.A.; Hurst, C.D. Molecular biology of bladder cancer: New insights into pathogenesis and clinical diversity. Nat. Rev. Cancer 2015, 15, 25–41. [Google Scholar] [CrossRef] [PubMed]
- Talukdar, S.; Emdad, L.; Das, S.K.; Sarkar, D.; Fisher, P.B. Noninvasive approaches for detecting and monitoring bladder cancer. Expert Rev. Anticancer Ther. 2014, 15, 283–294. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, A.; Dasari, S.; Long, W.; Mohan, C. Urine protein biomarkers for the detection, surveillance, and treatment response prediction of bladder cancer. Am. J. Cancer Res. 2019, 9, 1104–1117. [Google Scholar] [PubMed]
- Barbany, G.; Arthur, C.; Liedén, A.; Nordenskjöld, M.; Rosenquist, R.; Tesi, B. Cell-free tumour DNA testing for early detection of cancer—a potential future tool. J. Intern. Med. 2019, 286, 118–136. [Google Scholar] [PubMed] [Green Version]
- Geyer, P.E.; Kulak, N.A.; Pichler, G.; Holdt, L.M.; Teupser, D.; Mann, M. Plasma Proteome Profiling to Assess Human Health and Disease. Cell Syst. 2016, 2, 185–195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanash, S.M.; Pitteri, S.; Faça, V.M. Mining the plasma proteome for cancer biomarkers. Nat. Cell Biol. 2008, 452, 571–579. [Google Scholar] [CrossRef] [PubMed]
- Shukla, H.D.; Mahmood, J.; Vujaskovic, Z. Integrated proteo-genomic approach for early diagnosis and prognosis of cancer. Cancer Lett. 2015, 369, 28–36. [Google Scholar] [CrossRef]
- Zhou, Y.; Song, R.; Zhang, Z.; Lu, X.; Zeng, Z.; Hu, C.; Liu, X.; Li, Y.; Hou, J.; Sun, Y.; et al. The development of plasma pseudotargeted GC-MS metabolic profiling and its application in bladder cancer. Anal. Bioanal. Chem. 2016, 408, 6741–6749. [Google Scholar] [CrossRef] [PubMed]
- Lemańska-Perek, A.; Lis-Kuberka, J.; Lepczyński, A.; Dratwa-Chałupnik, A.; Tupikowski, K.; Katnik-Prastowska, I.; Ożgo, M. Potential plasma biomarkers of bladder cancer identified by proteomic analysis: A pilot study. Adv. Clin. Exp. Med. 2018, 28, 339–346. [Google Scholar] [CrossRef]
- Gupta, A.; Bansal, N.; Mitash, N.; Kumar, D.; Kumar, M.; Sankhwar, S.N.; Mandhani, A.; Singh, U.P. NMR-derived targeted serum metabolic biomarkers appraisal of bladder cancer: A pre- and post-operative evaluation. J. Pharm. Biomed. Anal. 2020, 183, 113134. [Google Scholar] [CrossRef]
- Cantiello, F.; Russo, G.I.; Vartolomei, M.D.; Abu Farhan, A.R.; Terracciano, D.; Musi, G.; Lucarelli, G.; Di Stasi, S.M.; Hurle, R.; Serretta, V.; et al. Systemic Inflammatory Markers and Oncologic Outcomes in Patients with High-risk Non–muscle-invasive Urothelial Bladder Cancer. Eur. Urol. Oncol. 2018, 1, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Bansal, N.; Gupta, A.K.; Gupta, A.; Sankhwar, S.N.; Mahdi, A.A. Serum-based protein biomarkers of bladder cancer: A pre- and post-operative evaluation. J. Pharm. Biomed. Anal. 2016, 124, 22–25. [Google Scholar] [CrossRef]
- Ferdosi, S.; Ho, T.H.; Castle, E.P.; Stanton, M.L.; Borges, C.R. Behavior of blood plasma glycan features in bladder cancer. PLoS ONE 2018, 13, e0201208. [Google Scholar] [CrossRef]
- Hristova, V.A.; Chan, D.W. Cancer biomarker discovery and translation: Proteomics and beyond. Expert Rev. Proteom. 2019, 16, 93–103. [Google Scholar] [CrossRef]
- Sanli, O.; Dobruch, J.; Knowles, M.A.; Burger, M.; Alemozaffar, M.; Nielsen, M.E.; Lotan, Y. Bladder cancer. Nat. Rev. Dis. Prim. 2017, 3, 17022. [Google Scholar] [CrossRef] [PubMed]
- Meleady, P. Two-Dimensional Gel Electrophoresis and 2D-DIGE. Methods Mol. Biol. 2018, 1664, 3–14. [Google Scholar] [PubMed]
- Kondo, T. Cancer biomarker development and two-dimensional difference gel electrophoresis (2D-DIGE). Biochim. Biophys. Acta Proteins Proteom. 2019, 1867, 2–8. [Google Scholar] [CrossRef]
- Nedjadi, T.; Benabdelkamal, H.; Albarakati, N.; Masood, A.; Al-Sayyad, A.; Alfadda, A.A.; Alanazi, I.O.; Al-Ammari, A.; Al-Maghrabi, J. Circulating proteomic signature for detection of biomarkers in bladder cancer patients. Sci. Rep. 2020, 10, 10999–11008. [Google Scholar] [CrossRef]
- Alfadda, A.A.; Benabdelkamel, H.; Masood, A.; Moustafa, A.; Sallam, R.; Bassas, A.; Duncan, M. Proteomic analysis of mature adipo cytes from obese patients in relation to aging. Exp. Gerontol. 2013, 48, 1196–1203. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Li, K.; Yin, X.; Li, H.; Li, Y.; Zhang, Q.; Wang, H.; Qiu, Y. Lower Expression of Gelsolin in Colon Cancer and Its Diagnostic Value in Colon Cancer Patients. J. Cancer 2019, 10, 1288–1296. [Google Scholar] [CrossRef] [PubMed]
- Jiang, G.-Y.; Zhang, Y.; Zhang, X.-P.; Lin, X.-Y.; Yu, J.-H.; Wang, E.-H. Inversin correlates with the malignant phenotype of non–small cell lung cancer and promotes the invasiveness of lung cancer cells. Tumor Biol. 2017, 39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, X.; Hong, S.; Huang, J.; Chen, Y.; Chen, Y.; Wu, Z. Plasma apolipoprotein A1 levels at diagnosis are independent prognostic factors in invasive ductal breast cancer. Discov. Med. 2017, 23, 247–258. [Google Scholar] [PubMed]
- Li, G.H.; Arora, P.D.; Chen, Y.; McCulloch, C.A.; Liu, P. Multifunctional roles of gelsolin in health and diseases. Med. Res. Rev. 2012, 32, 999–1025. [Google Scholar] [CrossRef] [PubMed]
- Ong, M.S.; Deng, S.; Halim, C.E.; Cai, W.; Tan, T.Z.; Huang, R.Y.-J.; Sethi, G.; Hooi, S.C.; Kumar, A.P.; Yap, C.T. Cytoskeletal Proteins in Cancer and Intracellular Stress: A Therapeutic Perspective. Cancers 2020, 12, 238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, C.S.; Furuya, F.; Ying, H.; Kato, Y.; Hanover, J.A.; Cheng, S.Y. Gelsolin: A novel thyroid hormone receptor-beta interacting protein that modulates tumor progression in a mouse model of follicular thyroid cancer. Endocrinology 2007, 148, 1306–1312. [Google Scholar] [CrossRef]
- Noske, A.; Denkert, C.; Schober, H.; Sers, C.; Zhumabayeva, B.; Weichert, W.; Dietel, M.; Wiechen, K. Loss of Gelsolin expression in human ovarian carcinomas. Eur. J. Cancer 2005, 41, 461–469. [Google Scholar] [CrossRef] [PubMed]
- Ni, X.G.; Zhou, L.; Wang, G.Q.; Liu, S.M.; Bai, X.F.; Liu, F. The ubiquitin-proteasome pathway mediates gelsolin protein downregulation in pancreatic cancer. Mol. Med. 2008, 14, 582–589. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Asch, H.L.; Ying, A.; Asch, B.B. Molecular Mechanism of Transcriptional Repression of Gelsolin in Human Breast Cancer Cells. Exp. Cell Res. 2002, 276, 328–336. [Google Scholar] [CrossRef]
- Chiu, C.-T.; Wang, P.-W.; Asare-Werehene, M.; Tsang, B.K.; Shieh, D.-B. Circulating Plasma Gelsolin: A Predictor of Favorable Clinical Outcomes in Head and Neck Cancer and Sensitive Biomarker for Early Disease Diagnosis Combined with Soluble Fas Ligand. Cancers 2020, 12, 1569. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.-L.; Wang, C.C.N.; Cai, J.-H.; Chou, C.-Y.; Lin, Y.-C.; Hung, C.-C. Identification of GSN and LAMC2 as Key Prognostic Genes of Bladder Cancer by Integrated Bioinformatics Analysis. Cancers 2020, 12, 1809. [Google Scholar] [CrossRef] [PubMed]
- Piktel, E.; Levental, I.; Durnaś, B.; Janmey, P.A.; Bucki, R. Plasma Gelsolin: Indicator of Inflammation and Its Potential as a Diagnostic Tool and Therapeutic Target. Int. J. Mol. Sci. 2018, 19, 2516. [Google Scholar] [CrossRef] [Green Version]
- Cohen, T.S.; Bucki, R.; Byfield, F.J.; Ciccarelli, N.J.; Rosenberg, B.; DiNubile, M.J. Therapeutic potential of plasma gelsolin administration in a rat model of sepsis. Cytokine 2011, 54, 235–238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, S.; Chen, R.; Brand, R.E.; Hawley, S.; Tamura, Y.; Gafken, P.R.; Milless, B.P.; Goodlett, D.R.; Rush, J.; Brentnall, T.A. Multiplex Targeted Proteomic Assay for Biomarker Detection in Plasma: A Pancreatic Cancer Biomarker Case Study. J. Proteome Res. 2012, 11, 1937–1948. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scumaci, D.; Tammè, L.; Fiumara, C.V.; Pappaianni, G.; Concolino, A.; Leone, E.; Faniello, M.C.; Quaresima, B.; Ricevuto, E.; Costanzo, F.S.; et al. Plasma Proteomic Profiling in Hereditary Breast Cancer Reveals a BRCA1-Specific Signature: Diagnostic and Functional Implications. PLoS ONE 2015, 10, e0129762. [Google Scholar] [CrossRef] [Green Version]
- Nürnberger, J.; Kribben, A.; Saez, A.O.; Heusch, G.; Philipp, T.; Phillips, C.L. The Invs gene encodes a microtubule-associated protein. J. Am. Soc. Nephrol. 2004, 15, 1700–1710. [Google Scholar] [CrossRef] [Green Version]
- Werner, M.E.; Ward, H.; Phillips, C.; Miller, C.; Gattone, V.H.; Bacallao, R. Inversin modulates the cortical actin network during mitosis. Am. J. Physiol. Physiol. 2013, 305, C36–C47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, B.; Zhuang, T.; Lin, Q.; Yang, B.; Xu, X.; Xin, G.; Zhu, S.; Wang, G.; Yu, B.; Zhang, T.; et al. Patched1–ArhGAP36–PKA–Inversin axis determines the ciliary translocation of Smoothened for Sonic Hedgehog pathway activation. Proc. Natl. Acad. Sci. USA 2019, 116, 874–879. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Srivastava, S.; Molinari, E.; Raman, S.; Sayer, J.A. Many Genes—One Disease? Genetics of Nephronophthisis (NPHP) and NPHP-Associated Disorders. Front. Pediatr. 2018, 5, 287. [Google Scholar] [CrossRef]
- Pisamai, S.; Roytrakul, S.; Phaonakrop, N.; Jaresitthikunchai, J.; Suriyaphol, G. Proteomic analysis of canine oral tumor tissues using MALDI-TOF mass spectrometry and in-gel digestion coupled with mass spectrometry (GeLC MS/MS) approaches. PLoS ONE 2018, 13, e0200619. [Google Scholar] [CrossRef] [PubMed]
- Suizu, F.; Hirata, N.; Kimura, K.; Edamura, T.; Tanaka, T.; Ishigaki, S.; Donia, T.; Noguchi, H.; Iwanaga, T.; Noguchi, M. Phosphorylation-dependent Akt–Inversin interaction at the basal body of primary cilia. EMBO J. 2016, 35, 1346–1363. [Google Scholar] [CrossRef]
- Mergen, M.; Engel, C.; Müller, B.; Follo, M.; Schäfer, T.; Jung, M.; Walz, G. The nephronophthisis gene product NPHP2/Inversin interacts with Aurora A and interferes with HDAC6-mediated cilia disassembly. Nephrol. Dial. Transplant. 2013, 28, 2744–2753. [Google Scholar] [CrossRef] [Green Version]
- Mangaraj, M.; Nanda, R.; Panda, S. Apolipoprotein A-I: A Molecule of Diverse Function. Indian J. Clin. Biochem. 2016, 31, 253–259. [Google Scholar] [CrossRef] [Green Version]
- Peng, M.; Zhang, Q.; Cheng, Y.; Fu, S.; Yang, H.; Guo, X.; Zhang, J.; Wang, L.; Zhang, L.; Xue, Z.; et al. Apolipoprotein A-I mimetic peptide 4F suppresses tumor-associated macrophages and pancreatic cancer progression. Oncotarget 2017, 8, 99693–99706. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chattopadhyay, A.; Yang, X.; Mukherjee, P.; Sulaiman, D.; Fogelman, H.R.; Grijalva, V.; Dubinett, S.; Wasler, T.C.; Paul, M.; Salehi-Rad, R.; et al. Treating the Intestine with Oral ApoA-I Mimetic Tg6F Reduces Tumor Burden in Mouse Models of Metastatic Lung Cancer. Sci. Rep. 2018, 8, 1–12. [Google Scholar]
- Wu, J.; Zhang, C.; Zhang, G.; Wang, Y.; Zhang, Z.; Su, W.; Lyu, J. Association between Pretreatment Serum Apolipoprotein A1 and Prognosis of Solid Tumors in Chinese Population: A Systematic Review and Meta-Analysis. Cell. Physiol. Biochem. 2018, 51, 575–588. [Google Scholar] [CrossRef] [PubMed]
- Sirniö, P.; Väyrynen, J.; Klintrup, K.; Mäkelä, J.; Mäkinen, M.J.; Karttunen, T.J.; Tuomisto, A. Decreased serum apolipoprotein A1 levels are associated with poor survival and systemic inflammatory response in colorectal cancer. Sci. Rep. 2017, 7, 5374. [Google Scholar] [CrossRef] [PubMed]
- Georgila, K.; Vyrla, D.; Drakos, E. Apolipoprotein A-I (ApoA-I), Immunity, Inflammation and Cancer. Cancers 2019, 11, 1097. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Li, C.; Wu, H.; Zhang, T.; Wang, J.; Wang, S.; Chang, J. Identification of Apo-A1 as a biomarker for early diagnosis of bladder transitional cell carcinoma. Proteome Sci. 2011, 9, 21. [Google Scholar] [CrossRef] [Green Version]
- Shang, Z.; Wang, J.; Wang, X.; Yan, H.; Cui, B.; Jia, C.; Wang, Q.; Cui, X.; Li, J.; Ou, T. Preoperative serum apolipoprotein A-I levels predict long-term survival in non-muscle-invasive bladder cancer patients. Cancer Manag. Res. 2018, 10, 1177–1190. [Google Scholar] [CrossRef] [Green Version]
- Zamanian-Daryoush, M.; DiDonato, J.A. Apolipoprotein A-I and Cancer. Front. Pharmacol. 2015, 6, 265–274. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Characteristics | MIBC Patients | Healthy Controls | |
|---|---|---|---|
| Number of subjects | 29 | 22 | |
| Age | (median/range) | 67/(51–91 y) | 51/(43–60 y) |
| Weight | (median/range) | 67/(51–94 y) | 74/(59–88 y) |
| Gender | Male | 92.0% | 83.3% |
| Female | 8.0% | 16.7% | |
| Stage | pT1 | 0% | N/A |
| pT2 | 96.5% | ||
| pT3 | 0% | ||
| pT4 | 3.5% | ||
| Tumor grade | High grade | 100.0% | N/A |
| Metastasis | Lymph node | 3.5% | N/A |
| Vascular Invasion | No | 70.0% | N/A |
| Yes | 30.0% | ||
| Status | Alive | 100.0% | N/A |
| Dead | 0.0% | ||
| Recurrence | No | 33.3% | N/A |
| Yes | 66.7% | ||
| Smoking | No | 100.0% | N/A |
| Spot No | Accession No | Protein Name | MASCOT ID | Pi a | MW b | Cov% c | Score d | p-Value (ANOVA) | Fold Change | Expression HG/Control |
|---|---|---|---|---|---|---|---|---|---|---|
| 350 | Q9Y283 | Inversin | INVS | 9.43 | 118837 | 61 | 58 | 0.002 | 1.5 | Down |
| 833 | P02743 | Serum amyloid P-component | SAMP | 6.10 | 25485 | 30 | 60 | 0.028 | 2 | Down |
| 889 | Q9H1K6 | Mesoderm development candidate 1 | MESD1 | 8.53 | 38533 | 34 | 58 | 0.030 | 2.8 | Down |
| 859 | P02647 | Apolipoprotein A-I | APOA1 | 5.56 | 30759 | 49 | 129 | 0.033 | 1.9 | Down |
| 866 | P02647 | Apolipoprotein A-I | APOA1 | 5.56 | 30759 | 56 | 145 | 0.044 | 1.7 | Down |
| 457 | P20020 | Plasma membrane calcium-transporting ATPase 1 | AT2B1 | 5.73 | 139637 | 19 | 66 | 0.030 | 1.7 | Up |
| 316 | P06396 | Gelsolin | GELS | 5.90 | 86043 | 51 | 62 | 0.052 | 1.5 | Down |
| 252 | P00747 | Plasminogen | PLMN | 7.04 | 93247 | 40 | 149 | 0.0259 | 1.6 | Down |
| 974 | P02647 | Apolipoprotein A-I | APOA1 | 5.56 | 30759 | 59 | 160 | 0.038 | 1.8 | Down |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nedjadi, T.; Albarakati, N.; Benabdelkamel, H.; Masood, A.; Alfadda, A.A.; Al-Maghrabi, J. Proteomic Profiling of Plasma-Derived Biomarkers in Patients with Bladder Cancer: A Step towards Clinical Translation. Life 2021, 11, 1294. https://doi.org/10.3390/life11121294
Nedjadi T, Albarakati N, Benabdelkamel H, Masood A, Alfadda AA, Al-Maghrabi J. Proteomic Profiling of Plasma-Derived Biomarkers in Patients with Bladder Cancer: A Step towards Clinical Translation. Life. 2021; 11(12):1294. https://doi.org/10.3390/life11121294
Chicago/Turabian StyleNedjadi, Taoufik, Nada Albarakati, Hicham Benabdelkamel, Afshan Masood, Assim A. Alfadda, and Jaudah Al-Maghrabi. 2021. "Proteomic Profiling of Plasma-Derived Biomarkers in Patients with Bladder Cancer: A Step towards Clinical Translation" Life 11, no. 12: 1294. https://doi.org/10.3390/life11121294
APA StyleNedjadi, T., Albarakati, N., Benabdelkamel, H., Masood, A., Alfadda, A. A., & Al-Maghrabi, J. (2021). Proteomic Profiling of Plasma-Derived Biomarkers in Patients with Bladder Cancer: A Step towards Clinical Translation. Life, 11(12), 1294. https://doi.org/10.3390/life11121294






