Immunity in Space: Prokaryote Adaptations and Immune Response in Microgravity
Abstract
1. Introduction
2. Microgravity Simulation and Applications
2.1. Microgravity Analogues—The Common Devices
2.2. Microgravity Analogues—History of Clinostats and Alternative Methods
3. Prokaryotic Responses to Microgravity
3.1. Cell Viability and Diversity
3.2. Overview of Previous Studies
3.3. Transcriptomic Changes
3.4. Antibiotic Resistance
3.5. Archaeal Responses to Microgravity
4. Immune Cell Responses to Microgravity
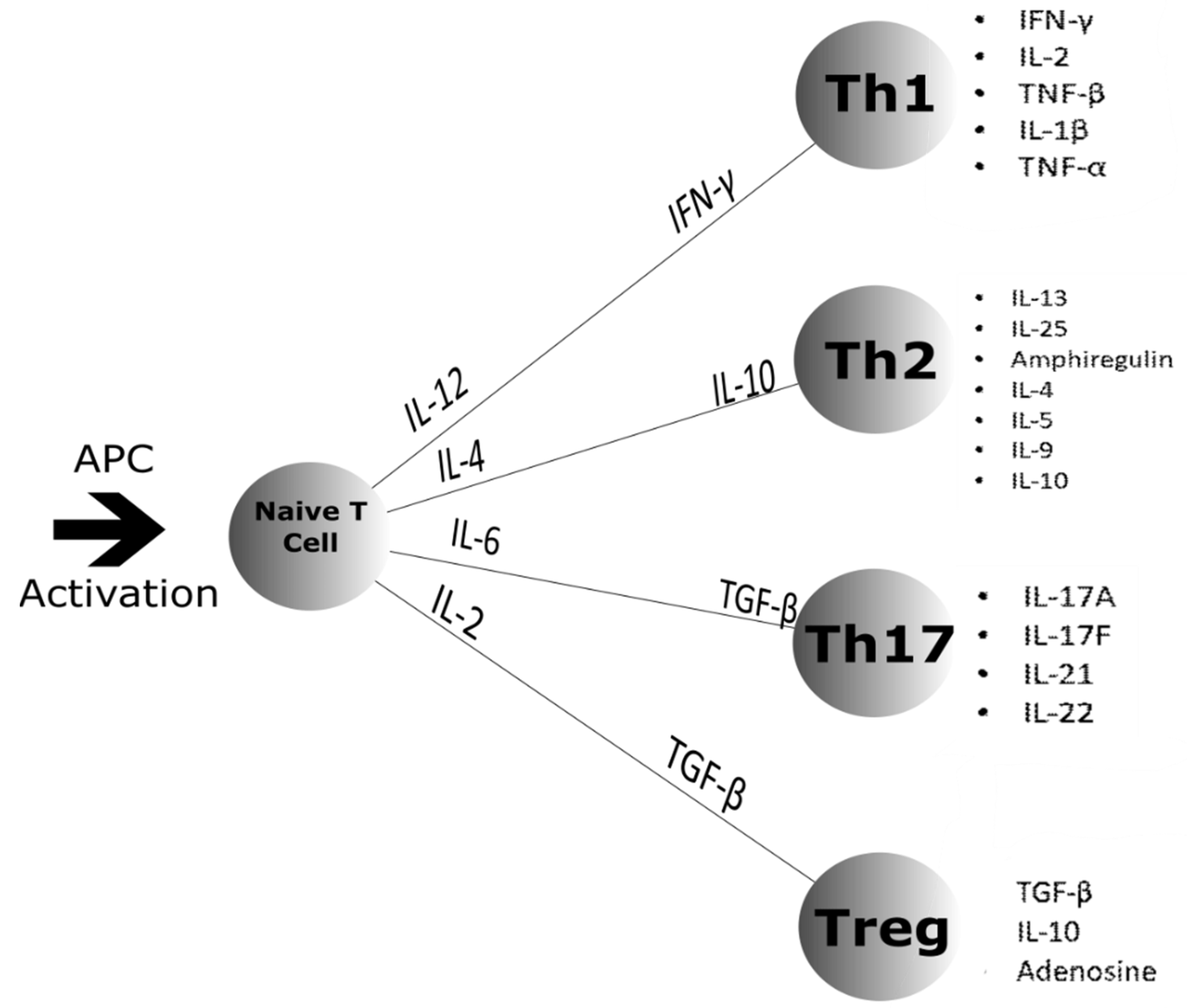
4.1. Cell Differentiation
4.2. Pathogen Recognition
4.3. Cell–Cell Interactions
4.4. Cytokines
5. Concluding Remarks
Author Contributions
Funding
Conflicts of Interest
References
- Crucian, B.E.; Chouker, A.; Simpson, R.J.; Mehta, S.; Marshall, G.; Smith, S.M.; Zwart, S.R.; Heer, M.; Ponomarev, S.; Whitmire, A.; et al. Immune system dysregulation during spaceflight: Potential countermeasures for deep space exploration missions. Front. Immunol. 2018, 9, 1437. [Google Scholar] [CrossRef] [PubMed]
- Crucian, B.; Stowe, R.P.; Mehta, S.; Quiriarte, H.; Pierson, D.; Sams, C. Alterations in adaptive immunity persist during long-duration spaceflight. NPJ Microgravity 2015, 1, 15013. [Google Scholar] [CrossRef] [PubMed]
- Kimzey, S.L.; Johnston, R.S.; Dietlein, L.F. Biomedical Results from Skylab; NASA Technical Reports Server; NASA: Washington, DC, USA, 1977. [Google Scholar]
- Taylor, P.W. Impact of space flight on bacterial virulence and antibiotic susceptibility. Infect. Drug Resist. 2015, 8, 249–262. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, R.; Torres, M.; Clemens, R.; Hateley, S.; Hosamani, R.; Wade, W.; Bhattacharya, S. Spaceflight and simulated microgravity conditions increase virulence of Serratia marcescens in the Drosophila melanogaster infection model. NPJ Microgravity 2020, 6, 1–9. [Google Scholar] [CrossRef]
- Rosenzweig, J.A.; Abogunde, O.; Thomas, K.; Lawal, A.; Nguyen, Y.-U.; Sodipe, A.; Jejelowo, O. Spaceflight and modeled microgravity effects on microbial growth and virulence. Appl. Microbiol. Biotechnol. 2009, 85, 885–891. [Google Scholar] [CrossRef]
- Wherry, E.J. T cell exhaustion. Nat. Immunol. 2011, 12, 492–499. [Google Scholar] [CrossRef]
- Salam, N.; Rane, S.; Das, R.; Faulkner, M.; Gund, R.; Kandpal, U.; Lewis, V.; Mattoo, H.; Prabhu, S.; Ranganathan, V.; et al. T cell ageing: Effects of age on development, survival & function. Indian J. Med. Res. 2013, 138, 595–608. [Google Scholar]
- Garrett-Bakelman, F.E.; Darshi, M.; Green, S.J.; Gur, R.C.; Lin, L.; Macias, B.R.; McKenna, M.J.; Meydan, C.; Mishra, T.; Nasrini, J.; et al. The NASA twins study: A multidimensional analysis of a year-long human spaceflight. Science 2019, 364, eaau8650. [Google Scholar]
- Gueguinou, N.; Huin-Schohn, C.; Bascove, M.; Bueb, J.L.; Tschirhart, E.; Legrand-Frossi, C.; Frippiat, J.P. Could spaceflight-associated immune system weakening preclude the expansion of human presence beyond Earth’s orbit? J. Leukoc. Biol. 2009, 86, 1027–1038. [Google Scholar] [CrossRef]
- Foster, J.S.; Wheeler, R.M.; Pamphile, R. Host-microbe interactions in microgravity: Assessment and implications. Life 2014, 4, 250–266. [Google Scholar] [CrossRef]
- Tian, Y.; Ma, X.; Yang, C.; Su, P.; Yin, C.; Qian, A.-R. The impact of oxidative stress on the bone system in response to the space special environment. Int. J. Mol. Sci. 2017, 18, 2132. [Google Scholar] [CrossRef] [PubMed]
- Synthecon. RCCS 3D Cell Culture Systems. 2020. Available online: https://synthecon.com/pages/rotary_cell_culture_systems_13.asp (accessed on 1 November 2020).
- Higginson, E.E.; Galen, J.E.; Levine, M.M.; Tennant, S.M. Microgravity as a biological tool to examine host–pathogen interactions and to guide development of therapeutics and preventatives that target pathogenic bacteria. Pathog. Dis. 2016, 74, ftw095. [Google Scholar] [CrossRef] [PubMed]
- Pardo, S.J.; Patel, M.J.; Sykes, M.C.; Platt, M.O.; Boyd, N.L.; Sorescu, G.P.; Xu, M.; Van Loon, J.J.; Wang, M.D.; Jo, H. Simulated microgravity using the Random Positioning Machine inhibits differentiation and alters gene expression profiles of 2T3 preosteoblasts. Am. J. Physiol. Physiol. 2005, 288, C1211–C1221. [Google Scholar] [CrossRef]
- Wuest, S.L.; Stern, P.; Casartelli, E.; Egli, M. Fluid dynamics appearing during simulated microgravity using random positioning machines. PLoS ONE 2017, 12, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Kiss, J.Z.; Wolverton, C.; Wyatt, S.E.; Hasenstein, K.H.; Van Loon, J.J. Comparison of microgravity analogs to spaceflight in studies of plant growth and development. Front. Plant Sci. 2019, 10, 1577. [Google Scholar] [CrossRef] [PubMed]
- Herranz, R.; Anken, R.; Boonstra, J.; Braun, M.; Christianen, P.C.; De Geest, M.; Hauslage, J.; Hilbig, R.; Hill, R.J.; Lebert, M.; et al. Ground-based facilities for simulation of microgravity: Organism-specific recommendations for their use, and recommended terminology. Astrobiology 2013, 13, 1–17. [Google Scholar] [CrossRef]
- Briegleb, W. Some qualitative and quantitative aspects of the fast-rotating clinostat as a research tool. ASGSB Bull. Publ. Am. Soc. Gravitational Space Biol. 1992, 5, 23–30. [Google Scholar]
- Herranz, R.; Larkin, O.J.; Dijkstra, C.E.; Hill, R.J.A.; Anthony, P.; Davey, M.R.; Eaves, L.; Van Loon, J.J.; Medina, F.J.; Marco, R. Microgravity simulation by diamagnetic levitation: Effects of a strong gradient magnetic field on the transcriptional profile of Drosophila melanogaster. BMC Genom. 2012, 13, 52. [Google Scholar] [CrossRef]
- Dijkstra, C.; Larkin, O.; Anthony, P.; Davey, M.; Eaves, L.; Rees, C.; Hill, R. Diamagnetic levitation enhances growth of liquid bacterial cultures by increasing oxygen availability. Nat. Précéd. 2010, 8, 334–344. [Google Scholar] [CrossRef]
- Moser, D.; Sun, S.J.; Li, N.; Biere, K.; Hoerl, M.; Matzel, S.; Feuerecker, M.; Buchheim, J.-I.; Strewe, C.; Thiel, C.S.; et al. Cells´ flow and immune cell priming under alternating g-forces in parabolic flight. Sci. Rep. 2019, 9, 1–11. [Google Scholar] [CrossRef]
- Stervbo, U.; Roch, T.; Kornprobst, T.; Sawitzki, B.; Grütz, G.; Wilhelm, A.; Lacombe, F.; Allou, K.; Kaymer, M.; Pacheco, A.; et al. Gravitational stress during parabolic flights reduces the number of circulating innate and adaptive leukocyte subsets in human blood. PLoS ONE 2018, 13, e0206272. [Google Scholar] [CrossRef] [PubMed]
- Hammond, T.G.; Benes, E.; O’Reilly, K.C.; Wolf, D.A.; Linnehan, R.M.; Taher, A.; Kaysen, J.H.; Allen, P.L.; Goodwin, T.J. Mechanical culture conditions effect gene expression: Gravity-induced changes on the space shuttle. Physiol. Genom. 2000, 3, 163–173. [Google Scholar] [CrossRef]
- Altermann, E. Tracing lifestyle adaptation in prokaryotic genomes. Front. Microbiol. 2012, 3, 48. [Google Scholar] [CrossRef] [PubMed]
- Thursby, E.; Juge, N. Introduction to the human gut microbiota. Biochem. J. 2017, 474, 1823–1836. [Google Scholar] [CrossRef] [PubMed]
- Cogen, A.L.; Nizet, V.; Gallo, R.L. Skin microbiota: A source of disease or defence? Br. J. Dermatol. 2008, 158, 442–455. [Google Scholar] [CrossRef]
- Yamaguchi, N.; Roberts, M.; Castro, S.; Oubre, C.; Makimura, K.; Leys, N.; Grohmann, E.; Sugita, T.; Ichijo, T.; Nasu, M. Microbial monitoring of crewed habitats in space—Current status and future perspectives. Microbes Environ. 2014, 29, 250–260. [Google Scholar] [CrossRef]
- Novikova, N.; De Boever, P.; Poddubko, S.; Deshevaya, E.; Polikarpov, N.; Rakova, N.; Coninx, I.; Mergeay, M. Survey of environmental biocontamination on board the International Space Station. Res. Microbiol. 2006, 157, 5–12. [Google Scholar] [CrossRef]
- Fergione, S. Effects of simulated microgravity on the microbial physiology of Ralstonia picketti isolates from the International Space Station. Biochem. Mol. Biol. 2016, 15, 35–41. [Google Scholar]
- Abshire, C.F.; Prasai, K.; Soto, I.; Shi, R.; Concha, M.; Baddoo, M.; Flemington, E.K.; Ennis, D.G.; Scott, R.S.; Harrison, L. Exposure of Mycobacterium marinum to low-shear modeled microgravity: Effect on growth, the transcriptome and survival under stress. NPJ Microgravity 2016, 2, 16038. [Google Scholar] [CrossRef]
- Wilson, J.W.; Ott, C.M.; Ramamurthy, R.; Porwollik, S.; McClelland, M.; Pierson, D.L.; Nickerson, C.A. Low-Shear modeled microgravity alters the Salmonella enterica serovar typhimurium stress response in an RpoS-independent manner. Appl. Environ. Microbiol. 2002, 68, 5408–5416. [Google Scholar] [CrossRef]
- Lynch, S.V.; Mukundakrishnan, K.; Benoit, M.R.; Ayyaswamy, P.S.; Matin, A. Escherichia coli biofilms formed under low-shear modeled microgravity in a ground-based system. Appl. Environ. Microbiol. 2006, 72, 7701–7710. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Wang, Y.; He, J.; Li, P.; Jin, R.; Wang, K.; Xu, X.; Hao, J.; Zhang, Y.; Liu, H.; et al. Intestinal microbiota contributes to colonic epithelial changes in simulated microgravity mouse model. FASEB J. 2017, 31, 3695–3709. [Google Scholar] [CrossRef] [PubMed]
- Soni, A.; O’Sullivan, L.; Quick, L.N.; Ott, C.M.; Nickerson, C.A.; Wilson, J.W. Conservation of the low-shear modeled microgravity response in enterobacteriaceae and analysis of the trp genes in this response. Open Microbiol. J. 2014, 8, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Wilson, J.W.; Ramamurthy, R.; Porwollik, S.; McClelland, M.; Hammond, T.; Allen, P.; Ott, C.M.; Pierson, D.L.; Nickerson, C.A. Microarray analysis identifies Salmonella genes belonging to the low-shear modeled microgravity regulon. Proc. Natl. Acad. Sci. USA 2002, 99, 13807–13812. [Google Scholar] [CrossRef]
- Orsini, S.S.; Lewis, A.M.; Rice, K.C. Investigation of simulated microgravity effects on Streptococcus mutans physiology and global gene expression. NPJ Microgravity 2017, 3, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Xu, X.; Chen, J.; Zhou, X.; Cheng, L.; Li, M.; Li, J.; Wang, R.; Jia, W.; Li, Y.Q. Effects of simulated microgravity on Streptococcus mutans physiology and biofilm structure. FEMS Microbiol. Lett. 2014, 359, 94–101. [Google Scholar] [CrossRef]
- Shao, D.; Yao, L.; Riaz, M.S.; Zhu, J.; Shi, J.; Jin, M.; Huang, Q.; Yang, H. Simulated microgravity affects some biological characteristics of Lactobacillus acidophilus. Appl. Microbiol. Biotechnol. 2016, 101, 3439–3449. [Google Scholar] [CrossRef]
- Morrison, M.D.; Fajardo-Cavazos, P.; Nicholson, W.L. Comparison of Bacillus subtilis transcriptome profiles from two separate missions to the International Space Station. NPJ Microgravity 2019, 5, 1–11. [Google Scholar] [CrossRef]
- Kim, W.; Tengra, F.K.; Shong, J.; Marchand, N.; Chan, H.K.; Young, Z.; Pangule, R.C.; Parra, M.; Dordick, J.S.; Plawsky, J.L.; et al. Effect of spaceflight on Pseudomonas aeruginosa final cell density is modulated by nutrient and oxygen availability. BMC Microbiol. 2013, 13, 241. [Google Scholar] [CrossRef]
- Kim, W.; Tengra, F.K.; Young, Z.; Shong, J.; Marchand, N.; Chan, H.K.; Pangule, R.C.; Parra, M.; Dordick, J.S.; Plawsky, J.L.; et al. Spaceflight promotes biofilm formation by Pseudomonas aeruginosa. PLoS ONE 2013, 8, e62437. [Google Scholar] [CrossRef]
- Crabbé, A.; Schurr, M.J.; Monsieurs, P.; Morici, L.; Schurr, J.; Wilson, J.W.; Ott, C.M.; Tsaprailis, G.; Pierson, D.L.; Stefanyshyn-Piper, H.; et al. Transcriptional and proteomic responses of Pseudomonas aeruginosaPAO1 to spaceflight conditions involve Hfq regulation and reveal a role for oxygen. Appl. Environ. Microbiol. 2010, 77, 1221–1230. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Yan, Y.; Rong, D.; Wang, J.; Wang, H.; Liu, Z.; Wang, J.; Yang, R.; Han, Y. Increased biofilm formation ability in Klebsiella pneumoniae after short-term exposure to a simulated microgravity environment. Microbiologyopen 2016, 5, 793–801. [Google Scholar] [CrossRef] [PubMed]
- Duscher, A.A.; Conesa, A.; Bishop, M.; Vroom, M.M.; Zubizarreta, S.D.; Foster, J.S. Transcriptional profiling of the mutualistic bacterium Vibrio fischeri and an hfq mutant under modeled microgravity. NPJ Microgravity 2018, 4, 25. [Google Scholar] [CrossRef] [PubMed]
- Rosado, H.; Doyle, M.; Hinds, J.; Taylor, P.W. Low-shear modelled microgravity alters expression of virulence determinants of Staphylococcus aureus. Acta Astronaut. 2010, 66, 408–413. [Google Scholar] [CrossRef]
- Mégroz, M.; Kleifeld, O.; Wright, A.; Powell, D.R.; Harrison, P.F.; Adler, B.; Harper, M.; Boyce, J.D. The RNA-binding chaperone Hfq is an important global regulator of gene expression in Pasteurella multocida and plays a crucial role in production of a number of virulence factors, including hyaluronic acid capsule. Infect. Immun. 2016, 84, 1361–1370. [Google Scholar] [CrossRef]
- Gottesman, S.; Storz, G. Bacterial small RNA regulators: Versatile roles and rapidly evolving variations. Cold Spring Harb. Perspect. Biol. 2010, 3, a003798. [Google Scholar] [CrossRef]
- Wagner, E.G.H.; Romby, P. Small RNAs in bacteria and archaea: Who they are, what they do, and how they do it. Adv. Genet. 2015, 90, 133–208. [Google Scholar]
- Troxell, B.; Hassan, H.M. Transcriptional regulation by Ferric Uptake Regulator (Fur) in pathogenic bacteria. Front. Cell. Infect. Microbiol. 2013, 3, 59. [Google Scholar] [CrossRef]
- Arunasri, K.; Adil, M.; Charan, K.V.; Suvro, C.; Reddy, S.H.; Shivaji, S. Effect of simulated microgravity on E. coli K12 MG1655 growth and gene expression. PLoS ONE 2013, 8, e57860. [Google Scholar] [CrossRef]
- Hengge-Aronis, R. Signal transduction and regulatory mechanisms involved in control of the sigma(S) (RpoS) subunit of RNA polymerase. Microbiol. Mol. Biol. Rev. 2002, 66, 373–395. [Google Scholar] [CrossRef]
- Repoila, F.; Gottesman, S. Signal transduction cascade for regulation of RpoS: Temperature regulation of DsrA. J. Bacteriol. 2001, 183, 2591–2596. [Google Scholar] [CrossRef] [PubMed]
- Mortazavi, S.M.J. Acquired antibiotic resistance in escherichia coli exposed to simulated microgravity: Possible role of other space stressors and adaptive responses. mBio 2019, 10, e00165-19. [Google Scholar] [CrossRef] [PubMed]
- AIP. Adaptive response of bacteria: Multiple hurdles, cross-tolerance and tools to illustrate underlying mechanisms. AIP Conf. Proc. 2015, 1642, 1. [Google Scholar]
- Mortazavi, S.M.J.; Zarei, S.; Taheri, M.; Tajbaksh, S.; Mortazavi, S.A.; Ranjbar, S.; Momeni, F.; Masoomi, S.; Ansari, L.; Movahedi, M.M.; et al. Sensitivity to Antibiotics of bacteria exposed to gamma radiation emitted from hot soils of the high background radiation areas of Ramsar, Northern Iran. Int. J. Occup. Environ. Med. 2017, 8, 80–84. [Google Scholar] [CrossRef] [PubMed]
- Taheri, M.; Mortazavi, S.M.J.; Moradi, M.; Mansouri, S.; Hatam, G.R.; Nouri, F. Evaluation of the effect of radiofrequency radiation emitted from wi-fi router and mobile phone simulator on the antibacterial susceptibility of pathogenic bacteria listeria monocytogenes and Escherichia coli. Dose-Response 2017, 15, 1559325816688527. [Google Scholar] [CrossRef]
- Schiwon, K.; Arends, K.; Rogowski, K.M.; Fürch, S.; Prescha, K.; Sakinc, T.; Van Houdt, R.; Werner, G.; Grohmann, E. Comparison of antibiotic resistance, biofilm formation and conjugative transfer of staphylococcus and enterococcus isolates from international space station and antarctic research station concordia. Microb. Ecol. 2013, 65, 638–651. [Google Scholar] [CrossRef] [PubMed]
- Tirumalai, M.R.; Karouia, F.; Tran, Q.; Stepanov, V.G.; Bruce, R.J.; Ott, C.M.; Pierson, D.L.; Fox, G.E. Evaluation of acquired antibiotic resistance in escherichia coliexposed to long-term low-shear modeled microgravity and background antibiotic exposure. mBio 2019, 10, e02637-18. [Google Scholar] [CrossRef]
- Casadevall, A. The pathogenic potential of a microbe. mSphere 2017, 2, e00015-17. [Google Scholar] [CrossRef]
- Lawal, A.; Kirtley, M.L.; van Lier, C.J.; Erova, T.E.; Kozlova, E.V.; Sha, J.; Chopra, A.K.; Rosenzweig, J.A. The effects of modeled microgravity on growth kinetics, antibiotic susceptibility, cold growth, and the virulence potential of a Yersinia pestis ymoA-deficient mutant and its isogenic parental strain. Astrobiology 2013, 13, 821–832. [Google Scholar] [CrossRef]
- Rosenzweig, J.A.; Ahmed, S.; Eunson, J.; Chopra, A.K. Low-shear force associated with modeled microgravity and spaceflight does not similarly impact the virulence of notable bacterial pathogens. Appl. Microbiol. Biotechnol. 2014, 98, 8797–8807. [Google Scholar] [CrossRef]
- Cavicchioli, R.; Curmi, P.M.G.; Saunders, N.F.W.; Thomas, T. Pathogenic archaea: Do they exist? BioEssays 2003, 25, 1119–1128. [Google Scholar] [CrossRef] [PubMed]
- Dornmayr-Pfaffenhuemer, M.; Legat, A.; Schwimbersky, K.; Fendrihan, S.; Stan-Lotter, H. Responses of haloarchaea to simulated microgravity. Astrobiology 2011, 11, 199–205. [Google Scholar] [CrossRef]
- Thombre, R.; Shinde, V.; Dixit, J.; Jagtap, S.; Vidyasagar, P.B. Response of extreme haloarchaeon Haloarcula argentinensis RR10 to simulated microgravity in clinorotation. 3 Biotech 2017, 7, 30. [Google Scholar] [CrossRef] [PubMed]
- Fuchsman, C.A.; Collins, R.E.; Rocap, G.; Brazelton, W.J. Effect of the environment on horizontal gene transfer between bacteria and archaea. PeerJ 2017, 5, e3865. [Google Scholar] [CrossRef]
- Nelson, K.E.; Clayton, R.A.; Gill, S.R.; Gwinn, M.L.; Dodson, R.J.; Haft, D.H.; Hickey, E.K.; Peterson, J.D.; Nelson, W.C.; Ketchum, K.A.; et al. Evidence for lateral gene transfer between Archaea and bacteria from genome sequence of Thermotoga maritima. Nature 1999, 399, 323–329. [Google Scholar] [CrossRef] [PubMed]
- Burmeister, A.R. Horizontal gene transfer. Evol. Med. Public Health 2015, 2015, 193–194. [Google Scholar] [CrossRef]
- Hato, T.; Dagher, P.C. How the innate immune system senses trouble and causes trouble. Clin. J. Am. Soc. Nephrol. 2014, 10, 1459–1469. [Google Scholar] [CrossRef]
- Gardiner, C.M.; Mills, K.H.G. The cells that mediate innate immune memory and their functional significance in inflammatory and infectious diseases. Semin. Immunol. 2016, 28, 343–350. [Google Scholar] [CrossRef]
- Chaplin, D.D. Overview of the immune response. J. Allergy Clin. Immunol. 2010, 125, S3–S23. [Google Scholar] [CrossRef]
- Akiyama, T.; Horie, K.; Hinoi, E.; Hiraiwa, M.; Kato, A.; Maekawa, Y.; Takahashi, A.; Furukawa, S. How does spaceflight affect the acquired immune system? NPJ Microgravity 2020, 6, 1–7. [Google Scholar] [CrossRef]
- Sundaresan, A.; Mann, V.; Mehta, S.K.; Crucian, B.; Doursout, M.F.; Devakottai, S. Effects of microgravity and other space stressors in immunosuppression and viral reactivation with potential nervous system involvement. Neurol. India 2019, 67, S198–S203. [Google Scholar] [CrossRef] [PubMed]
- Mehta, S.K.; Laudenslager, M.L.; Stowe, R.P.; Crucian, B.E.; Feiveson, A.H.; Sams, C.F.; Pierson, D.L. Latent virus reactivation in astronauts on the international space station. NPJ Microgravity 2017, 3, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Mehta, S.; Laudenslager, M.; Stowe, R.; Crucian, B.; Sams, C.; Pierson, D. Multiple latent viruses reactivate in astronauts during Space Shuttle missions. Brain Behav. Immun. 2014, 41, 210–217. [Google Scholar] [CrossRef]
- Frippiat, J.P.; Crucian, E.B.; De Quervain, D.J.-F.; Grimm, D.; Montano, N.; Praun, S.; Roozendaal, B.; Schelling, G.; Thiel, M.; Ullrich, O.; et al. Towards human exploration of space: The THESEUS review series on immunology research priorities. NPJ Microgravity 2016, 2, 16040. [Google Scholar] [CrossRef] [PubMed]
- Horie, K.; Sasanuma, H.; Kudo, T.; Fujita, S.-I.; Miyauchi, M.; Miyao, T.; Seki, T.; Akiyama, N.; Takakura, Y.; Shimbo, M.; et al. Down-regulation of GATA1-dependent erythrocyte-related genes in the spleens of mice exposed to a space travel. Sci. Rep. 2019, 9, 7654. [Google Scholar] [CrossRef]
- Bigley, A.B.; Agha, N.H.; Baker, F.L.; Spielmann, G.; Kunz, H.E.; Mylabathula, P.L.; Rooney, B.V.; Laughlin, M.S.; Mehta, S.K.; Bigley, A.; et al. NK cell function is impaired during long-duration spaceflight. J. Appl. Physiol. 2019, 126, 842–853. [Google Scholar] [CrossRef]
- Assaf, A.M.; Al-Abbassi, R.; Al-Binni, M. Academic stress-induced changes in Th1- and Th2-cytokine response. Saudi Pharm. J. 2017, 25, 1237–1247. [Google Scholar] [CrossRef]
- Guglani, L.; Khader, S.A. Th17 cytokines in mucosal immunity and inflammation. Curr. Opin. HIV AIDS 2010, 5, 120–127. [Google Scholar] [CrossRef]
- Hoeppli, R.E.; Wu, D.; Cook, L.; Levings, M.K. The environment of regulatory T cell biology: Cytokines, metabolites, and the microbiome. Front. Immunol. 2015, 6, 61. [Google Scholar] [CrossRef]
- Konstantinova, I.V.; Rykova, M.P.; Lesnyak, A.T.; Antropova, E.A. Immune changes during long-duration missions. J. Leukoc. Biol. 1993, 54, 189–201. [Google Scholar] [CrossRef]
- Rykova, M.; Antropova, E.; Larina, I.; Morukov, B. Humoral and cellular immunity in cosmonauts after the ISS missions. Acta Astronaut. 2008, 63, 697–705. [Google Scholar] [CrossRef]
- Tascher, G.; Gerbaix, M.; Maes, P.; Chazarin, B.; Ghislin, S.; Antropova, E.; Vassilieva, G.; Ouzren-Zarhloul, N.; Gauquelin-Koch, G.; Vico, L.; et al. Analysis of femurs from mice embarked on board BION-M1 biosatellite reveals a decrease in immune cell development, including B cells, after 1 wk of recovery on Earth. FASEB J. 2018, 33, 3772–3783. [Google Scholar] [CrossRef] [PubMed]
- Ward, C.; Rettig, T.A.; Hlavacek, S.; Bye, B.A.; Pecaut, M.J.; Chapes, S.K. Effects of spaceflight on the immunoglobulin repertoire of unimmunized C57BL/6 mice. Life Sci. Space Res. 2018, 16, 63–75. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.; Xie, Y.; He, J.; Zhou, J.; Gao, Y.; Wei, W.; Ding, N.; Ma, H.; Xian, C.J.; Chen, K.; et al. Microgravity induces inhibition of osteoblastic differentiation and mineralization through abrogating primary cilia. Sci. Rep. 2017, 7, 1866. [Google Scholar] [CrossRef] [PubMed]
- Gershovich, P.M.; Gershovich, I.G.; Buravkova, L.B. The effects of simulated microgravity on the pattern of gene expression in human bone marrow mesenchymal stem cells under osteogenic differentiation. Fiziol. Cheloveka 2013, 39, 105–111. [Google Scholar]
- Low, E.K.; Brudvik, E.; Kuhlman, B.M.; Wilson, P.F.; Almeida-Porada, G.; Porada, C.D. Microgravity impairs DNA damage repair in human hematopoietic stem/progenitor cells and inhibits their differentiation into dendritic cells. Stem Cells Dev. 2018, 27, 1257–1267. [Google Scholar] [CrossRef]
- Shi, L.; Tian, H.; Wang, P.; Li, L.; Zhang, Z.; Zhang, J.; Zhao, Y. Spaceflight and simulated microgravity suppresses macrophage development via altered RAS/ERK/NFkappaB and metabolic pathways. Cell. Mol. Immunol. 2020. [Google Scholar] [CrossRef]
- Xue, L.; Li, Y.; Chen, J. Duration of simulated microgravity affects the differentiation of mesenchymal stem cells. Mol. Med. Rep. 2017, 15, 3011–3018. [Google Scholar] [CrossRef]
- Yuge, L.; Sasaki, A.; Kawahara, Y.; Wu, S.-L.; Matsumoto, M.; Manabe, T.; Kajiume, T.; Takeda, M.; Magaki, T.; Takahashi, T.; et al. Simulated microgravity maintains the undifferentiated state and enhances the neural repair potential of bone marrow stromal cells. Stem Cells Dev. 2011, 20, 893–900. [Google Scholar] [CrossRef]
- Sundaresan, A.; Pellis, N.R. Cellular and genetic adaptation in low-gravity environments. Ann. N. Y. Acad. Sci. 2009, 1161, 135–146. [Google Scholar] [CrossRef]
- Ward, N.E.; Pellis, N.R.; Risin, S.A.; Risin, D. Gene expression alterations in activated human T-cells induced by modeled microgravity. J. Cell. Biochem. 2006, 99, 1187–1202. [Google Scholar] [CrossRef] [PubMed]
- Thiel, C.S.; Paulsen, K.; Bradacs, G.; Lust, K.; Tauber, S.; Dumrese, C.; Hilliger, A.; Schoppmann, K.; Biskup, J.; Golz, N.; et al. Rapid alterations of cell cycle control proteins in human T lymphocytes in microgravity. Cell Commun. Signal. 2012, 10, 1. [Google Scholar] [CrossRef] [PubMed]
- Martinez, E.M.; Yoshida, M.C.; Candelario, T.L.T.; Hughes-Fulford, M. Spaceflight and simulated microgravity cause a significant reduction of key gene expression in early T-cell activation. Am. J. Physiol. Integr. Comp. Physiol. 2015, 308, R480–R488. [Google Scholar] [CrossRef] [PubMed]
- Bradley, J.H.; Stein, R.; Randolph, B.; Molina, E.; Arnold, J.P.; Gregg, R.K. T cell resistance to activation by dendritic cells requires long-term culture in simulated microgravity. Life Sci. Space Res. 2017, 15, 55–61. [Google Scholar] [CrossRef] [PubMed]
- Kyurkchiev, D. Secretion of immunoregulatory cytokines by mesenchymal stem cells. World J. Stem Cells 2014, 6, 552–570. [Google Scholar] [CrossRef]
- Imura, T.; Nakagawa, K.; Kawahara, Y.; Yuge, L. Stem cell culture in microgravity and its application in cell-based therapy. Stem Cells Dev. 2018, 27, 1298–1302. [Google Scholar] [CrossRef]
- Monticone, M.; Liu, Y.; Pujic, N.; Cancedda, R. Activation of nervous system development genes in bone marrow derived mesenchymal stem cells following spaceflight exposure. J. Cell. Biochem. 2010, 111, 442–452. [Google Scholar] [CrossRef]
- Chopra, V.; Fadl, A.A.; Sha, J.; Chopra, S.; Galindo, C.L.; Chopra, A.K. Alterations in the virulence potential of enteric pathogens and bacterial–host cell interactions under simulated microgravity conditions. J. Toxicol. Environ. Health Part A 2006, 69, 1345–1370. [Google Scholar] [CrossRef]
- Kaur, I.; Simons, E.R.; Castro, V.A.; Ott, C.M.; Pierson, D.L. Changes in monocyte functions of astronauts. Brain Behav. Immun. 2005, 19, 547–554. [Google Scholar] [CrossRef]
- Alexander, C.; Rietschel, E.T. Bacterial lipopolysaccharides and innate immunity. J. Endotoxin Res. 2001, 7, 167–202. [Google Scholar] [CrossRef]
- Kasper, C.A.; Sorg, I.; Scmutz, C.; Tschon, T.; Wischnewski, H.; Kim, M.L.; Arrieumerlou, C. Cell-cell propagation of NF-kappaB transcription factor and MAP kinase activation amplifies innate immunity against bacterial infection. Immunity 2010, 33, 804–816. [Google Scholar] [CrossRef] [PubMed]
- Zarubin, T.; Han, J. Activation and signaling of the p38 MAP kinase pathway. Cell Res. 2005, 15, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, Y.H.; Wu, T.T.; Huang, C.Y.; Hsieh, Y.S.; Hwang, J.M.; Liu, J.Y. p38 mitogen-activated protein kinase pathway is involved in protein kinase Calpha-regulated invasion in human hepatocellular carcinoma cells. Cancer Res. 2007, 67, 4320–4327. [Google Scholar] [CrossRef] [PubMed]
- Verhaar, A.; Hoekstra, E.; Tjon, S.; Utomo, W.; Deuring, J.; Bakker, E.R.M.; Muncan, V.; Peppelenbosch, M.P. Dichotomal effect of space flight-associated microgravity on stress-activated protein kinases in innate immunity. Sci. Rep. 2014, 4, 5468. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Wang, D. Regulation of the response of caenorhabditis elegans to simulated microgravity by p38 mitogen-activated protein kinase signaling. Sci. Rep. 2018, 8, 1–10. [Google Scholar] [CrossRef]
- Zhang, Y.; Moreno-Villanueva, M.; Krieger, S.; Ramesh, G.T.; Neelam, S.; Wu, H. Transcriptomics, NF-κB pathway, and their potential spaceflight-related health consequences. Int. J. Mol. Sci. 2017, 18, 1166. [Google Scholar] [CrossRef]
- Tauber, S.; Hauschild, S.; Paulsen, K.; Gutewort, A.; Raig, C.; Hürlimann, E.; Biskup, J.; Philpot, C.; Lier, H.; Engelmann, F.; et al. Signal transduction in primary human T lymphocytes in altered gravity during parabolic flight and clinostat experiments. Cell. Physiol. Biochem. 2015, 35, 1034–1051. [Google Scholar] [CrossRef]
- Chang, T.T.; Walther, I.; Li, C.F.; Boonyaratanakornkit, J.; Galleri, G.; Meloni, M.A.; Pippia, P.; Cogoli, A.; Hughes-Fulford, M. The Rel/NF-kappaB pathway and transcription of immediate early genes in T cell activation are inhibited by microgravity. J. Leukoc. Biol. 2012, 92, 1133–1145. [Google Scholar] [CrossRef]
- Boonyaratanakornkit, J.B.; Cogoli, A.; Li, C.F.; Schopper, T.; Pippia, P.; Galleri, G.; Meloni, M.A.; Hughes-Fulford, M. Key gravity-sensitive signaling pathways drive T cell activation. FASEB J. 2005, 19, 2020–2022. [Google Scholar] [CrossRef]
- Sakai, J.; Cammarotta, E.; Wright, J.A.; Cicuta, P.; Gottschalk, R.A.; Li, N.; Fraser, I.D.C.; Bryant, C.E. Lipopolysaccharide-induced NF-kappaB nuclear translocation is primarily dependent on MyD88, but TNFalpha expression requires TRIF and MyD88. Sci. Rep. 2017, 7, 1428. [Google Scholar] [CrossRef]
- Chakraborty, N.; Gautam, A.; Muhie, S.; Miller, S.-A.; Jett, M.; Hammamieh, R. An integrated omics analysis: Impact of microgravity on host response to lipopolysaccharide in vitro. BMC Genom. 2014, 15, 659. [Google Scholar] [CrossRef] [PubMed]
- Brungs, S.; Kolanus, W.; Hemmersbach, R. Syk phosphorylation—A gravisensitive step in macrophage signalling. Cell Commun. Signal. 2015, 13, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Ran, F.; An, L.; Fan, Y.; Hang, H.; Wang, S. Simulated microgravity potentiates generation of reactive oxygen species in cells. Biophys. Rep. 2016, 2, 100–105. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Luo, H.; Zhu, L.; Yang, F.; Chu, Z.; Tian, H.; Feng, M.; Zhao, Y.; Shang, P. Microgravity inhibition of lipopolysaccharide-induced tumor necrosis factor-α expression in macrophage cells. Inflamm. Res. 2013, 63, 91–98. [Google Scholar] [CrossRef] [PubMed]
- Van Walleghem, M.; Tabury, K.; Fernandez-Gonzalo, R.; Janssen, A.; Buchheim, J.-I.; Choukér, A.; Baatout, S.; Moreels, M. Gravity-related immunological changes in human whole blood cultured under simulated microgravity using an in vitro cytokine release assay. J. Interferon Cytokine Res. 2017, 37, 531–540. [Google Scholar] [CrossRef] [PubMed]
- Tackett, N.; Bradley, J.H.; Moore, E.K.; Baker, S.H.; Minter, S.L.; DiGiacinto, B.; Arnold, J.P.; Gregg, R.K. Prolonged exposure to simulated microgravity diminishes dendritic cell immunogenicity. Sci. Rep. 2019, 9, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Hahn, C.; Schwartz, M.A. The role of cellular adaptation to mechanical forces in atherosclerosis. Arter. Thromb. Vasc. Biol. 2008, 28, 2101–2107. [Google Scholar] [CrossRef]
- Ratushnyy, A.; Yakubets, D.; Andreeva, E.; Buravkova, L. Simulated microgravity modulates the mesenchymal stromal cell response to inflammatory stimulation. Sci. Rep. 2019, 9, 1–9. [Google Scholar] [CrossRef]
- Buravkova, L.B.; Rudimov, E.G.; Andreeva, E.R.; Grigoriev, A.I. The ICAM-1 expression level determines the susceptibility of human endothelial cells to simulated microgravity. J. Cell. Biochem. 2018, 119, 2875–2885. [Google Scholar] [CrossRef]
- Zhang, J.-M.; An, J. Cytokines, inflammation, and pain. Int. Anesthesiol. Clin. 2007, 45, 27–37. [Google Scholar] [CrossRef]
- Smith, J.K. IL-6 and the dysregulation of immune, bone, muscle, and metabolic homeostasis during spaceflight. NPJ Microgravity 2018, 4, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Ponomarev, S.; Institute of Biomedical Problems of the Russian of the Russian Academy of Sciences; Berendeeva, T.; Kalinin, S.; Muranova, A. Status of the system of signaling pattern recognition receptors of monocytes and granulocytes in cosmonauts’’ peripheral blood before and after long-duration missions to the international space station. Aerosp. Environ. Med. 2016, 50, 18–23. [Google Scholar] [CrossRef] [PubMed]
- Rykova, M.P. Immune system in Russian cosmonauts after orbital space flights. Hum. Physiol. 2013, 39, 557–566. [Google Scholar] [CrossRef]
- Mehta, S.K.; Crucian, B.; Stowe, R.; Simpson, R.; Ott, C.; Sams, C.; Pierson, D. Reactivation of latent viruses is associated with increased plasma cytokines in astronauts. Cytokine 2013, 61, 205–209. [Google Scholar] [CrossRef]
- Novoselova, E.G.; Lunin, S.M.; Khrenov, M.O.; Parfenyuk, S.B.; Novoselova, T.V.; Shenkman, B.S.; Fesenko, E.E. Changes in immune cell signalling, apoptosis and stress response functions in mice returned from the BION-M1 mission in space. Immunobiology 2015, 220, 500–509. [Google Scholar] [CrossRef]
- Thiel, C.S.; Lauber, B.A.; Polzer, J.; Ullrich, O. Time course of cellular and molecular regulation in the immune system in altered gravity: Progressive damage or adaptation? REACH 2017, 5, 22–32. [Google Scholar] [CrossRef]
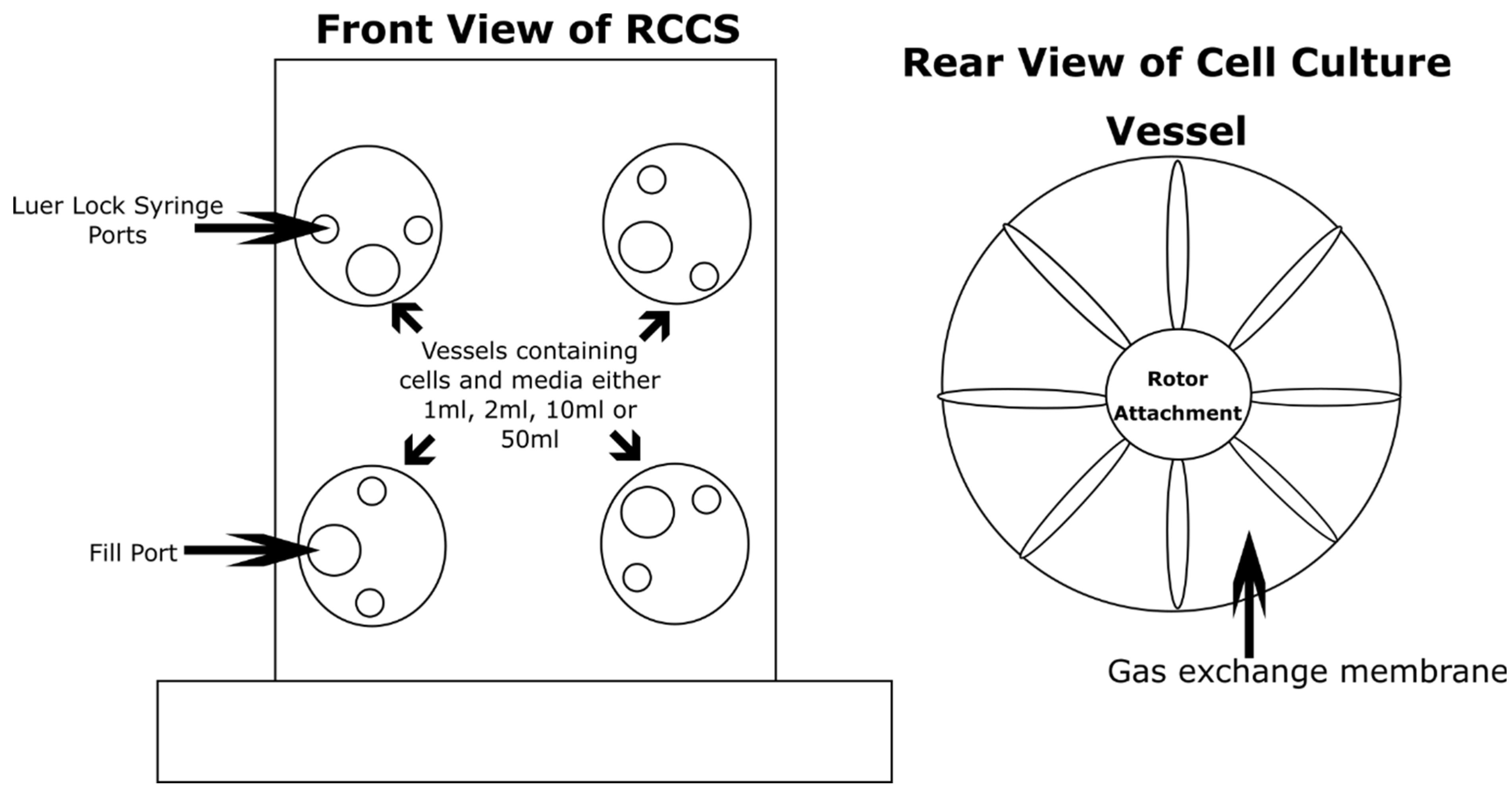
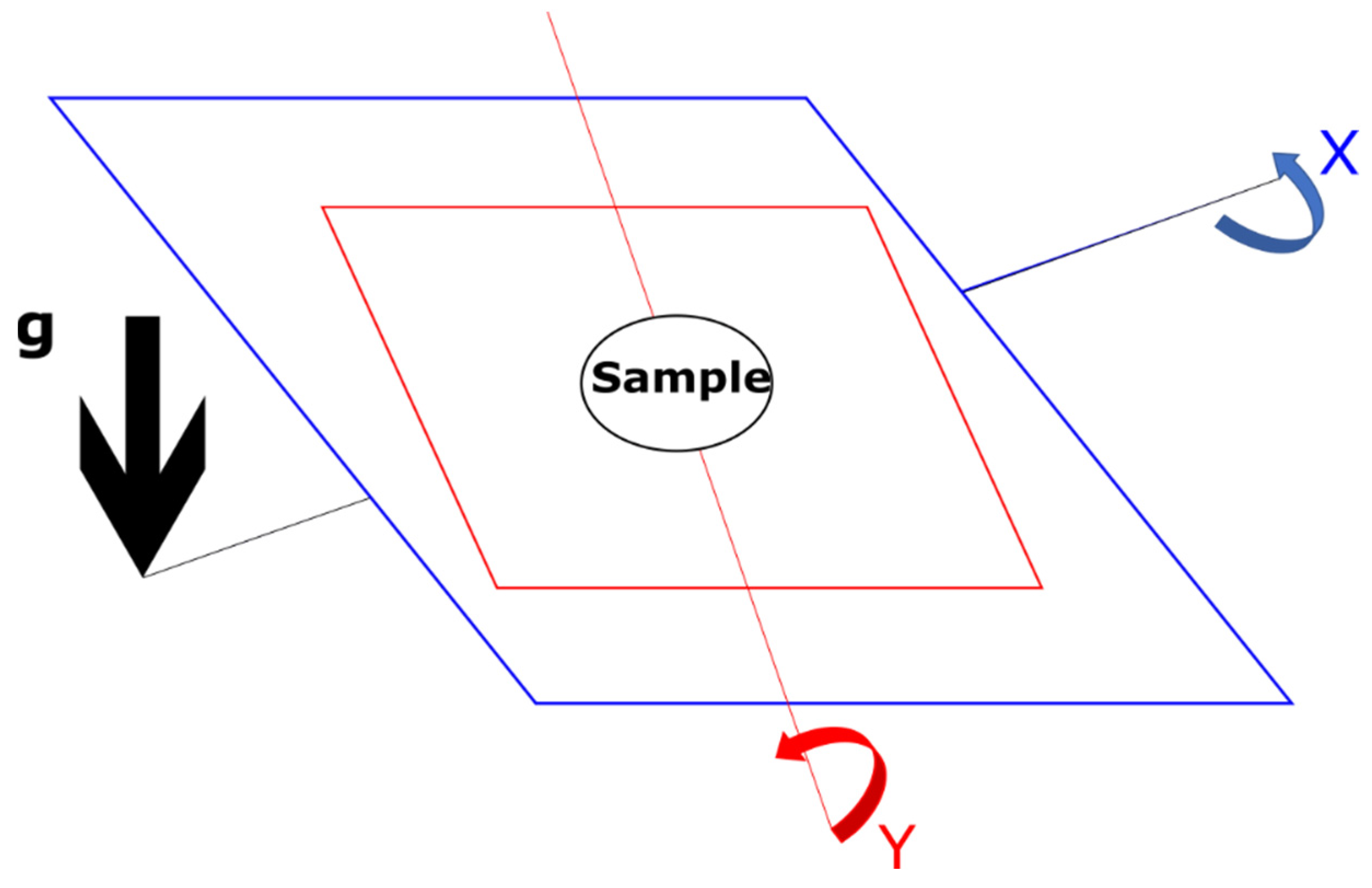
| Time Taken | Air | Surface | Water |
|---|---|---|---|
| Preflight | 300 CFU m−3 | 500 CFU 100 cm−2 | 50 CFU mL−1 |
| Inflight | 1000 CFU m−3 | 10,000 CFU 100 cm−2 | 50 CFU mL−1 |
| Name | Low-Shear Environment | Studies | Major Findings |
|---|---|---|---|
| Mycobacterium marinum | Rotating Cell Culture System | [31] | 562 genes altered transcription level after short growth, 328 after long growth periods. Downregulation of Metabolism. Increases sensitivity to hydrogen peroxide. |
| Ralstonia pickettii | Spaceflight samples in Rotating Cell Culture System | [30] | Increased growth rate |
| Escherichia coli | Rotating Cell Culture System | [32,33,34,35] | Shorter replication time, increased survivability in J774 macrophages, increased resistance to osmotic stress, heat and acid. Increase in biofilm thickness and biomass. |
| Salmonella enterica serovar typhimurium | Rotating Cell Culture System | [36] | Shorter replication time, increased survivability in J774 macrophages, increased resistance to osmotic stress, heat and acid. |
| Streptococcus mutans | Rotating Cell Culture System | [37,38] | 153 genes upregulated two-fold or more, 94 genes downregulated two-fold or more |
| Lactobacillus acidophilus | Rotating Cell Culture System | [39] | Shortened lag phase, increased growth rate, increased antibiotic resistance, increased acid and bile resistance. |
| Bacillus subtilis | Spaceflight | [40] | 55 genes upregulated (biofilm formation associated genes), 36 genes downregulated (anaerobic respiration associated genes). |
| Pseudomonas aeruginosa | Spaceflight | [41,42,43] | Different biofilm architecture to that formed under Earth gravity. |
| Klebsiella pneumoniae | Rotating Cell Culture System | [44] | Enhanced biofilm formation, thicker biofilms, increased cellulose production. |
| Vibrio fischeri | Rotating Cell Culture System | [45] | Hfq mutant studies. |
| Staphylococcus aureus | Rotating Cell Culture System Spaceflight | [46] [4] | Antibiotic resistance increases. Cell wall changes. |
| Name | Low-Shear Environment | Studies | Major Findings |
|---|---|---|---|
| Haloferax mediterranei | Rotary Cell Culture System | [64] | Increased resistance to bacitracin, rifampicin and erythromycin |
| Halococcus dombrowkskii | Rotary Cell Culture System | [64] | Reduced cell aggregation |
| Haloarcula argentinesis RR10 | Rotary Cell Culture System | [65] | Increased production of ribosomal proteins, became multi-drug resistant, evidence of antibiotic efflux pump |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Green, M.J.; Aylott, J.W.; Williams, P.; Ghaemmaghami, A.M.; Williams, P.M. Immunity in Space: Prokaryote Adaptations and Immune Response in Microgravity. Life 2021, 11, 112. https://doi.org/10.3390/life11020112
Green MJ, Aylott JW, Williams P, Ghaemmaghami AM, Williams PM. Immunity in Space: Prokaryote Adaptations and Immune Response in Microgravity. Life. 2021; 11(2):112. https://doi.org/10.3390/life11020112
Chicago/Turabian StyleGreen, Macauley J., Jonathan W. Aylott, Paul Williams, Amir M. Ghaemmaghami, and Philip M. Williams. 2021. "Immunity in Space: Prokaryote Adaptations and Immune Response in Microgravity" Life 11, no. 2: 112. https://doi.org/10.3390/life11020112
APA StyleGreen, M. J., Aylott, J. W., Williams, P., Ghaemmaghami, A. M., & Williams, P. M. (2021). Immunity in Space: Prokaryote Adaptations and Immune Response in Microgravity. Life, 11(2), 112. https://doi.org/10.3390/life11020112







