Transcriptomic Analysis of the Anthocyanin Biosynthetic Pathway Reveals the Molecular Mechanism Associated with Purple Color Formation in Dendrobium Nestor
Abstract
1. Introduction
2. Material and Methods
2.1. Plant Growth and Sample Preparation
2.2. RNA Extraction, Sequencing Libraries, and RNA-Seq
2.3. Sequenced Data Filtering, Assembly, and Functional Annotation
2.4. Detection of Differentially Expressed Genes and Functional Enrichment Analysis
2.5. Identification of Transcription Factors and Simple Sequence Repeats
2.6. Real-Time Quantitative Polymerase Chain Reaction Analysis
3. Results
3.1. Sequencing Summary, Assembly, and Unigene Annotation
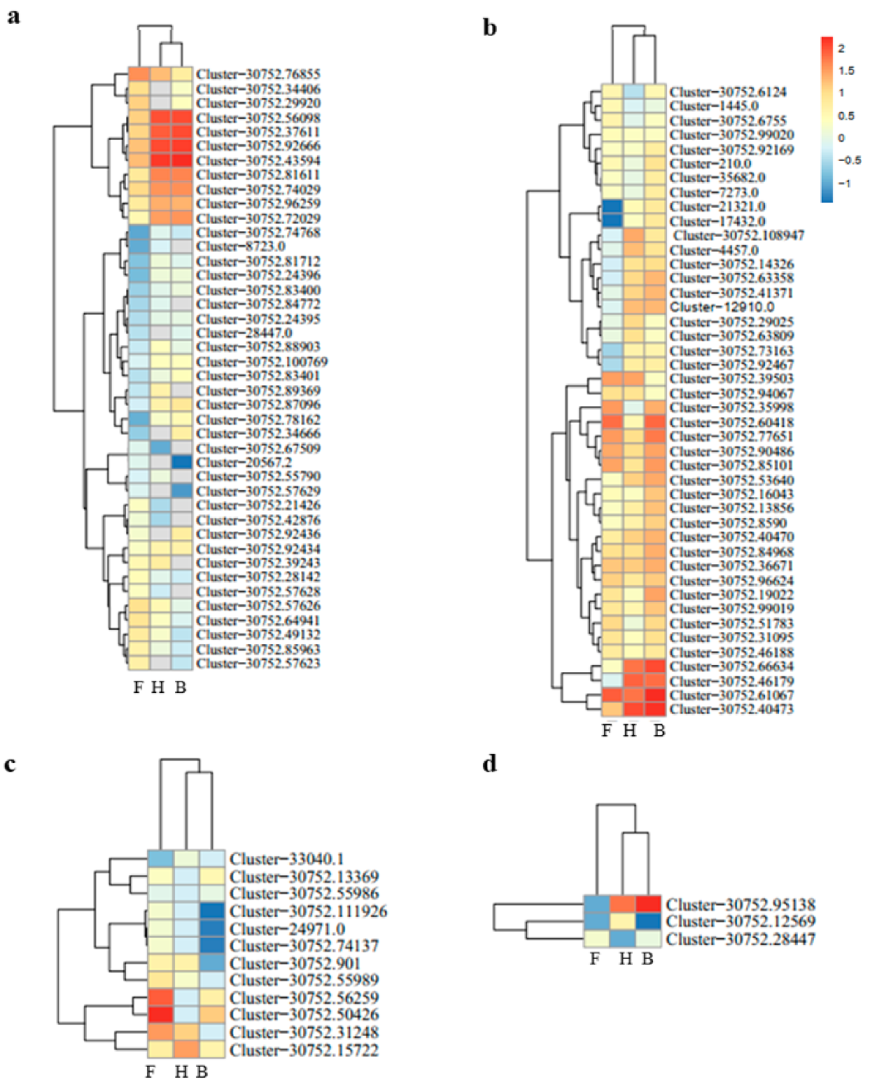
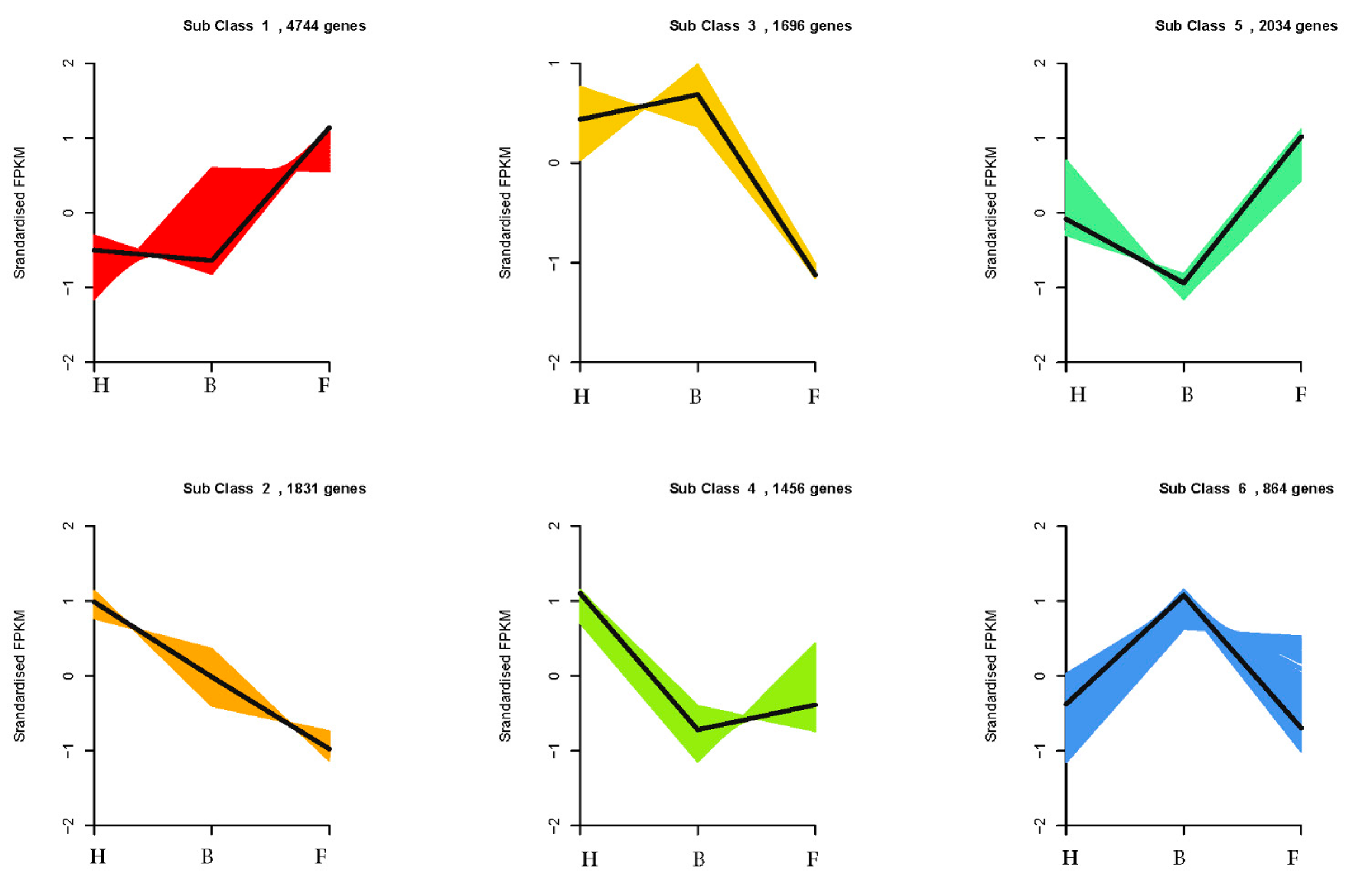
3.2. Differentially Expressed Genes and Functional Enrichment Analyses
3.3. DEGs Involved in Anthocyanins Biosynthesis Pathway
3.4. Plant Hormone Signal Transduction
3.5. Identification of Transcription Factors Regulating Color Formation in the Petals/Flowers of D. nestor
4. Discussion
4.1. Pathways Involved in Purple Color Formation in Petals of D. nestor at Different Stages of Development
4.2. Transcriptional Regulation of Purple Color Formation in D. nestor
4.3. SSR Markers Discovered for Practical D. nestor Breeding
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| COG | Clusters of orthologous groups of proteins |
| DEGs | Differentially expressed genes |
| FPKM | Fragments per kilobase of transcript per million fragments mapped |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| KOG | Eukaryotic Clusters of Orthologous Groups |
| MISA | MIcroSAtellite identification Single nucleotide polymorphisms |
| Nr | Non-redundant protein sequence database |
| ORF | Open reading frame |
| SSR | Simple sequence repeats |
| STEM | Short time-series expression miner |
| TFs | Transcription factors |
| TrEMBL | Translated European molecular biology laboratory |
References
- Yan, L.; Wang, X.; Liu, H.; Tian, Y.; Lian, J.; Yang, R.; Hao, S.; Wang, X.; Yang, S.; Li, Q.; et al. The genome of dendrobium officinale illuminates the biology of the important traditional Chinese orchid herb. Mol. Plant 2015, 8, 922–934. [Google Scholar]
- Chase, M.W.; Cameron, K.M.; Freudenstein, J.V.; Pridgeon, A.M.; Salazar, G.; van den Berg, C.; Schuiteman, A. An updated classification of Orchidaceae. Bot. J. Linn. Soc. 2015, 177, 151–174. [Google Scholar] [CrossRef]
- Tang, H.; Zhao, T.; Sheng, Y.; Zheng, T.; Fu, L.; Zhang, Y. Dendrobium officinale Kimura et Migo: A Review on Its Ethnopharmacology, Phytochemistry, Pharmacology, and Industrialization. Evid. based Complement. Altern. Med. 2017, 93, 1795–1803. [Google Scholar] [CrossRef] [PubMed]
- Zhitao, N.; Shuying, Z.; Jiajia, P.; Ludan, L.; Jing, S.; Xiaoyu, D. Comparative analysis of Dendrobium plastomes and utility of plastomic mutational hotspots. Sci. Rep. 2017, 7, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Riechmann, J.L.; Heard, J.; Martin, G.; Reuber, L.; Jiang, C.Z.; Keddie, J.; Adam, L.; Pineda, O.; Ratcliffe, O.J.; Samaha, R.R.; et al. Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 2000, 290, 2105–2110. [Google Scholar] [CrossRef]
- Cazzonelli, C.I.; Pogson, B.J. Source to sink: Regulation of carotenoid biosynthesis in plants. Trends Plant Sci. 2010, 15, 266–274. [Google Scholar] [CrossRef]
- Balilashaki, K.; Zakizadeh, H.; Olfati, J.A.; Vahedi, M.; Kumar, A.; Indracanti, M. Recent Advances in Phalaenopsis Orchid Improvement using Omics Approaches. Plant Tissue Cult. Biotechnol. 2019, 29, 133–149. [Google Scholar] [CrossRef]
- Rebecca, O.P.S.; Boyce, A.N.; Chandran, S. Pigment identification and antioxidant properties of red dragon fruit (Hylocereus polyrhizus). African J. Biotechnol. 2010, 9, 1450–1454. [Google Scholar]
- Zhang, S.; Zhang, A.; Wu, X.; Zhu, Z.; Yang, Z.; Zhu, Y.; Zha, D. Transcriptome analysis revealed expression of genes related to anthocyanin biosynthesis in eggplant (Solanum melongena L.) under high-temperature stress. BMC Plant Biol. 2019, 19, 1–13. [Google Scholar] [CrossRef]
- Yang, X.; Xia, X.; Zhang, Z.; Nong, B.; Zeng, Y.; Wu, Y.; Xiong, F.; Zhang, Y.; Liang, H.; Pan, Y.; et al. Identification of anthocyanin biosynthesis genes in rice pericarp using PCAMP. Plant Biotechnol. J. 2019, 17, 1700–1702. [Google Scholar] [CrossRef]
- Rameneni, J.J.; Choi, S.R.; Chhapekar, S.S.; Man-Sun, K.; Singh, S.; Yi, S.Y.; Heon, O.S.; Kim, H.; Lee, C.Y.; Man-Ho, O.; et al. Red Chinese cabbage transcriptome analysis reveals structural genes and multiple transcription factors regulating reddish purple color. Int. J. Mol. Sci. 2020, 21, 2901. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.S.; Shiraishi, A.; Hashimoto, F.; Aoki, N.; Shimizu, K.; Sakata, Y. Analysis of petal anthocyanins to investigate flower coloration of Zhongyuan (Chinese) and Daikon Island (Japanese) tree peony cultivars. J. Plant Res. 2001, 114, 33–43. [Google Scholar] [CrossRef]
- He, C.; Liu, X.; Teixeira da Silva, J.A.; Liu, N.; Zhang, M.; Duan, J. Transcriptome sequencing and metabolite profiling analyses provide comprehensive insight into molecular mechanisms of flower development in Dendrobium officinale (Orchidaceae). Plant Mol. Biol. 2020, 104, 529–548. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.H.; Yang, J.X.; Xiao, C.H.; Mao, T.Y.; Zhang, J.; Zhang, H.Y. Differences in flavonoid pathway metabolites and transcripts affect yellow petal colouration in the aquatic plant Nelumbo nucifera. BMC Plant Biol. 2019, 19, 277. [Google Scholar] [CrossRef]
- Yamagishi, M. How genes paint lily flowers: Regulation of colouration and pigmentation patterning. Sci. Hortic. 2013, 163, 27–36. [Google Scholar] [CrossRef]
- Koseki, M.; Goto, K.; Masuta, C.; Kanazawa, A. The star-type color pattern in Petunia hybrid ‘Red Star’ flowers is induced by sequence-specific degradation of chalcone synthase RNA. Plant Cell Psysiol. 2005, 46, 1879–1883. [Google Scholar] [CrossRef]
- Holton, T.A.; Brugliera, F.; Tanaka, Y. Cloning and expression of flavonol synthase from Petunia hybrida. Plant J. 1993, 4, 1003–1010. [Google Scholar] [CrossRef]
- Falcone Ferreyra, M.L.; Rius, S.P.; Casati, P. Flavonoids: Biosynthesis, biological functions, and biotechnological applications. Front. Plant Sci. 2012, 3, 1–15. [Google Scholar] [CrossRef]
- Stenbaek, A.; Jensen, P.E. Redox regulation of chlorophyll biosynthesis. Phytochemistry 2010, 71, 853–859. [Google Scholar] [CrossRef]
- Mudalige-Jayawickrama, R.G.; Champagne, M.M.; Hieber, A.D.; Kuehnle, A.R. Cloning and characterization of two anthocyanin biosynthetic genes from Dendrobium orchid. J. Am. Soc. Hortic. Sci. 2005, 130, 611–618. [Google Scholar] [CrossRef]
- Whang, S.S.; Um, W.S.; Song, I.J.; Lim, P.O.; Choi, K.; Park, K.W.; Kang, K.W.; Choi, M.S.; Koo, J.C. Molecular Analysis of Anthocyanin Biosynthetic Genes and Control of Flower Coloration by Flavonoid 3′,5′-Hydroxylase (F3′5′H) in Dendrobium moniliforme. J. Plant Biol. 2011, 54, 209–218. [Google Scholar] [CrossRef]
- Jaakola, L. New insights into the regulation of anthocyanin biosynthesis in fruits. Trends Plant Sci. 2013, 18, 477–483. [Google Scholar] [CrossRef] [PubMed]
- Winkel-Shirley, B. Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 2001, 126, 485–493. [Google Scholar] [CrossRef] [PubMed]
- Grotewold, E. The genetics and biochemistry of floral pigments. Annu. Rev. Plant Biol. 2006, 57, 761–780. [Google Scholar] [CrossRef] [PubMed]
- Davies, K.M.; Schwinn, K.E. Molecular biology and biotechnology of flower pigments. In Plant Developmental Biology; Pua, E.C., Davey, M.R., Eds.; Springer: Berlin/Heidelberg, Germany, 2010; pp. 161–187. ISBN 9783642046698. [Google Scholar]
- Quattrocchio, F.; Wing, J.F.; Van der Woude, K.; Mol, J.N.M.; Koes, R. Analysis of bHLH and MYB domain proteins: Species-specific regulatory differences are caused by divergent evolution of target anthocyanin genes. Plant J. 1998, 13, 475–488. [Google Scholar]
- Tian, J.; Chen, M.C.; Zhang, J.; Li, K.T.; Song, T.T.; Zhang, X.; Yao, Y.C. Characteristics of dihydroflavonol 4-reductase gene promoters from different leaf colored Malus crabapple cultivars. Hortic. Res. 2017, 4, 17070. [Google Scholar] [CrossRef]
- Arrom, L.; Munné-Bosch, S. Hormonal changes during flower development in floral tissues of Lilium. Planta 2012, 236, 343–354. [Google Scholar] [CrossRef]
- Eugster, C.H.; Märki-Fischer, E. The Chemistry of Rose Pigments. Angew. Chem. Int. Ed. Engl. 1991, 30, 654–672. [Google Scholar] [CrossRef]
- Lu, J.; Zhang, Q.; Lang, L.; Jiang, C.; Wang, X.; Sun, H. Integrated Metabolome and Transcriptome Analyses Reveal the Molecular Mechanism of a Color Mutation in Miniature Roses. BMC Plant Biol. 2019, 20, 611. [Google Scholar]
- Li, D.; Liu, X.; Shu, L.; Zhang, H.; Zhang, S.; Song, Y.; Zhang, Z. Global analysis of the AP2 / ERF gene family in rose ( Rosa chinensis ) genome unveils the role of RcERF099 in Botrytis resistance. BMC Plant Biol. 2020, 20, 1–15. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq-A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; He, Y.; Xia, R. TBtools, a Toolkit for Biologists integrating various biological data handling tools with a user-friendly interface. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. FASTQC A Quality Control tool for High Throughput Sequence Data. Babraham Inst. 2015, 12, 20–30. [Google Scholar]
- Galperin, M.Y.; Makarova, K.S.; Wolf YI, K.E. Expanded microbial genome coverage and improved protein family anno- tation in the COG database. Nucleic. Acids Res. 2014, 43, D261–D269. [Google Scholar] [CrossRef] [PubMed]
- Gene Ontology Consortium. Gene ontology consortium: Going forward. Nucleic Acids Res. 2014, 43, D1049–D1056. [Google Scholar]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016, 32, 277–280. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A.; et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016, 44, D279–D285. [Google Scholar]
- Bairoch, A.; Apweiler, R. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res. 2000, 28, 45–48. [Google Scholar] [CrossRef]
- Deng, Y.Y.; Li, J.Q.; Wu, S.F.; Zhu, Y.P.; Chen, Y.W.; He, F.C. Integrated nr database in protein annotation system and its localization. Nat. Commun. 2010, 32, 71–72. [Google Scholar]
- R Development Core Team R: A Language and Environment for Statistical Computing. R Found. Stat. Comput. 2013, 4, 2–7.
- Benjamini, Y.; Hocheberg, Y. Controlling the false discovery rate-A practical and powerful approach to multiple testing. J. Roy. Stat. Soc. Ser. 1995, B57, 289–300. [Google Scholar] [CrossRef]
- Ernst, J.; Bar-Joseph, Z. STEM: A tool for the analysis of short time series gene expression data. BMC Bioinform 2006, 7, 191. [Google Scholar] [CrossRef] [PubMed]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [PubMed]
- Rice, P.; Longden, L.; Bleasby, A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000, 16, 276–277. [Google Scholar] [CrossRef]
- Mistry, J.; Finn, R.D.; Eddy, S.R.; Bateman, A.; Punta, M. Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Res. 2013, 41, e121. [Google Scholar] [PubMed]
- Zhang, H.; Jin, J.; Tang, L.; Zhao, Y.; Gu, X.; Gao, G.; Luo, J. PlantTFDB 2.0: Update and improvement of the comprehensive plant transcription factor database. Nucleic Acids Res. 2011, 9, D1114–D1117. [Google Scholar] [CrossRef] [PubMed]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3-new capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef]
- KJ Livak, T.S. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Tanaka, Y.; Sasaki, N.; Ohmiya, A. Biosynthesis of plant pigments: Anthocyanins, betalains and carotenoids. Plant J. 2008, 54, 733–749. [Google Scholar] [CrossRef]
- Irani, N.G.; Hernandez, J.M.; Grotewold, E. Regulation of anthocyanin pigmentation. In Recent Advances in Phytochemistry; Springer: New York, NY, USA, 2003; pp. 59–78. [Google Scholar]
- Kolde, R. Pheatmap: Pretty Heatmaps, R package version 1.0.12, R Packag. version 1.0.8; 015. 2019.
- Rowan, D.D.; Cao, M.; Lin-Wang, K.; Cooney, J.M.; Jensen, D.J.; Austin, P.T.; Hunt, M.B.; Norling, C.; Hellens, R.P.; Schaffer, R.J.; et al. Environmental regulation of leaf colour in red 35S:PAP1 Arabidopsis thaliana. New Phytol. 2009, 182, 102–115. [Google Scholar] [CrossRef] [PubMed]
- Jeong, S.W.; Das, P.K.; Jeoung, S.C.; Song, J.Y.; Lee, H.K.; Kim, Y.K.; Kim, W.J.; Il Park, Y.; Yoo, S.D.; Choi, S.B.; et al. Ethylene suppression of sugar-induced anthocyanin pigmentation in arabidopsis. Plant Physiol. 2010, 154, 1514–1531. [Google Scholar] [CrossRef] [PubMed]
- Das, P.K.; Shin, D.H.; Choi, S.B.; Yoo, S.D.; Choi, G.; Park, Y. Il Cytokinins enhance sugar-induced anthocyanin biosynthesis in Arabidopsis. Mol. Cells 2012, 34, 93–101. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Zhao, J.; Qin, B.; Yin, Y.; An, W.; Mu, Z.; Cao, Y. ABA mediates development-dependent anthocyanin biosynthesis and fruit coloration in Lycium plants. BMC Plant Biol. 2019, 19, 317. [Google Scholar] [CrossRef]
- Gondor, O.K.; Janda, T.; Soós, V.; Pál, M.; Majláth, I.; Adak, M.K.; Balázs, E.; Szalai, G. Salicylic acid induction of flavonoid biosynthesis pathways in wheat varies by treatment. Front. Plant Sci. 2016, 7, 1447. [Google Scholar]
- Yuan, L.B.; Peng, Z.H.; Zhi, T.T.; Zho, Z.; Liu, Y.; Zhu, Q.; Xiong, X.Y.; Ren, C.M. Brassinosteroid enhances cytokinin-induced anthocyanin biosynthesis in Arabidopsis seedlings. Biol. Plant. 2014, 59, 99–105. [Google Scholar] [CrossRef]
- Mitsis, T.; Efthimiadou, A.; Bacopoulou, F.; Vlachakis, D.; Chrousos, G.; Eliopoulos, E. Transcription factors and evolution: An integral part of gene expression (Review). World Acad. Sci. J. 2020, 2, 3–8. [Google Scholar] [CrossRef]
- Handhayani, T.; Hiryanto, L. Intelligent Kernel K-Means for Clustering Gene Expression. In Proceedings of the Procedia Computer Science; Elsevier B.V.: Amsterdam, The Netherlands, 2015; pp. 171–177. [Google Scholar]
- Grotewold, E. Plant metabolic diversity: A regulatory perspective. Trends Plant Sci. 2005, 10, 57–62. [Google Scholar] [CrossRef]
- Petroni, K.; Tonelli, C. Recent advances on the regulation of anthocyanin synthesis in reproductive organs. Plant Sci. 2011, 181, 219–229. [Google Scholar] [CrossRef]
- Mol, J.; Grofewold, E.; Koes, R. How genes paint flowers and seeds. Trends Plant Sci. 1998, 3, 212–217. [Google Scholar] [CrossRef]
- Ben-Meir, H.; Zuker, A.; Weiss, D.; Vainstein, A. Molecular Control of Floral Pigmentation: Anthocyanins. In Breeding For Ornamentals: Classical and Molecular Approaches; Vainstein, A., Ed.; Springer: San Francisco, CA, USA, 2002; pp. 253–272. [Google Scholar]
- Sobel, J.M.; Streisfeld, M.A. Flower color as a model system for studies of plant evo-devo. Front. Plant Sci. 2013, 4, 321. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Yang, H.; Liu, H.; Yang, J.; Hu, Y. Extensive transcriptome changes underlying the flower color intensity variation in Paeonia ostii. Front. Plant Sci. 2016, 6, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Morita, Y.; Hoshino, A. Recent advances in flower color variation and patterning of Japanese morning glory and petunia. Breed. Sci. 2018, 68, 128–138. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Zhang, B.; Tang, X.; Zhang, J.; Lin, J. Comparative transcriptome analysis of different dendrobium species reveals active ingredients-related genes and pathways. Int. J. Mol. Sci. 2020, 21, 861. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; He, C.; Wu, K.; Teixeira da Silva, J.A.; Zeng, S.; Zhang, X.; Yu, Z.; Xia, H.; Duan, J. Transcriptome analysis of dendrobium officinale and its application to the identification of genes associated with polysaccharide synthesis. Front. Plant Sci. 2016, 7, 1–14. [Google Scholar] [CrossRef]
- Wang, Z.; Zhao, M.; Cui, H.; Li, J.; Wang, M. Transcriptomic Landscape of Medicinal Dendrobium Reveals Genes Associated with the Biosynthesis of Bioactive Components. Front. Plant Sci. 2020, 28, 391. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Zhang, J.; Liu, X.; Meng, M.; Wang, J.; Lin, J. Tissue-specific transcriptome for Dendrobium officinale reveals genes involved in flavonoid biosynthesis. Genomics 2020, 112, 1781–1794. [Google Scholar] [CrossRef]
- Ma, Z.H.; Li, W.F.; Mao, J.; Li, W.; Zuo, C.W.; Zhao, X.; Dawuda, M.M.; Shi, X.Y.; Chen, B.H. Synthesis of light-inducible and light-independent anthocyanins regulated by specific genes in grape “Marselan” (V. Vinifera L.). PeerJ 2019, 7, e6521. [Google Scholar]
- Luo, X.; Cao, D.; Li, H.; Zhao, D.; Xue, H.; Niu, J.; Chen, L.; Zhang, F.; Cao, S. Complementary iTRAQ-based proteomic and RNA sequencing-based transcriptomic analyses reveal a complex network regulating pomegranate (Punica granatum L.) fruit peel colour. Sci. Rep. 2018, 8, 1–13. [Google Scholar] [CrossRef]
- Liu, X.J.; Chuang, Y.N.; Chiou, C.Y.; Chin, D.C.; Shen, F.Q.; Yeh, K.W. Methylation effect on chalcone synthase gene expression determines anthocyanin pigmentation in floral tissues of two Oncidium orchid cultivars. Planta 2012, 236, 401–409. [Google Scholar] [CrossRef]
- Li, S.J.; Deng, X.M.; Mao, H.Z.; Hong, Y. Enhanced anthocyanin synthesis in foliage plant Caladium bicolor. Plant Cell Rep. 2005, 23, 716–720. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Li, G.; Gu, L.; Sun, Y.; Li, Z.; Liu, J.; Wu, X.; Dong, T.; Zhu, M. Comparative Metabolomic and Transcriptome Analysis Reveal Distinct Flavonoid Biosynthesis Regulation Between Petals of White and Purple Phalaenopsis amabilis. J. Plant Growth Regul. 2020, 39, 823–840. [Google Scholar] [CrossRef]
- Sapir, M.; Oren-Shamir, M.; Ovadia, R.; Reuveni, M.; Evenor, D.; Tadmor, Y.; Nahon, S.; Shlomo, H.; Chen, L.; Meir, A.; et al. Molecular aspects of Anthocyanin fruit tomato in relation to high pigment-1. J. Hered. 2008, 99, 292–303. [Google Scholar] [CrossRef] [PubMed]
- Stommel, J.R.; Lightbourn, G.J.; Winkel, B.S.; Griesbach, R.J. Transcription factor families regulate the anthocyanin biosynthetic pathway in capsicum annuum. J. Am. Soc. Hortic. Sci. 2009, 134, 244–251. [Google Scholar] [CrossRef]
- Xie, D.Y.; Sharma, S.B.; Paiva, N.L.; Ferreira, D.; Dixon, R.A. Role of anthocyanidin reductase, encoded by BANYULS in plant flavonoid biosynthesis. Science 2003, 299, 396–399. [Google Scholar] [CrossRef]
- Jiao, F.; Zhao, L.; Wu, X.; Song, Z.; Li, Y. Metabolome and transcriptome analyses of the molecular mechanisms of flower color mutation in tobacco. BMC Genom. 2020, 21, 1–10. [Google Scholar] [CrossRef]
- Murthy, H.N.; Lee, E.J.; Paek, K.Y. Production of secondary metabolites from cell and organ cultures: Strategies and approaches for biomass improvement and metabolite accumulation. Plant Cell. Tissue Organ. Cult. 2014, 118, 1–16. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, N.; Xu, H.; Jiang, S.H.; Fang, H.C.; Su, M.Y.; Zhang, Z.Y.; Zhang, T.L.; Chen, X.S. Auxin regulates anthocyanin biosynthesis through the Aux/IAA–ARF signaling pathway in apple. Hortic. Res. 2018, 5, 1–11. [Google Scholar] [CrossRef]
- Taylor, J.B.; Triggle, D.J. Comprehensive Medicinal Chemistry II; Elsevier Ltd.: Amsterdam, The Netherlands, 2006; ISBN 9780080450445. [Google Scholar]
- Cheatle Jarvela, A.M.; Hinman, V.F. Evolution of transcription factor function as a mechanism for changing metazoan developmental gene regulatory networks. Evodevo 2015, 6, 3–9. [Google Scholar] [CrossRef]
- Allan, A.C.; Hellens, R.P.; Laing, W.A. MYB transcription factors that colour our fruit. Trends Plant Sci. 2008, 13, 99–102. [Google Scholar] [CrossRef]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef] [PubMed]
- Takos, A.M.; Jaffé, F.W.; Jacob, S.R.; Bogs, J.; Robinson, S.P.; Walker, A.R. Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol. 2006, 142, 1216–1232. [Google Scholar] [CrossRef] [PubMed]
- Ban, Y.; Honda, C.; Hatsuyama, Y.; Igarashi, M.; Bessho, H.; Moriguchi, T. Isolation and functional analysis of a MYB transcription factor gene that is a key regulator for the development of red coloration in apple skin. Plant Cell Physiol. 2007, 48, 958–970. [Google Scholar] [CrossRef] [PubMed]
- Espley, R.V.; Hellens, R.P.; Putterill, J.; Stevenson, D.E.; Kutty-Amma, S.; Allan, A.C. Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J. 2007, 49, 414–427. [Google Scholar] [CrossRef] [PubMed]
- An, J.P.; Wang, X.F.; Li, Y.Y.; Song, L.Q.; Zhao, L.L.; You, C.X.; Hao, Y.J. EIN3-LIKE1, MYB1, and ethylene response factor3 act in a regulatory loop that synergistically modulates ethylene biosynthesis and anthocyanin accumulation1 [OPEN]. Plant Physiol. 2018, 178, 808–823. [Google Scholar] [CrossRef]
- An, J.P.; Wang, X.F.; Zhang, X.W.; Bi, S.Q.; You, C.X.; Hao, Y.J. MdBBX22 regulates UV-B-induced anthocyanin biosynthesis through regulating the function of MdHY5 and is targeted by MdBT2 for 26S proteasome-mediated degradation. Plant Biotechnol. J. 2019, 17, 2231–2233. [Google Scholar] [CrossRef]
- An, J.P.; Liu, Y.J.; Zhang, X.W.; Bi, S.Q.; Wang, X.F.; You, C.X.; Hao, Y.J.; Vicente, A. Dynamic regulation of anthocyanin biosynthesis at different light intensities by the BT2-TCP46-MYB1 module in apple. J. Exp. Bot. 2020, 71, 3094–3109. [Google Scholar] [CrossRef]
- An, J.P.; Zhang, X.W.; Bi, S.Q.; You, C.X.; Wang, X.F.; Hao, Y.J. The ERF transcription factor MdERF38 promotes drought stress-induced anthocyanin biosynthesis in apple. Plant J. 2020, 101, 573–589. [Google Scholar] [CrossRef]
- Lloyd, A.; Brockman, A.; Aguirre, L.; Campbell, A.; Bean, A.; Cantero, A.; Gonzalez, A. Advances in the MYB-bHLH-WD Repeat (MBW) pigment regulatory model: Addition of a WRKY factor and co-option of an anthocyanin MYB for betalain regulation. Plant Cell Physiol. 2017, 58, 1431–1441. [Google Scholar] [CrossRef]
- Verweij, W.; Spelt, C.E.; Bliek, M.; de Vries, M.; Wit, N.; Faraco, M.; Koes, R.; Quattrocchio, F.M. Functionally similar WRKY proteins regulate vacuolar acidification in petunia and hair development in arabidopsis. Plant Cell 2016, 28, 786–803. [Google Scholar] [CrossRef]
- Zhou, H.; Lin-Wang, K.; Wang, H.; Gu, C.; Dare, A.P.; Espley, R.V.; He, H.; Allan, A.C.; Han, Y. Molecular genetics of blood-fleshed peach reveals activation of anthocyanin biosynthesis by NAC transcription factors. Plant. J. 2015, 82, 105–121. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Chen, Y.; Zhao, L.; Li, C.; Yu, J.; Li, T.; Yang, W.; Zhang, S.; Su, H.; Wang, L. A novel NAC transcription factor, MdNAC42, regulates anthocyanin accumulation in red-fleshed apple by interacting with MdMYB10. Tree Physiol. 2020, 40, 413–423. [Google Scholar] [CrossRef] [PubMed]
- Perez-de-Castro, A.M.; Vilanova, S.; Canizares, J.; Pascual, L.; Blanca, M.J.; Diez, M.J.; Prohens, J.; Pico, B. Application of Genomic Tools in Plant Breeding. Curr. Genom. 2012, 13, 179–195. [Google Scholar] [CrossRef] [PubMed]
- Singh, V.K.; Singh, A.K.; Singh, S.; Singh, B.D. Next-generation sequencing (NGS) tools and impact in plant breeding. In Advances in Plant Breeding Strategies: Breeding, Biotechnology and Molecular Tools; Al-Khayri, J.M., Jain, S.M., Johnson, D.V., Eds.; Springer: Cham, Switzerland, 2016; pp. 563–612. ISBN 9783319225210. [Google Scholar]

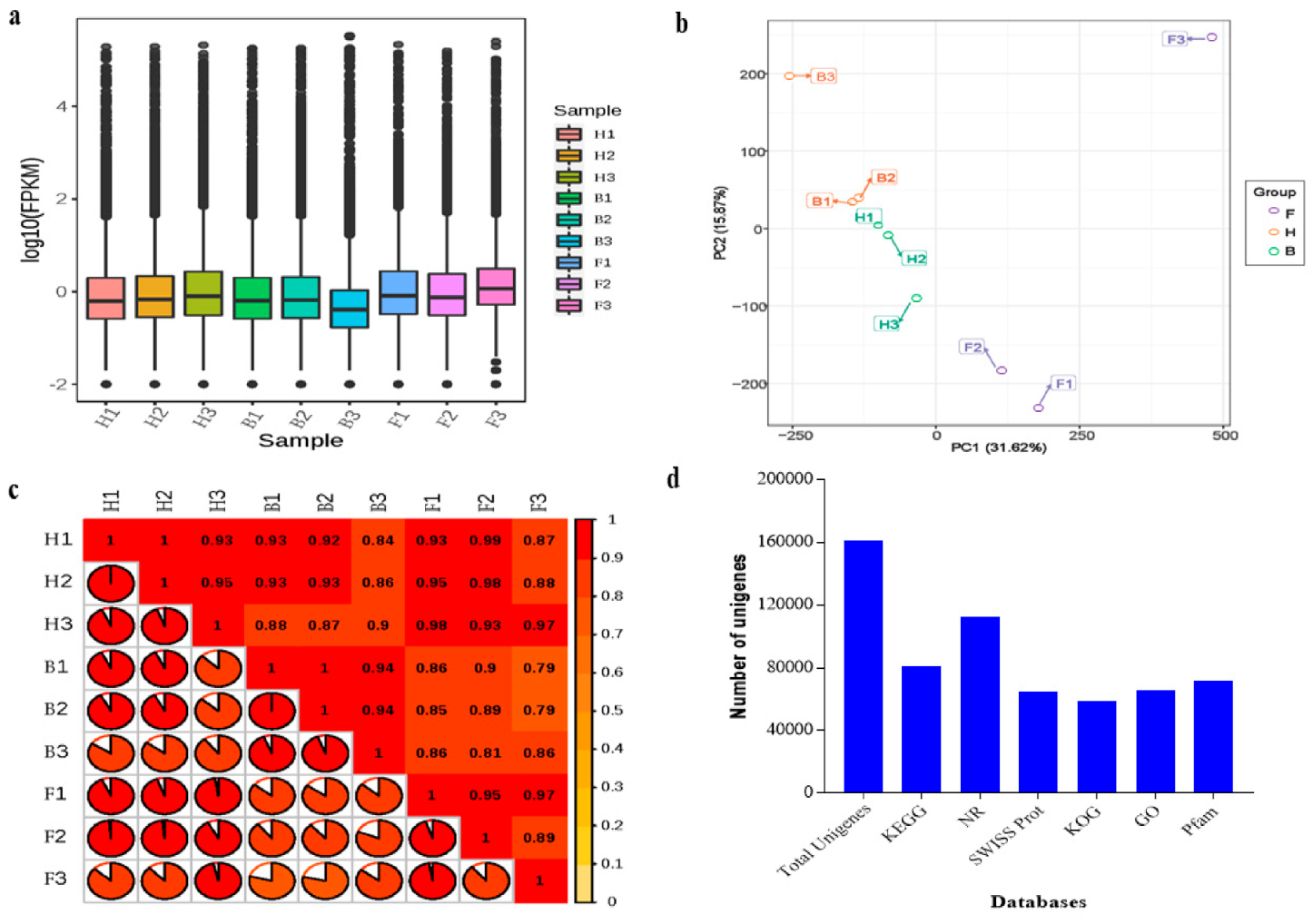
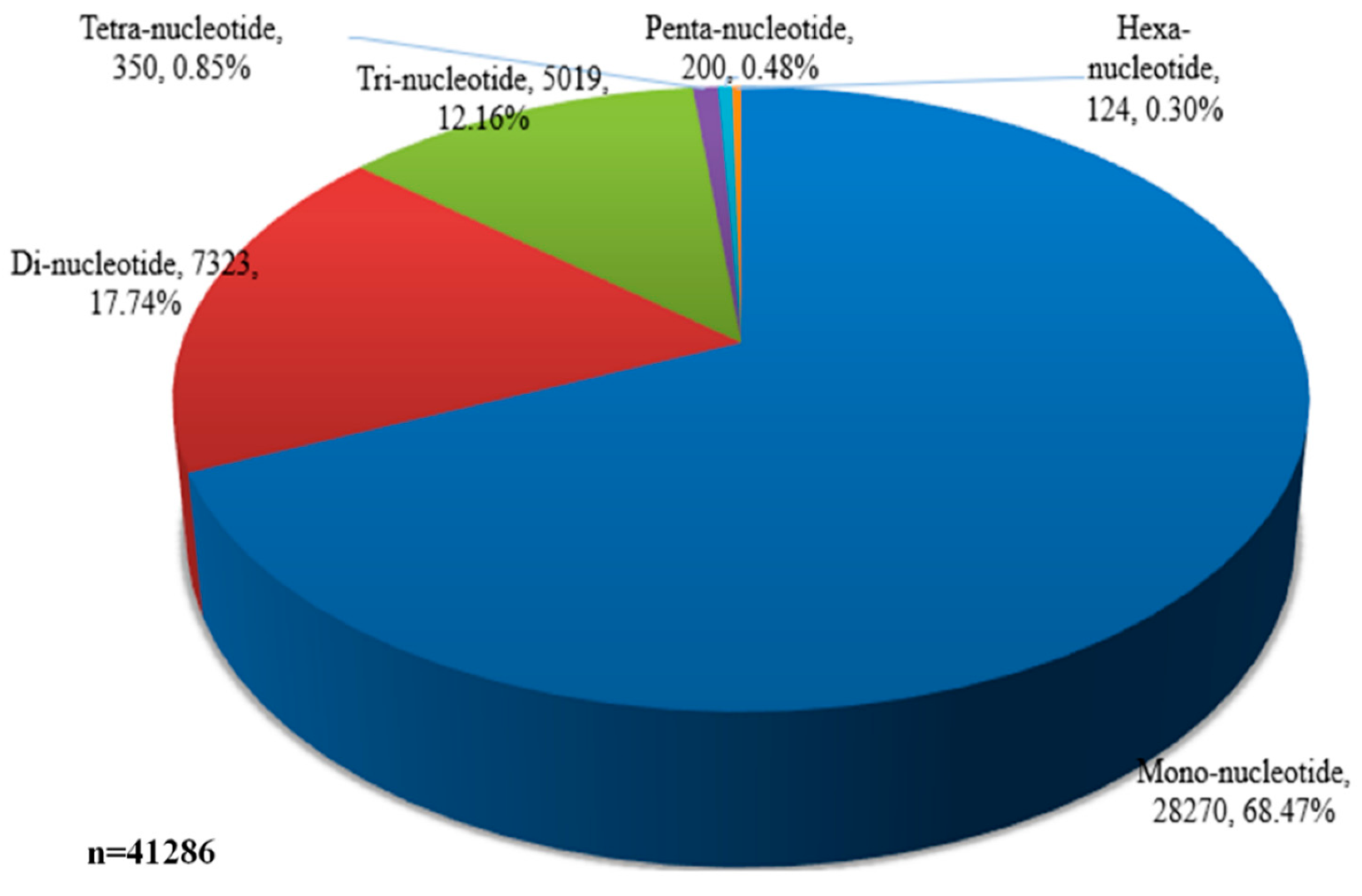
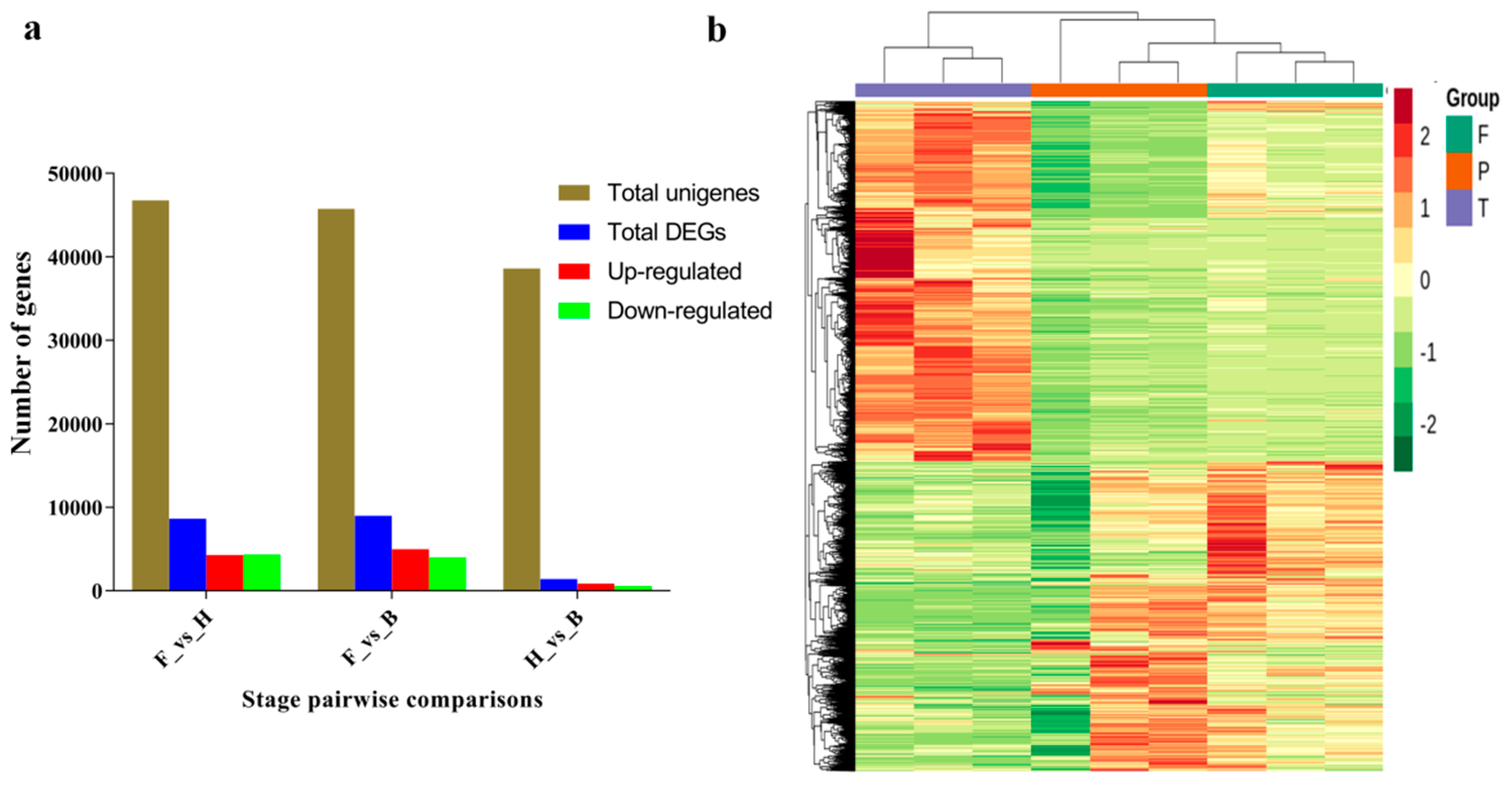
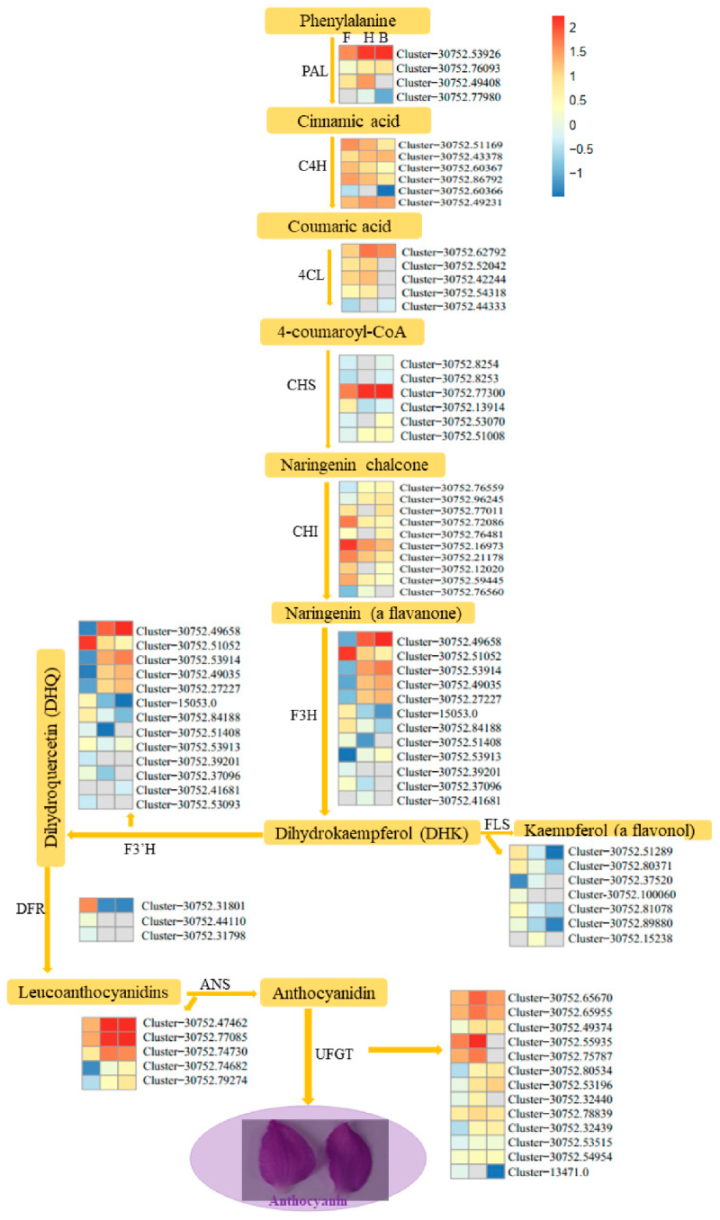


| Sample a | Raw Reads | Clean Reads b | Clean Base (G) | Q30 (%) | GC Content (%) |
|---|---|---|---|---|---|
| F1 | 53327576 | 52337746 (98.14%) | 7.85 | 93.58 | 47.24 |
| F2 | 52995426 | 49011814 (92.48%) | 7.35 | 94.70 | 47.50 |
| F3 | 47190160 | 46301832 (98.12%) | 6.95 | 93.43 | 46.36 |
| Average F | 50047108 | 47538849 (96.25%) | 7.13 | 94.32 | 47.72 |
| H1 | 53098924 | 49694824 (93.59%) | 7.45 | 94.00 | 47.98 |
| H2 | 51764262 | 49406926 (95.45%) | 7.41 | 94.53 | 47.91 |
| H3 | 45278138 | 43514798 (96.11%) | 6.53 | 94.42 | 47.26 |
| Average H | 50047108 | 47538849 (94.98%) | 7.03 | 94.07 | 47.96 |
| B1 | 52422668 | 50153474 (95.67%) | 7.5248 | 94.30 | 47.81 |
| B2 | 50331532 | 47943212 (95.25%) | 7.19 | 94.17 | 47.74 |
| B3 | 50758962 | 49698100 (97.91%) | 6.39 | 93.73 | 48.32 |
| Average P | 51171054 | 48879555 (95.52%) | 7.38 | 93.90 | 47.03 |
| TF a | F_vs_H | F_vs_B | H_vs_B | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Total b | Up c | Down d | Total | Up | Down | Total | Up | Down | |
| AP2/ERF | 44 | 40 | 4 | 59 | 37 | 22 | 16 | 1 | 15 |
| AUX/IAA | 10 | 6 | 4 | 13 | 3 | 10 | 8 | - | 8 |
| B3 | 29 | 20 | 6 | 29 | 13 | 16 | 3 | - | 3 |
| bHLH | 56 | 42 | 24 | 38 | 9 | 29 | 8 | 2 | 6 |
| bZIP | 19 | 12 | 7 | 24 | 17 | 7 | 2 | - | 2 |
| C2C2 | 30 | 8 | 22 | 37 | 5 | 32 | 4 | 1 | 3 |
| C2H2 | 25 | 17 | 8 | 14 | 14 | - | 1 | - | 1 |
| C3H | 26 | 20 | 6 | 23 | 17 | 6 | 3 | 2 | 1 |
| GARP | 21 | 8 | 13 | 17 | 7 | 10 | - | - | - |
| GNAT | 8 | 2 | 6 | 2 | 1 | 1 | - | - | - |
| GRAS | 14 | 12 | 2 | 12 | 7 | 5 | 2 | 1 | 1 |
| HB | 31 | 17 | 14 | 39 | 25 | 14 | 6 | 2 | 4 |
| MADS | 11 | 5 | 6 | 9 | 5 | 4 | 1 | - | 1 |
| MYB | 44 | 17 | 27 | 35 | 13 | 22 | 7 | - | 7 |
| NAC | 33 | 30 | 3 | 25 | 22 | 3 | 6 | 2 | 4 |
| PHD | 8 | 4 | 4 | 10 | 1 | 9 | - | - | - |
| PLATZ | 11 | 11 | - | 12 | 12 | - | 5 | 5 | - |
| Tify | 16 | 5 | 11 | 11 | - | 11 | 6 | - | 6 |
| Trihelix | 6 | 3 | 3 | 9 | 6 | 3 | 3 | 3 | - |
| WRKY | 37 | 32 | 5 | 24 | 18 | 6 | 13 | 6 | 7 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cui, X.; Deng, J.; Huang, C.; Tang, X.; Li, X.; Li, X.; Lu, J.; Zhang, Z. Transcriptomic Analysis of the Anthocyanin Biosynthetic Pathway Reveals the Molecular Mechanism Associated with Purple Color Formation in Dendrobium Nestor. Life 2021, 11, 113. https://doi.org/10.3390/life11020113
Cui X, Deng J, Huang C, Tang X, Li X, Li X, Lu J, Zhang Z. Transcriptomic Analysis of the Anthocyanin Biosynthetic Pathway Reveals the Molecular Mechanism Associated with Purple Color Formation in Dendrobium Nestor. Life. 2021; 11(2):113. https://doi.org/10.3390/life11020113
Chicago/Turabian StyleCui, Xueqiang, Jieling Deng, Changyan Huang, Xuan Tang, Xianmin Li, Xiuling Li, Jiashi Lu, and Zibin Zhang. 2021. "Transcriptomic Analysis of the Anthocyanin Biosynthetic Pathway Reveals the Molecular Mechanism Associated with Purple Color Formation in Dendrobium Nestor" Life 11, no. 2: 113. https://doi.org/10.3390/life11020113
APA StyleCui, X., Deng, J., Huang, C., Tang, X., Li, X., Li, X., Lu, J., & Zhang, Z. (2021). Transcriptomic Analysis of the Anthocyanin Biosynthetic Pathway Reveals the Molecular Mechanism Associated with Purple Color Formation in Dendrobium Nestor. Life, 11(2), 113. https://doi.org/10.3390/life11020113





