De Novo Transcriptome Profiling of Brain Tissue from the Annual Killifish Nothobranchius guentheri
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fish Diet and Maintenance
2.2. RNA Isolation, Library Preparation, and Transcriptome Sequencing
2.3. NGS Data Processing
3. Results
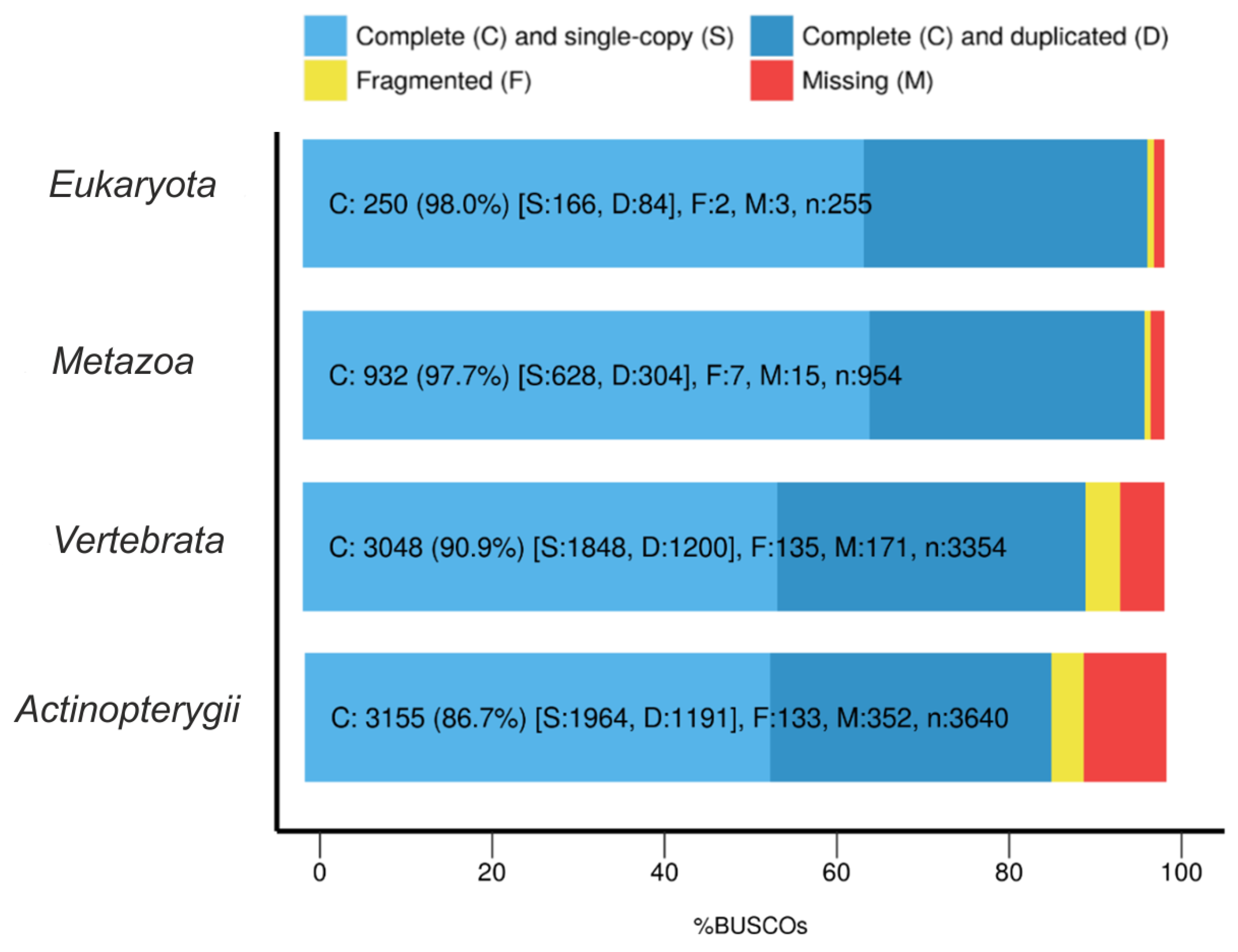
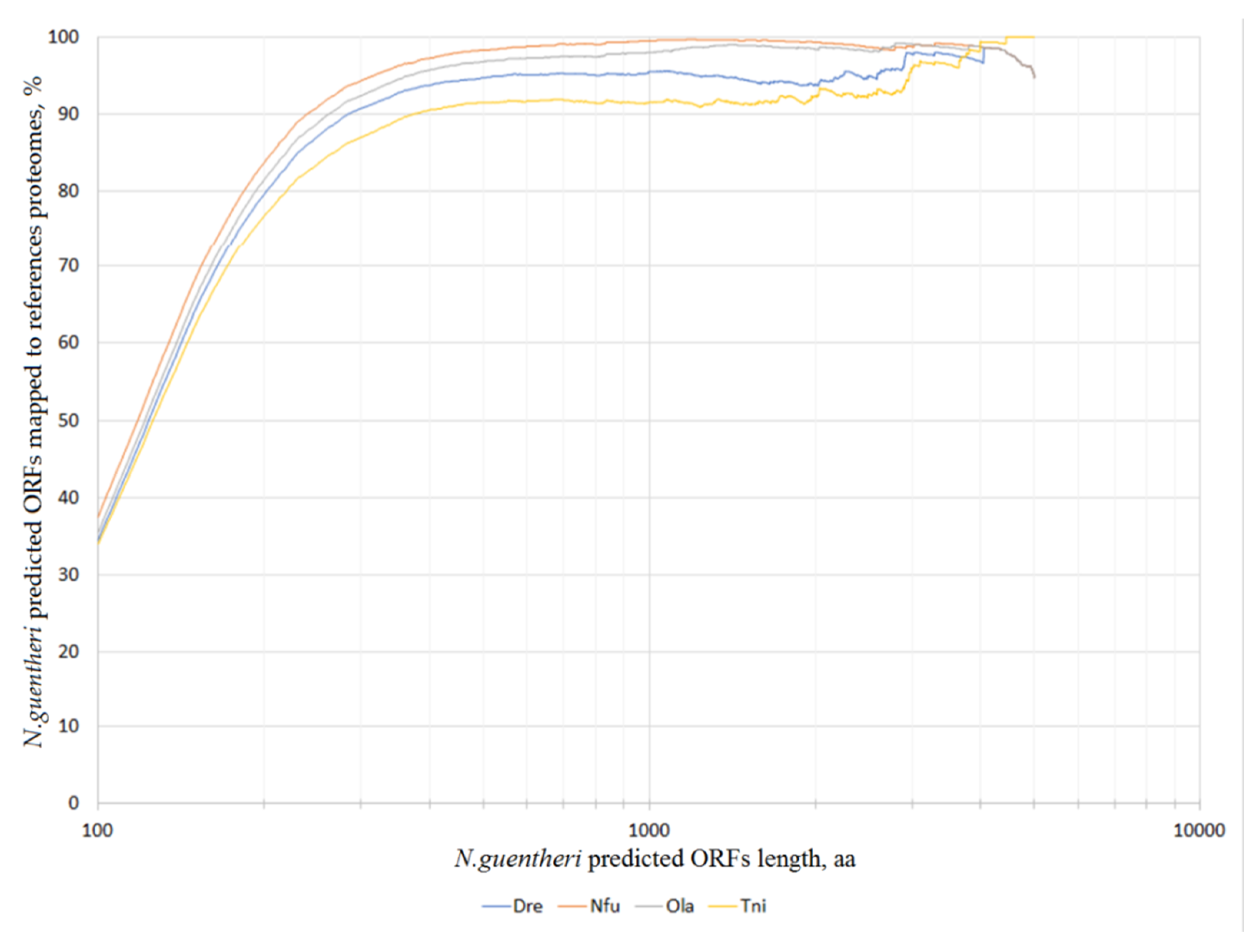
3.1. De Novo Transcriptome Assembly
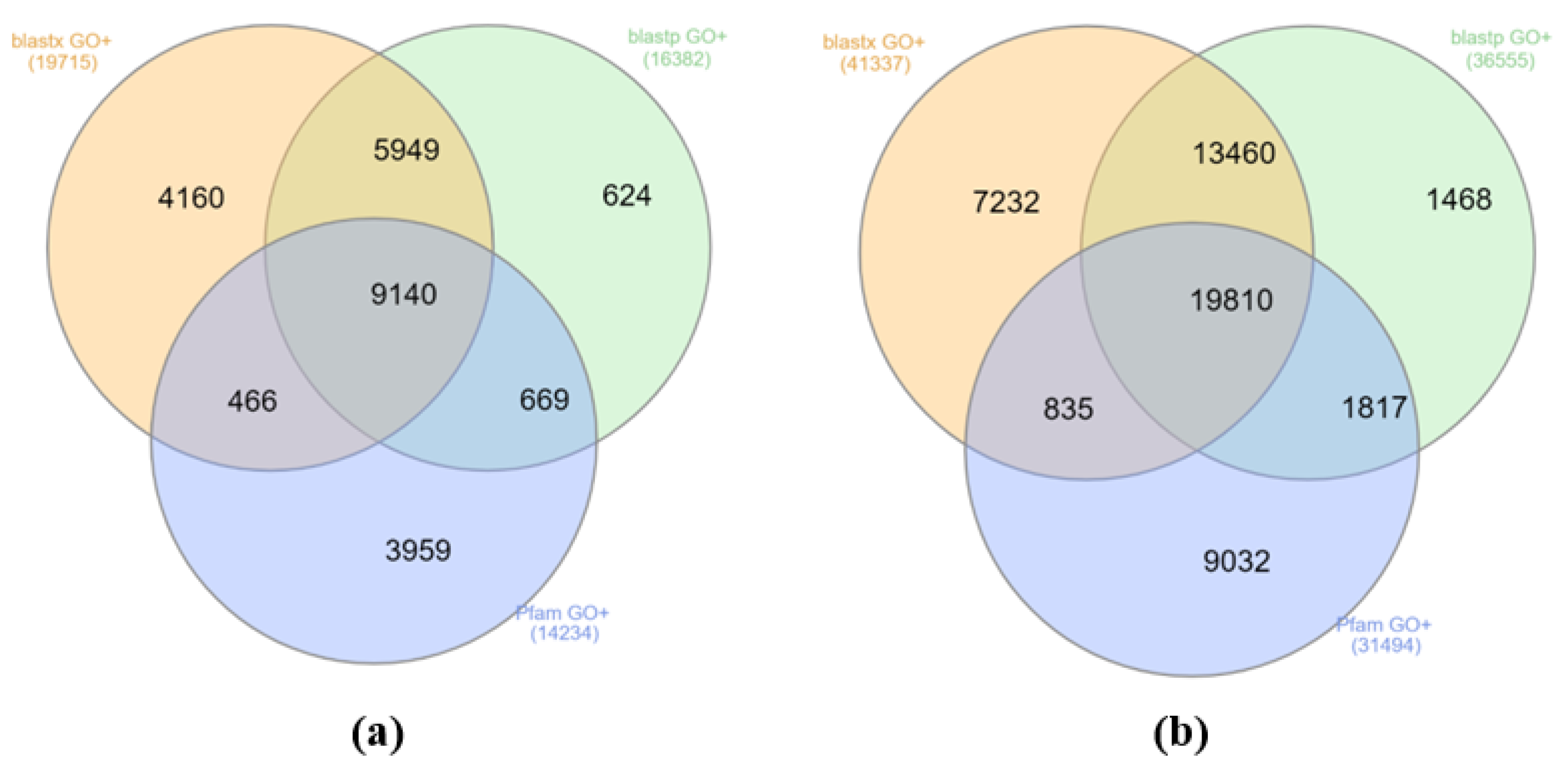
3.2. Transcriptome Annotation
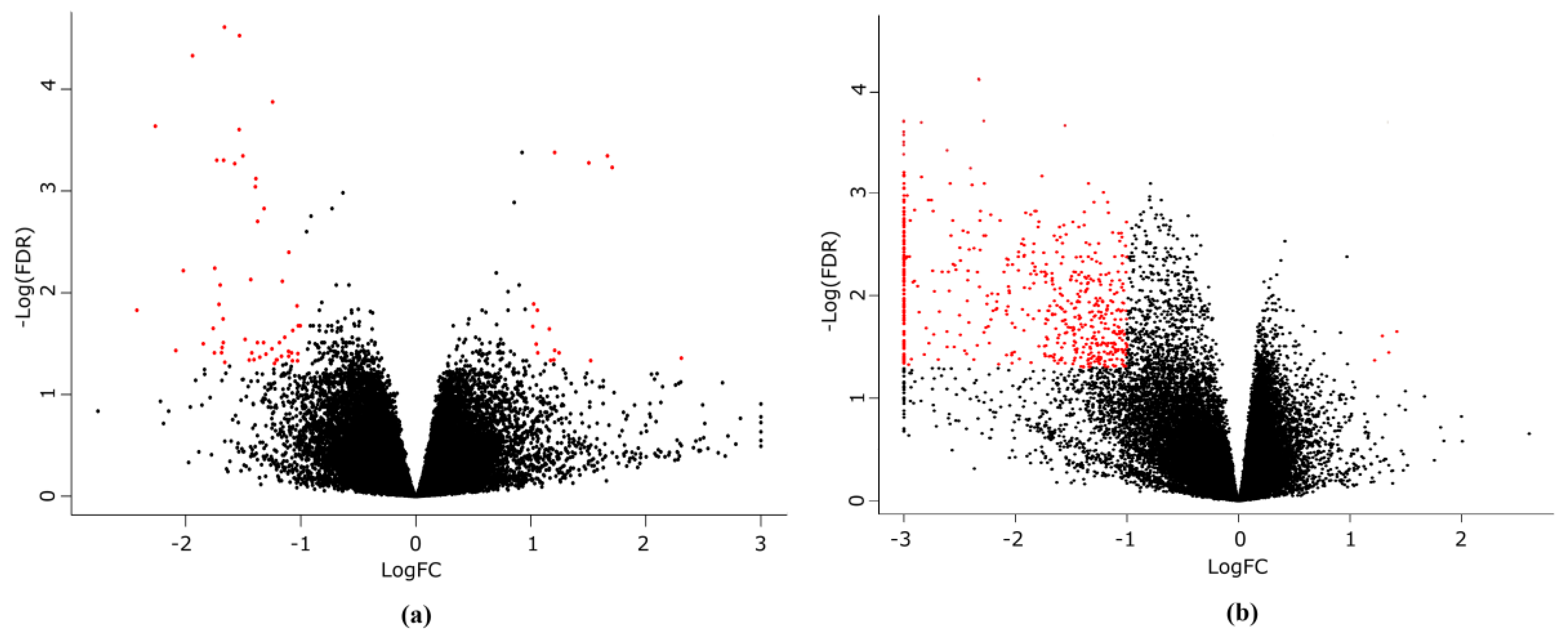
3.3. Gene Expression Changes Associated with the Torin 2 Treatment
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dilan Celebi-Birand, B.E.; Ozdemir, A.T.; Kafaligonul, H.; Adams, M. Zebra Aging Models and Possible Interventions. In Recent Advances in Zebrafish Researches; Bozkurt, Y., Ed.; IntechOpen: London, UK, 2018; Volume 1. [Google Scholar]
- Ding, L.; Kuhne, W.W.; Hinton, D.E.; Song, J.; Dynan, W.S. Quantifiable biomarkers of normal aging in the Japanese medaka fish (Oryzias latipes). PLoS ONE 2010, 5, e13287. [Google Scholar] [CrossRef] [PubMed]
- Conn, P.M. Conn’s Handbook of Models for Human Aging SECOND EDITION Preface. In Conn’s Handbook of Models for Human Aging, 2nd ed.; Academic Press Inc.: Cambridge, MA, USA, 2018; p. Xvii. [Google Scholar]
- Matrone, G.; Tucker, C.S.; Denvir, M.A. Cardiomyocyte proliferation in zebrafish and mammals: Lessons for human disease. Cell. Mol. Life Sci. Cmls 2017, 74, 1367–1378. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, R.; Strahle, U.; Scholpp, S. Neurogenesis in zebrafish—From embryo to adult. Neural Dev. 2013, 8, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kelmer Sacramento, E.; Kirkpatrick, J.M.; Mazzetto, M.; Baumgart, M.; Bartolome, A.; Di Sanzo, S.; Caterino, C.; Sanguanini, M.; Papaevgeniou, N.; Lefaki, M.; et al. Reduced proteasome activity in the aging brain results in ribosome stoichiometry loss and aggregation. Mol. Syst. Biol. 2020, 16, e9596. [Google Scholar] [CrossRef]
- Baumgart, M.; Groth, M.; Priebe, S.; Savino, A.; Testa, G.; Dix, A.; Ripa, R.; Spallotta, F.; Gaetano, C.; Ori, M.; et al. RNA-seq of the aging brain in the short-lived fish N. furzeri—conserved pathways and novel genes associated with neurogenesis. Aging Cell 2014, 13, 965–974. [Google Scholar] [CrossRef] [PubMed]
- Lucas-Sanchez, A.; Almaida-Pagan, P.F.; Mendiola, P.; de Costa, J. Nothobranchius as a model for aging studies. A review. Aging Dis. 2014, 5, 281–291. [Google Scholar] [CrossRef]
- Reichard, M.; Polacik, M. Nothobranchius furzeri, an ‘instant’ fish from an ephemeral habitat. eLife 2019, 8. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Shang, X.; Luan, J.; Zhang, S. Identification, expression and function of apolipoprotein E in annual fish Nothobranchius guentheri: Implication for an aging marker. Biogerontology 2014, 15, 233–243. [Google Scholar] [CrossRef]
- Liu, C.; Wang, X.; Feng, W.; Li, G.; Su, F.; Zhang, S. Differential expression of aging biomarkers at different life stages of the annual fish Nothobranchius guentheri. Biogerontology 2012, 13, 501–510. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Li, G. Effects of resveratrol on longevity, cognitive ability and aging-related histological markers in the annual fish Nothobranchius guentheri. Exp. Gerontol. 2012, 47, 940–949. [Google Scholar] [CrossRef]
- Dong, Y.; Cui, P.; Li, Z.; Zhang, S. Aging asymmetry: Systematic survey of changes in age-related biomarkers in the annual fish Nothobranchius guentheri. Fish Physiol. Biochem. 2017, 43, 309–319. [Google Scholar] [CrossRef] [PubMed]
- Saxton, R.A.; Sabatini, D.M. mTOR Signaling in Growth, Metabolism, and Disease. Cell 2017, 168, 960–976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bockaert, J.; Marin, P. mTOR in Brain Physiology and Pathologies. Physiol. Rev. 2015, 95, 1157–1187. [Google Scholar] [CrossRef] [PubMed]
- Ryskalin, L.; Lazzeri, G.; Flaibani, M.; Biagioni, F.; Gambardella, S.; Frati, A.; Fornai, F. mTOR-Dependent Cell Proliferation in the Brain. Biomed. Res. Int. 2017, 2017, 7082696. [Google Scholar] [CrossRef]
- Schenone, S.; Brullo, C.; Musumeci, F.; Radi, M.; Botta, M. ATP-competitive inhibitors of mTOR: An update. Curr. Med. Chem. 2011, 18, 2995–3014. [Google Scholar] [CrossRef]
- Bonazzi, S.; Goold, C.P.; Gray, A.; Thomsen, N.M.; Nunez, J.; Karki, R.G.; Gorde, A.; Biag, J.D.; Malik, H.A.; Sun, Y.; et al. Discovery of a Brain-Penetrant ATP-Competitive Inhibitor of the Mechanistic Target of Rapamycin (mTOR) for CNS Disorders. J. Med. Chem. 2020, 63, 1068–1083. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Wang, X.; Su, Z.; Fei, H.; Liu, X.; Pan, Q. The novel mTOR inhibitor Torin-2 induces autophagy and downregulates the expression of UHRF1 to suppress hepatocarcinoma cell growth. Oncol. Rep. 2015, 34, 1708–1716. [Google Scholar] [CrossRef] [Green Version]
- Garber, J.C.; Barbee, R.W. Guide for the Care and Use of Laboratory Animalsl: Eighth Edition; The National Academies Press: Washington, DC, USA, 2011. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [Green Version]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [Green Version]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [Green Version]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [Green Version]
- Waterhouse, R.M.; Seppey, M.; Simao, F.A.; Manni, M.; Ioannidis, P.; Klioutchnikov, G.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO Applications from Quality Assessments to Gene Prediction and Phylogenomics. Mol. Biol. Evol. 2018, 35, 543–548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sayers, E.W.; Agarwala, R.; Bolton, E.E.; Brister, J.R.; Canese, K.; Clark, K.; Connor, R.; Fiorini, N.; Funk, K.; Hefferon, T.; et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2019, 47, D23–D28. [Google Scholar] [CrossRef] [Green Version]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef] [PubMed]
- UniProt, C. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [Google Scholar] [CrossRef] [Green Version]
- Madden, T.L. The BLAST sequence analysis tool. In NCBI Handbook; McEntyre, J.O., Ed.; National Library of Medicine: Bethesda, MD, USA, 2005. [Google Scholar]
- Kroj, T.; Chanclud, E.; Michel-Romiti, C.; Grand, X.; Morel, J.B. Integration of decoy domains derived from protein targets of pathogen effectors into plant immune receptors is widespread. New Phytol. 2016, 210, 618–626. [Google Scholar] [CrossRef]
- Hayward, A.; Ghazal, A.; Andersson, G.; Andersson, L.; Jern, P. ZBED evolution: Repeated utilization of DNA transposons as regulators of diverse host functions. PLoS ONE 2013, 8, e59940. [Google Scholar] [CrossRef] [Green Version]
- Jha, M.K.; Morrison, B.M. Glia-neuron energy metabolism in health and diseases: New insights into the role of nervous system metabolic transporters. Exp. Neurol. 2018, 309, 23–31. [Google Scholar] [CrossRef]
- Wallace, R. High metabolic demand in neural tissues: Information and control theory perspectives on the synergism between rate and stability. J. Theor. Biol. 2016, 409, 86–96. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Guan, K.L. mTOR as a central hub of nutrient signalling and cell growth. Nat. Cell Biol. 2019, 21, 63–71. [Google Scholar] [CrossRef]
- LiCausi, F.; Hartman, N.W. Role of mTOR Complexes in Neurogenesis. Int. J. Mol. Sci. 2018, 19, 1544. [Google Scholar] [CrossRef] [Green Version]
- Swiech, L.; Perycz, M.; Malik, A.; Jaworski, J. Role of mTOR in physiology and pathology of the nervous system. Biochim. Et Biophys. Acta 2008, 1784, 116–132. [Google Scholar] [CrossRef] [PubMed]
- Perez-Alvarez, M.J.; Villa Gonzalez, M.; Benito-Cuesta, I.; Wandosell, F.G. Role of mTORC1 Controlling Proteostasis after Brain Ischemia. Front. Neurosci. 2018, 12, 60. [Google Scholar] [CrossRef] [PubMed]
- Oddo, S. The role of mTOR signaling in Alzheimer disease. Front. Biosci. 2012, 4, 941–952. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.G.; Du, J.H.; Lu, Y.; Lin, X.J. Neuroprotective effects of rapamycin on spinal cord injury in rats by increasing autophagy and Akt signaling. Neural Regen. Res. 2019, 14, 721–727. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Yang, C.; Iyaswamy, A.; Krishnamoorthi, S.; Sreenivasmurthy, S.G.; Liu, J.; Wang, Z.; Tong, B.C.; Song, J.; Lu, J.; et al. Balancing mTOR Signaling and Autophagy in the Treatment of Parkinson’s Disease. Int. J. Mol. Sci. 2019, 20, 728. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dunlop, E.A.; Tee, A.R. mTOR and autophagy: A dynamic relationship governed by nutrients and energy. Semin. Cell Dev. Biol. 2014, 36, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Bromme, D.; Panwar, P.; Turan, S. Cathepsin K osteoporosis trials, pycnodysostosis and mouse deficiency models: Commonalities and differences. Expert Opin. Drug Discov. 2016, 11, 457–472. [Google Scholar] [CrossRef] [PubMed]
- Haeckel, C.; Krueger, S.; Buehling, F.; Broemme, D.; Franke, K.; Schuetze, A.; Roese, I.; Roessner, A. Expression of cathepsin K in the human embryo and fetus. Dev. Dyn. Off. Publ. Am. Assoc. Anat. 1999, 216, 89–95. [Google Scholar] [CrossRef]
- Bernstein, H.G.; Bukowska, A.; Dobrowolny, H.; Bogerts, B.; Lendeckel, U. Cathepsin K and schizophrenia. Synapse 2007, 61, 252–253. [Google Scholar] [CrossRef]
- Dauth, S.; Sirbulescu, R.F.; Jordans, S.; Rehders, M.; Avena, L.; Oswald, J.; Lerchl, A.; Saftig, P.; Brix, K. Cathepsin K deficiency in mice induces structural and metabolic changes in the central nervous system that are associated with learning and memory deficits. BMC Neurosci. 2011, 12, 74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wendt, W.; Lubbert, H.; Stichel, C.C. Upregulation of cathepsin S in the aging and pathological nervous system of mice. Brain Res. 2008, 1232, 7–20. [Google Scholar] [CrossRef]
- Stolp, H.B.; Liddelow, S.A.; Sa-Pereira, I.; Dziegielewska, K.M.; Saunders, N.R. Immune responses at brain barriers and implications for brain development and neurological function in later life. Front. Integr. Neurosci. 2013, 7, 61. [Google Scholar] [CrossRef] [Green Version]
- Powell, J.D.; Pollizzi, K.N.; Heikamp, E.B.; Horton, M.R. Regulation of immune responses by mTOR. Annu. Rev. Immunol. 2012, 30, 39–68. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Araki, K.; Turner, A.P.; Shaffer, V.O.; Gangappa, S.; Keller, S.A.; Bachmann, M.F.; Larsen, C.P.; Ahmed, R. mTOR regulates memory CD8 T-cell differentiation. Nature 2009, 460, 108–112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pearce, E.L.; Walsh, M.C.; Cejas, P.J.; Harms, G.M.; Shen, H.; Wang, L.S.; Jones, R.G.; Choi, Y. Enhancing CD8 T-cell memory by modulating fatty acid metabolism. Nature 2009, 460, 103–107. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Araki, K.; Li, S.; Han, J.H.; Ye, L.; Tan, W.G.; Konieczny, B.T.; Bruinsma, M.W.; Martinez, J.; Pearce, E.L.; et al. Autophagy is essential for effector CD8(+) T cell survival and memory formation. Nat. Immunol. 2014, 15, 1152–1161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W.; Prazak, L.; Chatterjee, N.; Gruninger, S.; Krug, L.; Theodorou, D.; Dubnau, J. Activation of transposable elements during aging and neuronal decline in Drosophila. Nat. Neurosci. 2013, 16, 529–531. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jonsson, M.E.; Garza, R.; Johansson, P.A.; Jakobsson, J. Transposable Elements: A Common Feature of Neurodevelopmental and Neurodegenerative Disorders. Trends Genet. Tig. 2020, 36, 610–623. [Google Scholar] [CrossRef]
- Suarez, N.A.; Macia, A.; Muotri, A.R. LINE-1 retrotransposons in healthy and diseased human brain. Dev. Neurobiol. 2018, 78, 434–455. [Google Scholar] [CrossRef] [PubMed]
- Bodea, G.O.; McKelvey, E.G.Z.; Faulkner, G.J. Retrotransposon-induced mosaicism in the neural genome. Open Biol. 2018, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Woolfrey, K.M.; Srivastava, D.P. Control of Dendritic Spine Morphological and Functional Plasticity by Small GTPases. Neural Plast. 2016, 2016, 3025948. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Weiner, J.A.; Levi, S.; Craig, A.M.; Bradley, A.; Sanes, J.R. Gamma protocadherins are required for survival of spinal interneurons. Neuron 2002, 36, 843–854. [Google Scholar] [CrossRef] [Green Version]
- Prasad, T.; Wang, X.; Gray, P.A.; Weiner, J.A. A differential developmental pattern of spinal interneuron apoptosis during synaptogenesis: Insights from genetic analyses of the protocadherin-gamma gene cluster. Development 2008, 135, 4153–4164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weiner, J.A.; Wang, X.; Tapia, J.C.; Sanes, J.R. Gamma protocadherins are required for synaptic development in the spinal cord. Proc. Natl. Acad. Sci. USA 2005, 102, 8–14. [Google Scholar] [CrossRef] [Green Version]
- Garrett, A.M.; Schreiner, D.; Lobas, M.A.; Weiner, J.A. gamma-protocadherins control cortical dendrite arborization by regulating the activity of a FAK/PKC/MARCKS signaling pathway. Neuron 2012, 74, 269–276. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Feature | Value |
|---|---|
| total length, bp | 285,906,387 |
| total length, bp (only transcripts > 500 bp) | 217,410,128 |
| genes (any length) | 288,989 |
| genes (transcripts > 500 bp) | 104,271 |
| transcripts (any length) | 352,297 |
| transcripts > 500 bp | 127,262 |
| transcripts > 1000 bp | 66,471 |
| transcripts > 5000 bp | 6322 |
| transcripts > 10,000 bp | 604 |
| transcripts > 25,000 bp | 5 |
| largest transcript, bp | 27,376 |
| N50, bp | 2539 |
| N75, bp | 1231 |
| L50, bp | 24,581 |
| L75, bp | 55,134 |
| GC, % | 47.07 |
| Gene ID | Top BLAST Hit in UniProt | Gene Name | Control | Torin 2 | LogFC | LogCPM | FDR | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TRINITY_DN217860_c0_g1 | CATK | Cathepsin K | 1.05 | 4.1 | 0.032 | |||||||||||
| TRINITY_DN6195_c0_g1 | CATS | Cathepsin S | 1.20 | 4.8 | 0.046 | |||||||||||
| TRINITY_DN369_c0_g1 | HMR1 | Major histocompatibility complex class I-related gene protein | 1.16 | 6.6 | 0.023 | |||||||||||
| TRINITY_DN118978_c0_g1 | HG2A | HLA class II histocompatibility antigen gamma chain | 1.06 | 4.6 | 0.039 | |||||||||||
| TRINITY_DN217842_c0_g1 | DRA | Mamu class II histocompatibility antigen, DR alpha chain | 1.02 | 5.1 | 0.021 | |||||||||||
| TRINITY_DN6030_c0_g1 | CDN1B | Cyclin-dependent kinase inhibitor 1B | 1.21 | 4.9 | 0.037 | |||||||||||
| TRINITY_DN3092_c0_g1 | HEBP2 | Heme-binding protein 2 | −1.03 | 5.1 | 0.013 | |||||||||||
| TRINITY_DN17955_c0_g1 | PROD | Proline dehydrogenase 1 | −0.85 | 4.3 | 0.046 | |||||||||||
| TRINITY_DN121258_c0_g1 | PROF1 | Profilin-1 | 1.25 | 5.3 | 0.039 | |||||||||||
| TRINITY_DN10729_c0_g1 | SMU1 | WD40 repeat-containing protein SMU1 | −1.57 | 3.8 | 0.001 | |||||||||||
| TRINITY_DN151812_c0_g1 | TEF | Transcription factor VBP | −1.53 | 4.8 | 0.000 | |||||||||||
| TRINITY_DN918_c0_g1 | BHE41 | Class E basic helix-loop-helix protein 41 | −1.50 | 5.2 | 0.000 | |||||||||||
| TRINITY_DN217450_c0_g1 | CIART | Circadian-associated transcriptional repressor | −1.54 | 3.2 | 0.000 | |||||||||||
| TRINITY_DN6901_c0_g1 | CIPC | CLOCK-interacting pacemaker | −1.11 | 3.2 | 0.038 | |||||||||||
| TRINITY_DN13536_c0_g1 | DBP | D site-binding protein | −1.25 | 4.8 | 0.000 | |||||||||||
| TRINITY_DN3883_c0_g1 | NFIL3 | Nuclear factor interleukin-3-regulated protein | 1.71 | 4.4 | 0.001 | |||||||||||
| TRINITY_DN221983_c0_g1 | NR1D1 | Nuclear receptor subfamily 1 group D member 1 | −1.67 | 4.0 | 0.000 | |||||||||||
| TRINITY_DN2234_c0_g1 | NR1D2 | Nuclear receptor subfamily 1 group D member 2 | −0.91 | 5.9 | 0.002 | |||||||||||
| TRINITY_DN13399_c0_g1 | PER1 | Period circadian protein homolog 1 | −1.39 | 4.2 | 0.001 | |||||||||||
| TRINITY_DN12790_c0_g1 | PER2 | Period circadian protein homolog 2 | −1.32 | 4.5 | 0.001 | |||||||||||
| TRINITY_DN2073_c0_g1 | RORB | Nuclear receptor ROR-beta | 1.21 | 5.5 | 0.000 | |||||||||||
| Gene ID | Top BLAST Hit in UniProt | Gene Name | Control | Torin 2 | LogFC | LogCPM | FDRvalue | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TRINITY_DN9769_c0_g1 | PCDB1 | Protocadherin beta 1 | −1.01 | 3.9 | 0.003 | ||||||||||
| TRINITY_DN30433_c0_g1 | PCDHGB | Protocadherin gamma subfamily B | −0.92 | 2.9 | 0.002 | ||||||||||
| TRINITY_DN29438_c0_g1 | PCDHGA | Protocadherin gamma subfamily A | −1.85 | 1.8 | 0.005 | ||||||||||
| TRINITY_DN221187_c0_g1 | PCDHD2 | Protocadherin delta 2 | −1.79 | 2.2 | 0.002 | ||||||||||
| TRINITY_DN10099_c0_g1 | PCDHGC | Protocadherin gamma subfamily C | −1.43 | 3.1 | 0.002 | ||||||||||
| TRINITY_DN5709_c0_g3 | ZBED1 | Zinc finger BED domain-containing protein 1 | −1.21 | 4.4 | 0.0010 | ||||||||||
| TRINITY_DN32433_c0_g1 | ZBED4 | Zinc finger BED domain-containing protein 4 | −1.53 | 1.6 | 0.01 | ||||||||||
| TRINITY_DN11566_c0_g2 | ZBED9 | SCAN domain-containing protein 3 | −1.22 | 4.8 | 0.01 | ||||||||||
| TRINITY_DN28479_c0_g1 | LITD1 | LINE-1 type transposase domain-containing protein 1 | −2.22 | 1.5 | 0.002 | ||||||||||
| TRINITY_DN5063_c0_g1 | PEG10 | Retrotransposon-derived protein PEG10 | −1.37 | 4.0 | 0.009 | ||||||||||
| TRINITY_DN6282_c0_g1 | RTL1 | Retrotransposon-like protein 1 | −1.45 | 4.2 | 0.007 | ||||||||||
| TRINITY_DN4053_c0_g1 | YTX1 | Transposon TX1 uncharacterized 149 kDa protein | −1.37 | 3.7 | 0.003 | ||||||||||
| TRINITY_DN18778_c0_g1 | PO22 | Retrovirus-related Pol polyprotein from type-1 retrotransposable element R2 | −0.91 | 2.8 | 0.05 | ||||||||||
| TRINITY_DN20585_c0_g1 | FMRF | FMRF-amide neuropeptides | −2.88 | 4.5 | 0.008 | ||||||||||
| TRINITY_DN28618_c0_g1 | NEFH | Neurofilament heavy polypeptide | −1.05 | 2.8 | 0.002 | ||||||||||
| TRINITY_DN36218_c0_g1 | CSMD1 | CUB and sushi domain-containing protein 1 | −1.83 | 2.2 | 0.006 | ||||||||||
| TRINITY_DN20186_c0_g1 | DYH10 | Dynein heavy chain 10, axonemal | −1.32 | 2.4 | 0.004 | ||||||||||
| TRINITY_DN23456_c0_g1 | FR1L6 | Fer-1-like protein 6 | 1.35 | 2.2 | 0.04 | ||||||||||
| TRINITY_DN20670_c1_g1 | NMDE2 | Glutamate receptor ionotropic, NMDA 2B | −1.18 | 2.5 | 0.01 | ||||||||||
| TRINITY_DN19802_c0_g1 | SRFBP1 | Serum response factor-binding protein 1 | −3.95 | 2.5 | 0.01 | ||||||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guvatova, Z.G.; Fedorova, M.S.; Vershinina, Y.S.; Pudova, E.A.; Lipatova, A.V.; Volodin, V.V.; Gladysh, N.S.; Tokarev, A.T.; Kornev, A.B.; Pavlov, V.S.; et al. De Novo Transcriptome Profiling of Brain Tissue from the Annual Killifish Nothobranchius guentheri. Life 2021, 11, 137. https://doi.org/10.3390/life11020137
Guvatova ZG, Fedorova MS, Vershinina YS, Pudova EA, Lipatova AV, Volodin VV, Gladysh NS, Tokarev AT, Kornev AB, Pavlov VS, et al. De Novo Transcriptome Profiling of Brain Tissue from the Annual Killifish Nothobranchius guentheri. Life. 2021; 11(2):137. https://doi.org/10.3390/life11020137
Chicago/Turabian StyleGuvatova, Zulfiia G., Maria S. Fedorova, Yulia S. Vershinina, Elena A. Pudova, Anastasiya V. Lipatova, Vsevolod V. Volodin, Natalya S. Gladysh, Artemiy T. Tokarev, Alexey B. Kornev, Vladislav S. Pavlov, and et al. 2021. "De Novo Transcriptome Profiling of Brain Tissue from the Annual Killifish Nothobranchius guentheri" Life 11, no. 2: 137. https://doi.org/10.3390/life11020137
APA StyleGuvatova, Z. G., Fedorova, M. S., Vershinina, Y. S., Pudova, E. A., Lipatova, A. V., Volodin, V. V., Gladysh, N. S., Tokarev, A. T., Kornev, A. B., Pavlov, V. S., Bakhtogarimov, I. R., Krysanov, E. Y., Moskalev, A. A., Krasnov, G. S., & Kudryavtseva, A. V. (2021). De Novo Transcriptome Profiling of Brain Tissue from the Annual Killifish Nothobranchius guentheri. Life, 11(2), 137. https://doi.org/10.3390/life11020137






