The Emerging Physiological Role of AGMO 10 Years after Its Gene Identification
Abstract
:1. Introduction
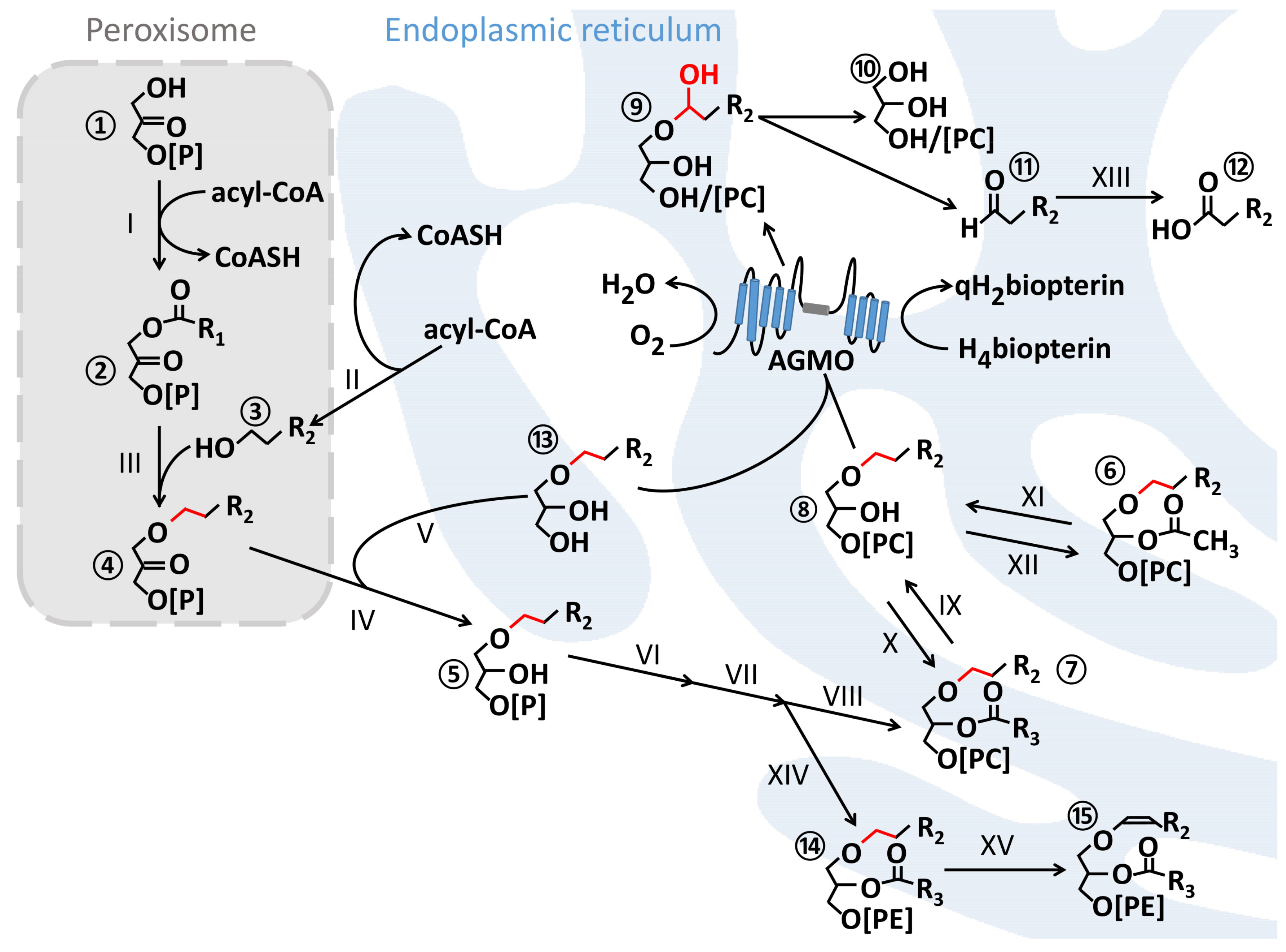
1.1. Ether Lipids and Their Biosynthesis
1.2. The AGMO Reaction
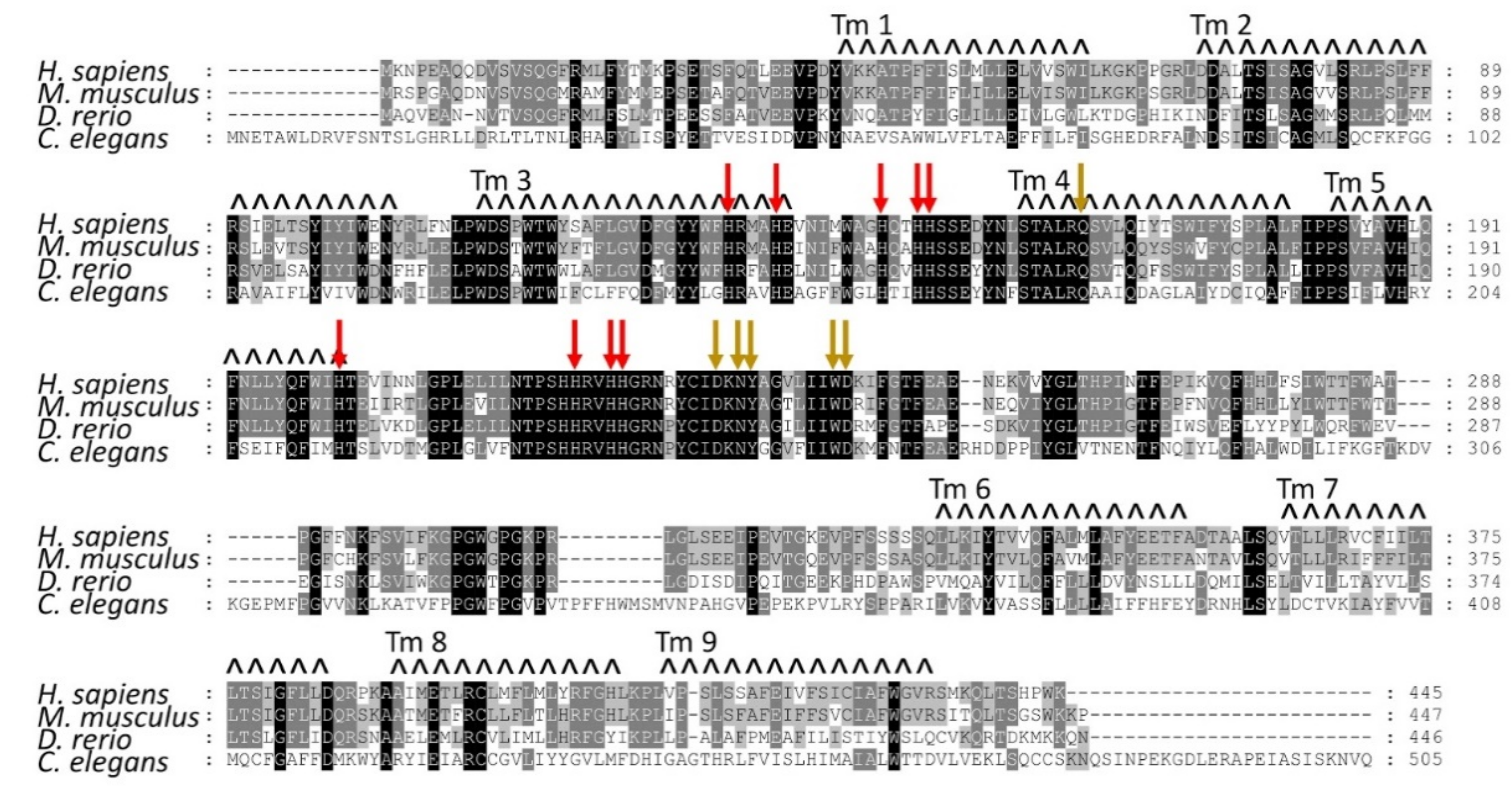
1.3. Structural Features of the AGMO Protein
2. Alkylglycerols
2.1. Occurrence and Properties of Alkylglycerols
2.2. Dietary Supplementation of Alkylglycerols
2.3. Interdependence of Ether Lipid and Sphingolipid Metabolism
2.4. Analytics of Ether Lipids
2.5. Discrimination of 1-O-Alkyl- and 1-O-Alk-1′-Enyl-Lipids
3. AGMO in Human Diseases
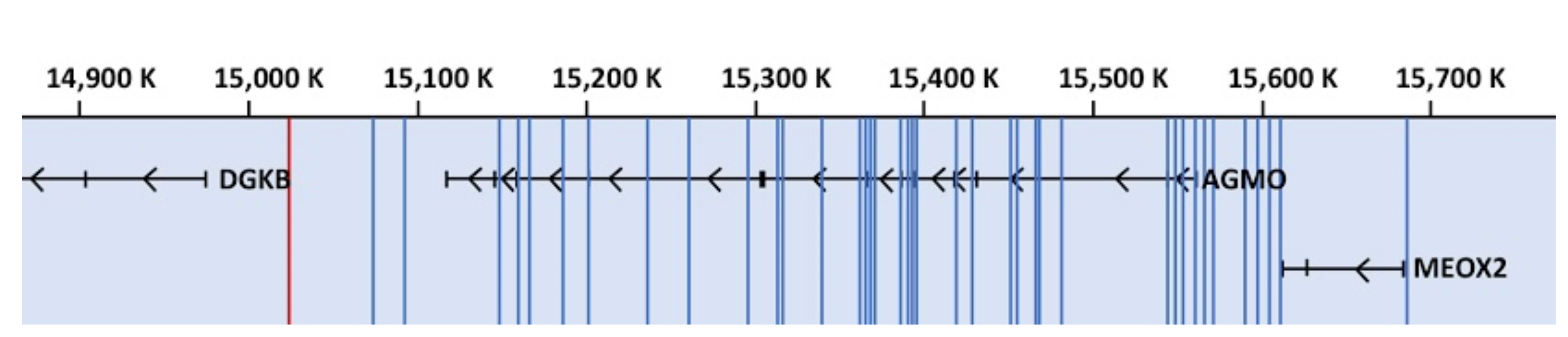
3.1. Genome-Wide Association Studies
3.2. Mutations, Copy Number Variations, and Deletions in the AGMO Gene
3.2.1. Autism Spectrum Disorders
3.2.2. Microcephaly
3.2.3. Neurodevelopmental Disorders
3.2.4. Inflammation
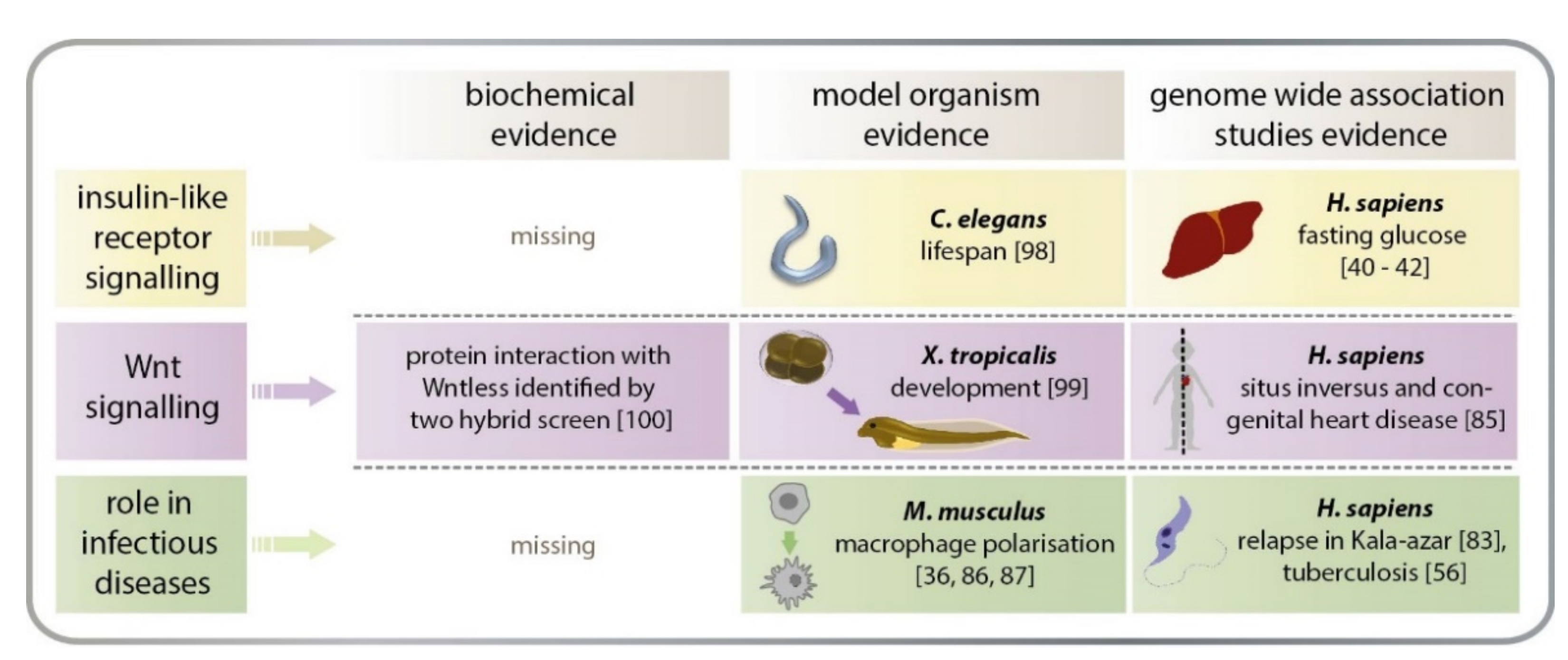
3.2.5. Heterotaxy
4. AGMO in Model Organisms
4.1. Mouse (Mus Musculus)
4.1.1. AGMO in Macrophage Polarisation
4.1.2. AGMO in Experimental Colitis
4.1.3. Mouse Models Deficient in the AGMO Cofactor Tetrahydrobiopterin
4.2. The Nematode Caenorhabditis Elegans
4.2.1. Importance of AGMO for Cuticle Stability
4.2.2. Role of AGMO in Insulin-Like Signalling
4.3. The Clawed Frog Xenopus tropicalis
4.4. Chicken (Gallus Gallus)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Tietz, A.; Lindberg, M.; Kennedy, E.P. A new pteridine-requiring enzyme system for the oxidation of glyceryl ethers. J. Biol. Chem. 1964, 239, 4081–4090. [Google Scholar] [CrossRef]
- Taguchi, H.; Kosar-Hashemi, B.; Paal, B.; Yang, N.; Armarego, W.L. Glyceryl-Ether Monooxygenase (EC 1.14.16.5): Nature of the Glyceryl-Ether Lipid Substrates in Aqueous Buffer. Biol. Chem. Hoppe Seyler 1994, 375, 329–334. [Google Scholar] [CrossRef]
- Taguchi, H.; Paal, B.; Armarego, W.L.F. Glyceryl-Ether Monooxygenase [EC 1.14.16.5J Part VIII. Probing the Nature of the Active Site. Pteridines 1995, 6, 45–57. [Google Scholar] [CrossRef]
- Taguchi, H.; Armarego, W.L. Glyceryl-ether monooxygenase [EC 1.14.16.5]. A microsomal enzyme of ether lipid metabolism. Med. Res. Rev. 1998, 18, 43–89. [Google Scholar] [CrossRef]
- Soodsma, J.F.; Piantadosi, C.; Snyder, F. Partial characterization of the alkylglycerol cleavage enzyme system of rat liver. J. Biol. Chem. 1972, 247, 3923–3929. [Google Scholar] [CrossRef]
- Snyder, F.; Malone, B.; Piantadosi, C. Tetrahydropteridine-dependent cleavage enzyme for O-alkyl lipids: Substrate specificity. Biochim. Biophys. Acta 1973, 316, 259–265. [Google Scholar] [CrossRef]
- Rock, C.O.; Baker, R.C.; Fitzgerald, V.; Snyder, F. Stimulation of the microsomal alkylglycerol monooxygenase by catalase. Biochim. Biophys. Acta 1976, 450, 469–473. [Google Scholar] [CrossRef]
- Taguchi, H.; Paal, B.; Armarego, W.L.F. Glyceryl-ether monooxygenase [EC 1.14.16.5]. Part 9. Stereospecificity of the oxygenase reaction. J. Chem. Soc. Perkin Trans. 1 1997. [Google Scholar] [CrossRef]
- Watschinger, K.; Keller, M.A.; Golderer, G.; Hermann, M.; Maglione, M.; Sarg, B.; Lindner, H.H.; Hermetter, A.; Werner-Felmayer, G.; Konrat, R.; et al. Identification of the gene encoding alkylglycerol monooxygenase defines a third class of tetrahydrobiopterin-dependent enzymes. Proc. Natl. Acad. Sci. USA 2010, 107, 13672–13677. [Google Scholar]
- Dorninger, F.; Forss-Petter, S.; Wimmer, I.; Bergera, J. Plasmalogens, platelet-activating factor and beyond—Ether lipids in signaling and neurodegeneration. Neurobiol. Dis. 2020, 145, 105061. [Google Scholar] [CrossRef]
- Dean, J.M.; Lodhi, I.J. Structural and functional roles of ether lipids. Protein Cell 2018, 9, 196–206. [Google Scholar] [CrossRef]
- Gallego-García, A.; Monera-Girona, A.J.; Pajares-Martínez, E.; Bastida-Martínez, E.; Pérez-Castaño, R.; Iniesta, A.A.; Fontes, M.; Padmanabhan, S.; Elías-Arnanz, M. A bacterial light response reveals an orphan desaturase for human plasmalogen synthesis. Science 2019, 366, 128–132. [Google Scholar] [CrossRef]
- Werner, E.R.; Keller, M.A.; Sailer, S.; Lackner, K.; Koch, J.; Hermann, M.; Coassin, S.; Golderer, G.; Werner-Felmayer, G.; Zoeller, R.A.; et al. The gene encodes plasmanylethanolamine desaturase which introduces the characteristic vinyl ether double bond into plasmalogens. Proc. Natl. Acad. Sci. USA 2020, 117, 7792–7798. [Google Scholar]
- De Rond, T.; Stow, P.; Eigl, I.; Johnson, R.E.; Chan, L.J.G.; Goyal, G.; Baidoo, E.E.K.; Hillson, N.J.; Petzold, C.J.; Sarpong, R.; et al. Oxidative cyclization of prodigiosin by an alkylglycerol monooxygenase-like enzyme. Nat. Chem. Biol. 2017, 13, 1155–1157. [Google Scholar] [CrossRef] [Green Version]
- Keller, M.A.; Zander, U.; Fuchs, J.E.; Kreutz, C.; Watschinger, K.; Mueller, T.; Golderer, G.; Liedl, K.R.; Ralser, M.; Kräutler, B.; et al. A gatekeeper helix determines the substrate specificity of Sjögren–Larsson Syndrome enzyme fatty aldehyde dehydrogenase. Nat. Commun. 2014, 5, 4439. [Google Scholar] [CrossRef] [Green Version]
- Watschinger, K.; Fuchs, J.E.; Yarov-Yarovoy, V.; Keller, M.A.; Golderer, G.; Hermetter, A.; Werner-Felmayer, G.; Hulo, N.; Werner, E.R. Catalytic residues and a predicted structure of tetrahydrobiopterin-dependent alkylglycerol mono-oxygenase. Biochem. J. 2012, 443, 279–286. [Google Scholar] [CrossRef] [Green Version]
- Ishibashi, T.; Imai, Y. Solubilization and partial characterization of alkylglycerol monooxygenase from rat liver microsomes. Eur. J. Biochem. 1983, 132, 23–27. [Google Scholar] [CrossRef]
- Watschinger, K.; Werner, E.R. Alkylglycerol monooxygenase. IUBMB Life 2013, 65, 366–372. [Google Scholar] [CrossRef] [Green Version]
- Magnusson, C.D.; Haraldsson, G.G. Ether lipids. Chem. Phys. Lipids 2011, 164, 315–340. [Google Scholar] [CrossRef]
- Jiménez-Rojo, N.; Riezman, H. On the road to unraveling the molecular functions of ether lipids. FEBS Lett. 2019, 593, 2378–2389. [Google Scholar] [CrossRef] [Green Version]
- Bada Juarez, J.F.; O’Rourke, D.; Judge, P.J.; Liu, L.C.; Hodgkin, J.; Watts, A. Lipodisqs for eukaryote lipidomics with retention of viability: Sensitivity and resistance to Leucobacter infection linked to C.elegans cuticle composition. Chem. Phys. Lipids 2019, 222, 51–58. [Google Scholar] [CrossRef]
- Iannitti, T.; Palmieri, B. An Update on the Therapeutic Role of Alkylglycerols. Mar. Drugs 2010, 8, 2267–2300. [Google Scholar] [CrossRef] [Green Version]
- Yamamoto, N.; Ngwenya, B.Z. Activation of mouse peritoneal macrophages by lysophospholipids and ether derivatives of neutral lipids and phospholipids. Cancer Res. 1987, 47, 2008–2013. Available online: https://www.ncbi.nlm.nih.gov/pubmed/2950993 (accessed on 24 January 2021).
- Pédrono, F.; Martin, B.; LeDuc, C.; Le Lan, J.; Saiag, B.; Legrand, P.; Moulinoux, J.-P.; Legrand, A.B. Natural Alkylglycerols Restrain Growth and Metastasis of Grafted Tumors in Mice. Nutr. Cancer 2004, 48, 64–69. [Google Scholar] [CrossRef]
- Erdlenbruch, B.; Alipour, M.; Fricker, G.; Miller, D.S.; Kugler, W.; Eibl, H.; Lakomek, M. Alkylglycerol opening of the blood-brain barrier to small and large fluorescence markers in normal and C6 glioma-bearing rats and isolated rat brain capillaries. Br. J. Pharmacol. 2003, 140, 1201–1210. [Google Scholar] [CrossRef] [Green Version]
- Haynes, M.P.; Buckley, H.R.; Higgins, M.L.; Pieringer, R.A. Synergism between the antifungal agents amphotericin B and alkyl glycerol ethers. Antimicrob. Agents Chemother. 1994, 38, 1523–1529. [Google Scholar] [CrossRef] [Green Version]
- Zhang, M.; Sun, S.; Tang, N.; Cai, W.; Qian, L. Oral Administration of Alkylglycerols Differentially Modulates High-Fat Diet-Induced Obesity and Insulin Resistance in Mice. Evid. Based. Complement. Altern. Med. 2013, 2013, 1–11. [Google Scholar] [CrossRef]
- Parri, A.; Fitó, M.; Torres, C.F.; Muñoz-Aguayo, D.; Schröder, H.; Cano, J.F.; Vázquez, L.; Reglero, G.; Covas, M.-I. Alkylglycerols reduce serum complement and plasma vascular endothelial growth factor in obese individuals. Inflammopharmacology 2016, 24, 127–131. [Google Scholar] [CrossRef] [Green Version]
- Yu, H.; Dilbaz, S.; Coßmann, J.; Hoang, A.C.; Diedrich, V.; Herwig, A.; Harauma, A.; Hoshi, Y.; Moriguchi, T.; Landgraf, K.; et al. Breast milk alkylglycerols sustain beige adipocytes through adipose tissue macrophages. J. Clin. Investig. 2019, 129, 2485–2499. [Google Scholar] [CrossRef] [Green Version]
- Jiménez-Rojo, N.; Leonetti, M.D.; Zoni, V.; Colom, A.; Feng, S.; Iyengar, N.R.; Matile, S.; Roux, A.; Vanni, S.; Weissman, J.S.; et al. Conserved Functions of Ether Lipids and Sphingolipids in the Early Secretory Pathway. Curr. Biol. 2020, 30, 3775–3787. [Google Scholar] [CrossRef]
- Liaw, L.; Prudovsky, I.; Koza, R.A.; Anunciado-Koza, R.V.; Siviski, M.E.; Lindner, V.; Friesel, R.E.; Rosen, C.J.; Baker, P.R.S.; Simons, B.; et al. Lipid Profiling of In Vitro Cell Models of Adipogenic Differentiation: Relationships with Mouse Adipose Tissues. J. Cell. Biochem. 2016, 117, 2182–2193. [Google Scholar] [CrossRef] [Green Version]
- Bartz, R.; Li, W.-H.; Venables, B.; Zehmer, J.K.; Roth, M.R.; Welti, R.; Anderson, R.G.W.; Liu, P.; Chapman, K.D. Lipidomics reveals that adiposomes store ether lipids and mediate phospholipid traffic. J. Lipid Res. 2007, 48, 837–847. [Google Scholar] [CrossRef] [Green Version]
- Werner, E.R.; Hermetter, A.; Prast, H.; Golderer, G.; Werner-Felmayer, G. Widespread occurrence of glyceryl ether monooxygenase activity in rat tissues detected by a novel assay. J. Lipid Res. 2007, 48, 1422–1427. [Google Scholar] [CrossRef] [Green Version]
- Owens, K. A two-dimensional thin-layer chromatographic procedure for the estimation of plasmalogens. Biochem. J. 1966, 100, 354–361. [Google Scholar] [CrossRef] [Green Version]
- Holčapek, M.; Liebisch, G.; Ekroos, K. Lipidomic Analysis. Anal. Chem. 2018, 90, 4249–4257. [Google Scholar] [CrossRef] [Green Version]
- Watschinger, K.; Keller, M.A.; McNeill, E.; Alam, M.T.; Lai, S.; Sailer, S.; Rauch, V.; Patel, J.; Hermetter, A.; Golderer, G.; et al. Tetrahydrobiopterin and alkylglycerol monooxygenase substantially alter the murine macrophage lipidome. Proc. Natl. Acad. Sci. USA 2015, 112, 2431–2436. [Google Scholar]
- Hsu, F.-F.; Turk, J.; Thukkani, A.K.; Messner, M.C.; Wildsmith, K.R.; Ford, D.A. Characterization of alkylacyl, alk-1-enylacyl and lyso subclasses of glycerophosphocholine by tandem quadrupole mass spectrometry with electrospray ionization. J. Mass Spectrom. 2003, 38, 752–763. [Google Scholar] [CrossRef]
- Hsu, F.-F.; Turk, J. Differentiation of 1-O-alk-1′-enyl-2-acyl and 1-O-alkyl-2-acyl Glycerophospholipids by Multiple-Stage Linear Ion-Trap Mass Spectrometry with Electrospray Ionization. J. Am. Soc. Mass Spectrom. 2007, 18, 2065–2073. [Google Scholar] [CrossRef] [Green Version]
- Koch, J.; Lackner, K.; Wohlfarter, Y.; Sailer, S.; Zschocke, J.; Werner, E.R.; Watschinger, K.; Keller, M.A. Unequivocal Mapping of Molecular Ether Lipid Species by LC–MS/MS in Plasmalogen-Deficient Mice. Anal. Chem. 2020, 92, 11268–11276. [Google Scholar] [CrossRef]
- Dupuis, J.; Langenberg, C.; Prokopenko, I.; Saxena, R.; Soranzo, N.; Jackson, A.U.; Wheeler, E.; Glazer, N.L.; Bouatia-Naji, N.; Gloyn, A.L.; et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat. Genet. 2010, 42, 105–116. [Google Scholar] [CrossRef]
- Boesgaard, T.W.; Meta-Analysis of Glucose and Insulin-Related Trait Consortium (MAGIC); Grarup, N.; Jørgensen, T.; Borch-Johnsen, K.; Hansen, T.; Pedersen, O. Variants at DGKB/TMEM195, ADRA2A, GLIS3 and C2CD4B loci are associated with reduced glucose-stimulated beta cell function in middle-aged Danish people. Diabetologia 2010, 53, 1647–1655. [Google Scholar] [CrossRef] [Green Version]
- Fontaine-Bisson, B.; The MAGIC investigators; Renström, F.; Rolandsson, O.; Payne, F.; Hallmans, G.; Barroso, I.; Franks, P.W. Evaluating the discriminative power of multi-trait genetic risk scores for type 2 diabetes in a northern Swedish population. Diabetologia 2010, 53, 2155–2162. [Google Scholar] [CrossRef] [Green Version]
- Jung, S.Y.; Sobel, E.M.; Papp, J.C.; Zhang, Z.-F. Effect of genetic variants and traits related to glucose metabolism and their interaction with obesity on breast and colorectal cancer risk among postmenopausal women. BMC Cancer 2017, 17, 290. [Google Scholar] [CrossRef]
- Manning, A.K.; Hivert, M.-F.; Scott, R.A.; Grimsby, J.L.; Bouatia-Naji, N.; Chen, H.; Rybin, D.; Liu, C.-T.; Bielak, L.F.; Prokopenko, I.; et al. A genome-wide approach accounting for body mass index identifies genetic variants influencing fasting glycemic traits and insulin resistance. Nat. Genet. 2012, 44, 659–669. [Google Scholar] [CrossRef]
- Wessel, J.; Chu, A.Y.; Willems, S.M.; Wang, S.; Yaghootkar, H.; Brody, J.A.; Dauriz, M.; Hivert, M.-F.; Raghavan, S.; Lipovich, L.; et al. Low-frequency and rare exome chip variants associate with fasting glucose and type 2 diabetes susceptibility. Nat. Commun. 2015, 6, 5897. [Google Scholar] [CrossRef] [Green Version]
- Wheeler, E.; Leong, A.; Liu, C.-T.; Hivert, M.-F.; Strawbridge, R.J.; Podmore, C.; Li, M.; Yao, J.; Sim, X.; Hong, J.; et al. Impact of common genetic determinants of Hemoglobin A1c on type 2 diabetes risk and diagnosis in ancestrally diverse populations: A transethnic genome-wide meta-analysis. PLoS Med. 2017, 14, e1002383. [Google Scholar] [CrossRef] [Green Version]
- Zhao, W.; Rasheed, A.; Tikkanen, E.; Lee, J.-J.; Butterworth, A.S.; Howson, J.M.M.; Assimes, T.L.; Chowdhury, R.; Orho-Melander, M.; Damrauer, S.; et al. Identification of new susceptibility loci for type 2 diabetes and shared etiological pathways with coronary heart disease. Nat. Genet. 2017, 49, 1450–1457. [Google Scholar] [CrossRef]
- Wojcik, G.L.; Graff, M.; Nishimura, K.K.; Tao, R.; Haessler, J.; Gignoux, C.R.; Highland, H.M.; Patel, Y.M.; Sorokin, E.P.; Avery, C.L.; et al. Genetic analyses of diverse populations improves discovery for complex traits. Nature 2019, 570, 514–518. [Google Scholar] [CrossRef]
- Vujkovic, M.; Keaton, J.M.; Lynch, J.A.; Miller, D.R.; Zhou, J.; Tcheandjieu, C.; Huffman, J.E.; Assimes, T.L.; Lorenz, K.; Zhu, X.; et al. Discovery of 318 new risk loci for type 2 diabetes and related vascular outcomes among 1.4 million participants in a multi-ancestry meta-analysis. Nat. Genet. 2020, 52, 680–691. [Google Scholar] [CrossRef]
- Akiyama, K.; Narita, A.; Nakaoka, H.; Cui, T.; Takahashi, T.; Yasuno, K.; Tajima, A.; Krischek, B.; Yamamoto, K.; Kasuya, H.; et al. Genome-wide association study to identify genetic variants present in Japanese patients harboring intracranial aneurysms. J. Hum. Genet. 2010, 55, 656–661. [Google Scholar] [CrossRef]
- Comuzzie, A.G.; Cole, S.A.; Laston, S.L.; Voruganti, V.S.; Haack, K.; Gibbs, R.A.; Butte, N.F. Novel Genetic Loci Identified for the Pathophysiology of Childhood Obesity in the Hispanic Population. PLoS ONE 2012, 7, e51954. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Z.; Guo, Y.; Shi, H.; Liu, C.-L.; Panganiban, R.A.; Chung, W.; O’Connor, L.J.; Himes, B.E.; Gazal, S.; Hasegawa, K.; et al. Shared genetic and experimental links between obesity-related traits and asthma subtypes in UK Biobank. J. Allergy Clin. Immunol. 2020, 145, 537–549. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Sun, M.; Adeyemo, A.; Pirie, F.; Carstensen, T.; Pomilla, C.; Doumatey, A.P.; Chen, G.; Young, E.H.; Sandhu, M.; et al. Genome-wide association study of type 2 diabetes in Africa. Diabetologia 2019, 62, 1204–1211. [Google Scholar] [CrossRef] [Green Version]
- Iyengar, S.K.; Sedor, J.R.; Freedman, B.I.; Kao, W.H.L.; Kretzler, M.; Keller, B.J.; Abboud, H.E.; Adler, S.G.; Best, L.G.; Bowden, D.W.; et al. Genome-Wide Association and Trans-ethnic Meta-Analysis for Advanced Diabetic Kidney Disease: Family Investigation of Nephropathy and Diabetes (FIND). PLoS Genet. 2015, 11, e1005352. [Google Scholar] [CrossRef]
- Mahajan, A.; Taliun, D.; Thurner, M.; Robertson, N.R.; Torres, J.M.; Rayner, N.W.; Payne, A.J.; Steinthorsdottir, V.; Scott, R.A.; Grarup, N.; et al. Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat. Genet. 2018, 50, 1505–1513. [Google Scholar] [CrossRef] [Green Version]
- Grant, A.V.; Sabri, A.; Abid, A.; Abderrahmani Rhorfi, I.; Benkirane, M.; Souhi, H.; Naji Amrani, H.; Alaoui-Tahiri, K.; Gharbaoui, Y.; Lazrak, F.; et al. A genome-wide association study of pulmonary tuberculosis in Morocco. Hum. Genet. 2016, 135, 299–307. [Google Scholar] [CrossRef] [Green Version]
- Shrine, N.; Guyatt, A.L.; Erzurumluoglu, A.M.; Jackson, V.E.; Hobbs, B.D.; Melbourne, C.A.; Batini, C.; Fawcett, K.A.; Song, K.; Sakornsakolpat, P.; et al. New genetic signals for lung function highlight pathways and chronic obstructive pulmonary disease associations across multiple ancestries. Nat. Genet. 2019, 51, 481–493. [Google Scholar] [CrossRef] [Green Version]
- Kichaev, G.; Bhatia, G.; Loh, P.-R.; Gazal, S.; Burch, K.; Freund, M.K.; Schoech, A.; Pasaniuc, B.; Price, A.L. Leveraging Polygenic Functional Enrichment to Improve GWAS Power. Am. J. Hum. Genet. 2019, 104, 65–75. [Google Scholar] [CrossRef] [Green Version]
- Wyss, A.B.; Sofer, T.; Lee, M.K.; Terzikhan, N.; Nguyen, J.N.; LaHousse, L.; Latourelle, J.C.; Smith, A.V.; Bartz, T.M.; Feitosa, M.F.; et al. Multiethnic meta-analysis identifies ancestry-specific and cross-ancestry loci for pulmonary function. Nat. Commun. 2018, 9, 2976. [Google Scholar] [CrossRef] [Green Version]
- Koster, R.; Panagiotou, O.A.; Wheeler, W.A.; Karlins, E.; Gastier-Foster, J.M.; De Toledo, S.R.C.; Petrilli, A.S.; Flanagan, A.M.; Tirabosco, R.; Andrulis, I.L.; et al. Genome-wide association study identifies theGLDC/IL33locus associated with survival of osteosarcoma patients. Int. J. Cancer 2018, 142, 1594–1601. [Google Scholar] [CrossRef] [Green Version]
- Kerns, S.L.; Fachal, L.; Dorling, L.; Barnett, G.C.; Baran, A.; Peterson, D.R.; Hollenberg, M.; Hao, K.; Narzo, A.D.; Ahsen, M.E.; et al. Radiogenomics Consortium Genome-Wide Association Study Meta-Analysis of Late Toxicity After Prostate Cancer Radiotherapy. J. Natl. Cancer Inst. 2020, 112, 179–190. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.K. Identification of 613 new loci associated with heel bone mineral density and a polygenic risk score for bone mineral density, osteoporosis and fracture. PLoS ONE 2018, 13, e0200785. [Google Scholar] [CrossRef] [Green Version]
- Morris, J.A.; Kemp, J.P.; Youlten, S.E.; Laurent, L.; Logan, J.G.; Chai, R.C.; Vulpescu, N.A.; Forgetta, V.; Kleinman, A.; Mohanty, S.T.; et al. An atlas of genetic influences on osteoporosis in humans and mice. Nat. Genet. 2019, 51, 258–266. [Google Scholar] [CrossRef]
- Kou, I.; Otomo, N.; Takeda, K.; Momozawa, Y.; Lu, H.-F.; Kubo, M.; Kamatani, Y.; Ogura, Y.; Takahashi, Y.; Nakajima, M.; et al. Genome-wide association study identifies 14 previously unreported susceptibility loci for adolescent idiopathic scoliosis in Japanese. Nat. Commun. 2019, 10, 3685. [Google Scholar] [CrossRef]
- Rhee, E.P.; Ho, J.E.; Chen, M.-H.; Shen, D.; Cheng, S.; Larson, M.G.; Ghorbani, A.; Shi, X.; Helenius, I.T.; O’Donnell, C.J.; et al. A Genome-wide Association Study of the Human Metabolome in a Community-Based Cohort. Cell Metab. 2013, 18, 130–143. [Google Scholar] [CrossRef] [Green Version]
- Kawafune, K.; Hachiya, T.; Nogawa, S.; Takahashi, S.; Jia, H.; Saito, K.; Kato, H. Strong association between the 12q24 locus and sweet taste preference in the Japanese population revealed by genome-wide meta-analysis. J. Hum. Genet. 2020, 939–947. [Google Scholar] [CrossRef]
- Evans, D.M.; Zhu, G.; Dy, V.; Heath, A.C.; Madden, P.A.F.; Kemp, J.P.; McMahon, G.; Pourcain, B.S.; Timpson, N.J.; Golding, J.; et al. Genome-wide association study identifies loci affecting blood copper, selenium and zinc. Hum. Mol. Genet. 2013, 22, 3998–4006. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Yang, J.; Schneider, J.A.; De Jager, P.L.; Bennett, D.A.; Zhang, H.-Y. Genome-wide interaction analysis of pathological hallmarks in Alzheimer’s disease. Neurobiol. Aging 2020, 93, 61–68. [Google Scholar] [CrossRef]
- Coleman, J.R.I.; Peyrot, W.J.; Purves, K.L.; Davis, K.A.S.; Rayner, C.; Choi, S.W.; Hübel, C.; Gaspar, H.A.; Kan, C.; Van der Auwera, S.; et al. Genome-wide gene-environment analyses of major depressive disorder and reported lifetime traumatic experiences in UK Biobank. Mol. Psychiatry 2020, 25, 1430–1446. [Google Scholar] [CrossRef]
- Leandro-García, L.J.; Inglada-Pérez, L.; Pita, G.; Hjerpe, E.; Leskelä, S.; Jara, C.; Mielgo, X.; González-Neira, A.; Robledo, M.; Åvall-Lundqvist, E.; et al. Genome-wide association study identifies ephrin type A receptors implicated in paclitaxel induced peripheral sensory neuropathy. J. Med Genet. 2013, 50, 599–605. [Google Scholar] [CrossRef]
- Zhou, H.; Cheng, Z.; Bass, N.; Krystal, J.H.; Farrer, L.A.; Kranzler, H.R.; Gelernter, J. Genome-wide association study identifies glutamate ionotropic receptor GRIA4 as a risk gene for comorbid nicotine dependence and major depression. Transl. Psychiatry 2018, 8, 208. [Google Scholar] [CrossRef] [Green Version]
- Van Der Meer, D.; Frei, O.; Kaufmann, T.; Shadrin, A.A.; Devor, A.; Smeland, O.B.; Thompson, W.K.; Fan, C.C.; Holland, D.; Westlye, L.T.; et al. Understanding the genetic determinants of the brain with MOSTest. Nat. Commun. 2020, 11, 3512. [Google Scholar] [CrossRef]
- Zhao, B.; Luo, T.; Li, T.; Li, Y.; Zhang, J.; Shan, Y.; Wang, X.; Yang, L.; Zhou, F.; Zhu, Z.; et al. Genome-wide association analysis of 19,629 individuals identifies variants influencing regional brain volumes and refines their genetic co-architecture with cognitive and mental health traits. Nat. Genet. 2019, 51, 1637–1644. [Google Scholar] [CrossRef]
- Sebat, J.; Lakshmi, B.; Malhotra, D.; Troge, J.; Lese-Martin, C.; Walsh, T.; Yamrom, B.; Yoon, S.; Krasnitz, A.; Kendall, J.; et al. Strong Association of De Novo Copy Number Mutations with Autism. Science 2007, 316, 445–449. [Google Scholar] [CrossRef] [Green Version]
- Awadalla, P.; Gauthier, J.; Myers, R.A.; Casals, F.; Hamdan, F.F.; Griffing, A.R.; Côté, M.; Henrion, E.; Spiegelman, D.; Tarabeux, J.; et al. Direct Measure of the De Novo Mutation Rate in Autism and Schizophrenia Cohorts. Am. J. Hum. Genet. 2010, 87, 316–324. [Google Scholar] [CrossRef] [Green Version]
- Prasad, A.; Merico, D.; Thiruvahindrapuram, B.; Wei, J.; Lionel, A.C.; Sato, D.; Rickaby, J.; Lu, C.; Szatmari, P.; Roberts, W.; et al. A Discovery Resource of Rare Copy Number Variations in Individuals with Autism Spectrum Disorder. G3 2012, 2, 1665–1685. [Google Scholar] [CrossRef] [Green Version]
- De Rubeis, S.; He, X.; Goldberg, A.P.; Poultney, C.S.; Samocha, K.; Cicek, A.E.; Kou, Y.; Liu, L.; Fromer, M.; Walker, S.; et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature 2014, 515, 209–215. [Google Scholar] [CrossRef]
- Doan, R.N.; Bae, B.-I.; Cubelos, B.; Chang, C.; Hossain, A.A.; Al-Saad, S.; Mukaddes, N.M.; Oner, O.; Al-Saffar, M.; Balkhy, S.; et al. Mutations in Human Accelerated Regions Disrupt Cognition and Social Behavior. Cell 2016, 167, 341–354. [Google Scholar] [CrossRef] [Green Version]
- Ruzzo, E.K.; Pérez-Cano, L.; Jung, J.-Y.; Wang, L.-K.; Kashef-Haghighi, D.; Hartl, C.; Singh, C.; Xu, J.; Hoekstra, J.N.; Leventhal, O.; et al. Inherited and De Novo Genetic Risk for Autism Impacts Shared Networks. Cell 2019, 178, 850–866. [Google Scholar] [CrossRef] [Green Version]
- Woodbury-Smith, M.; Zarrei, M.; Wei, J.; Thiruvahindrapuram, B.; O’Connor, I.; Paterson, A.D.; Yuen, R.K.C.; Dastan, J.; Stavropoulos, D.J.; Howe, J.L.; et al. Segregating patterns of copy number variations in extended autism spectrum disorder (ASD) pedigrees. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2020, 183, 268–276. [Google Scholar] [CrossRef]
- Alrayes, N.; Mohamoud, H.S.A.; Ahmed, S.; Almramhi, M.M.; Shuaib, T.M.; Wang, J.; Al-Aama, J.Y.; Everett, K.; Nasir, J.; Jelani, M. The alkylglycerol monooxygenase (AGMO) gene previously involved in autism also causes a novel syndromic form of primary microcephaly in a consanguineous Saudi family. J. Neurol. Sci. 2016, 363, 240–244. [Google Scholar] [CrossRef]
- Okur, V.; Watschinger, K.; Niyazov, D.; McCarrier, J.; Basel, D.; Hermann, M.; Werner, E.R.; Chung, W.K. Biallelic variants in AGMO with diminished enzyme activity are associated with a neurodevelopmental disorder. Hum. Genet. 2019, 138, 1259–1266. [Google Scholar] [CrossRef]
- Marquet, S.; Bucheton, B.; Reymond, C.; Argiro, L.; El-Safi, S.H.; Kheir, M.M.; Desvignes, J.P.; Béroud, C.; Mergani, A.; Hammad, A.; et al. Exome Sequencing Identifies Two Variants of the Alkylglycerol Monooxygenase Gene as a Cause of Relapses in Visceral Leishmaniasis in Children, in Sudan. J. Infect. Dis. 2017, 216, 22–28. [Google Scholar] [CrossRef] [Green Version]
- Watschinger, K.; Keller, M.A.; Golderer, G.; Coassin, S.; Zschocke, J.; Werner, E.R. Biochemical Characterization of AGMO Variants Implicated in Relapses in Visceral Leishmaniasis. J. Infect. Dis. 2018, 217, 1846–1847. [Google Scholar] [CrossRef]
- Fakhro, K.A.; Choi, M.; Ware, S.M.; Belmont, J.W.; Towbin, J.A.; Lifton, R.P.; Khokha, M.K.; Brueckner, M. Rare copy number variations in congenital heart disease patients identify unique genes in left-right patterning. Proc. Natl. Acad. Sci. USA 2011, 108, 2915–2920. [Google Scholar]
- Tokuoka, S.M.; Kita, Y.; Shindou, H.; Shimizu, T. Alkylglycerol monooxygenase as a potential modulator for PAF synthesis in macrophages. Biochem. Biophys. Res. Commun. 2013, 436, 306–312. [Google Scholar] [CrossRef]
- Waqas, S.F.H.; Hoang, A.C.; Lin, Y.-T.; Ampem, G.; Azegrouz, H.; Balogh, L.; Thuróczy, J.; Chen, J.-C.; Gerling, I.C.; Nam, S.; et al. Neuropeptide FF increases M2 activation and self-renewal of adipose tissue macrophages. J. Clin. Investig. 2017, 127, 2842–2854. [Google Scholar] [CrossRef]
- Zschiebsch, K.; Fischer, C.; Pickert, G.; Häeussler, A.; Radeke, H.; Grösch, S.; Ferreirós, N.; Geisslinger, G.; Werner, E.R.; Tegeder, I. Tetrahydrobiopterin Attenuates DSS-evoked Colitis in Mice by Rebalancing Redox and Lipid Signalling. J. Crohns. Colitis 2016, 10, 965–978. [Google Scholar] [CrossRef] [Green Version]
- Werner, E.R.; Blau, N.; Thöny, B. Tetrahydrobiopterin: Biochemistry and pathophysiology. Biochem. J. 2011, 438, 397–414. [Google Scholar] [CrossRef] [Green Version]
- Douglas, G.; Hale, A.B.; Crabtree, M.J.; Ryan, B.J.; Hansler, A.; Watschinger, K.; Gross, S.S.; Lygate, C.A.; Alp, N.J.; Channon, K.M. A requirement for Gch1 and tetrahydrobiopterin in embryonic development. Dev. Biol. 2015, 399, 129–138. [Google Scholar] [CrossRef] [Green Version]
- Elzaouk, L.; Leimbacher, W.; Turri, M.; Ledermann, B.; Bürki, K.; Blau, N.; Thöny, B. Dwarfism and Low Insulin-like Growth Factor-1 Due to Dopamine Depletion in Pts –/– Mice Rescued by Feeding Neurotransmitter Precursors and H4-biopterin. J. Biol. Chem. 2003, 278, 28303–28311. [Google Scholar] [CrossRef] [Green Version]
- Korner, G.; Scherer, T.; Adamsen, D.; Rebuffat, A.; Crabtree, M.J.; Rassi, A.; Scavelli, R.; Homma, D.; Ledermann, B.; Konrad, D.; et al. Mildly compromised tetrahydrobiopterin cofactor biosynthesis due to Pts variants leads to unusual body fat distribution and abdominal obesity in mice. J. Inherit. Metab. Dis. 2016, 39, 309–319. [Google Scholar] [CrossRef] [Green Version]
- Alp, N.J.; Mussa, S.; Khoo, J.; Cai, S.; Guzik, T.; Jefferson, A.; Goh, N.; Rockett, K.A.; Channon, K.M. Tetrahydrobiopterin-dependent preservation of nitric oxide–mediated endothelial function in diabetes by targeted transgenic GTP–cyclohydrolase I overexpression. J. Clin. Investig. 2003, 112, 725–735. [Google Scholar] [CrossRef]
- Ihlemann, N.; Rask-Madsen, C.; Perner, A.; Dominguez, H.; Hermann, T.; Køber, L.; Torp-Pedersen, C. Tetrahydrobiopterin restores endothelial dysfunction induced by an oral glucose challenge in healthy subjects. Am. J. Physiol. Heart Circ. Physiol. 2003, 285, H875–H882. [Google Scholar] [CrossRef] [Green Version]
- Meininger, C.; Cai, S.; Parker, J.L.; Channon, K.M.; Kelly, K.A.; Becker, E.J.; Wood, M.K.; Wade, L.A.; Wu, G. GTP cyclohydrolase I gene transfer reverses tetrahydrobiopterin deficiency and increases nitric oxide synthesis in endothelial cells and isolated vessels from diabetic rats. FASEB J. 2004, 18, 1900–1902. [Google Scholar] [CrossRef]
- Nyström, T.; Nygren, A.; Sjöholm, A. Tetrahydrobiopterin increases insulin sensitivity in patients with type 2 diabetes and coronary heart disease. Am. J. Physiol. Endocrinol. Metab. 2004, 287, E919–E925. [Google Scholar] [CrossRef]
- Loer, C.M.; Calvo, A.C.; Watschinger, K.; Werner-Felmayer, G.; O’Rourke, D.; Stroud, D.; Tong, A.; Gotenstein, J.R.; Chisholm, A.D.; Hodgkin, J.; et al. Cuticle Integrity and Biogenic Amine Synthesis in Caenorhabditis elegans Require the Cofactor Tetrahydrobiopterin (BH4). Genetics 2015, 200, 237–253. [Google Scholar] [CrossRef] [Green Version]
- Samuelson, A.V.; Carr, C.E.; Ruvkun, G. Gene activities that mediate increased life span of C. elegans insulin-like signaling mutants. Genes Dev. 2007, 21, 2976–2994. [Google Scholar] [CrossRef] [Green Version]
- Duncan, A.R.; González, D.P.; Del Viso, F.; Robson, A.; Khokha, M.K.; Griffin, J.N. Alkylglycerol monooxygenase, a heterotaxy candidate gene, regulates left-right patterning via Wnt signaling. Dev. Biol. 2019, 456, 1–7. [Google Scholar] [CrossRef]
- Petko, J.; Thileepan, M.; Sargen, M.; Canfield, V.; Levenson, R. Alternative splicing of the Wnt trafficking protein, Wntless and its effects on protein-protein interactions. BMC Mol. Cell Biol. 2019, 20, 22. [Google Scholar] [CrossRef] [Green Version]
- Korošec, T.; Tomažin, U.; Horvat, S.; Keber, R.; Salobir, J. The diverse effects of α- and γ-tocopherol on chicken liver transcriptome. Poult. Sci. 2017, 96, 667–680. [Google Scholar] [CrossRef]
- Luo, W.; Luo, C.; Wang, M.; Guo, L.; Chen, X.; Li, Z.; Zheng, M.; Folaniyi, B.S.; Luo, W.; Shu, D.; et al. Genome diversity of Chinese indigenous chicken and the selective signatures in Chinese gamecock chicken. Sci. Rep. 2020, 10, 14532. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sailer, S.; Keller, M.A.; Werner, E.R.; Watschinger, K. The Emerging Physiological Role of AGMO 10 Years after Its Gene Identification. Life 2021, 11, 88. https://doi.org/10.3390/life11020088
Sailer S, Keller MA, Werner ER, Watschinger K. The Emerging Physiological Role of AGMO 10 Years after Its Gene Identification. Life. 2021; 11(2):88. https://doi.org/10.3390/life11020088
Chicago/Turabian StyleSailer, Sabrina, Markus A. Keller, Ernst R. Werner, and Katrin Watschinger. 2021. "The Emerging Physiological Role of AGMO 10 Years after Its Gene Identification" Life 11, no. 2: 88. https://doi.org/10.3390/life11020088
APA StyleSailer, S., Keller, M. A., Werner, E. R., & Watschinger, K. (2021). The Emerging Physiological Role of AGMO 10 Years after Its Gene Identification. Life, 11(2), 88. https://doi.org/10.3390/life11020088






