Compensatory Base Changes Reveal Sexual Incompatibility among Members of the Anopheles subpictus Sensu Lato (Diptera: Culicidae) Species Complex in Sri Lanka
Abstract
:1. Introduction
2. Materials and Methods
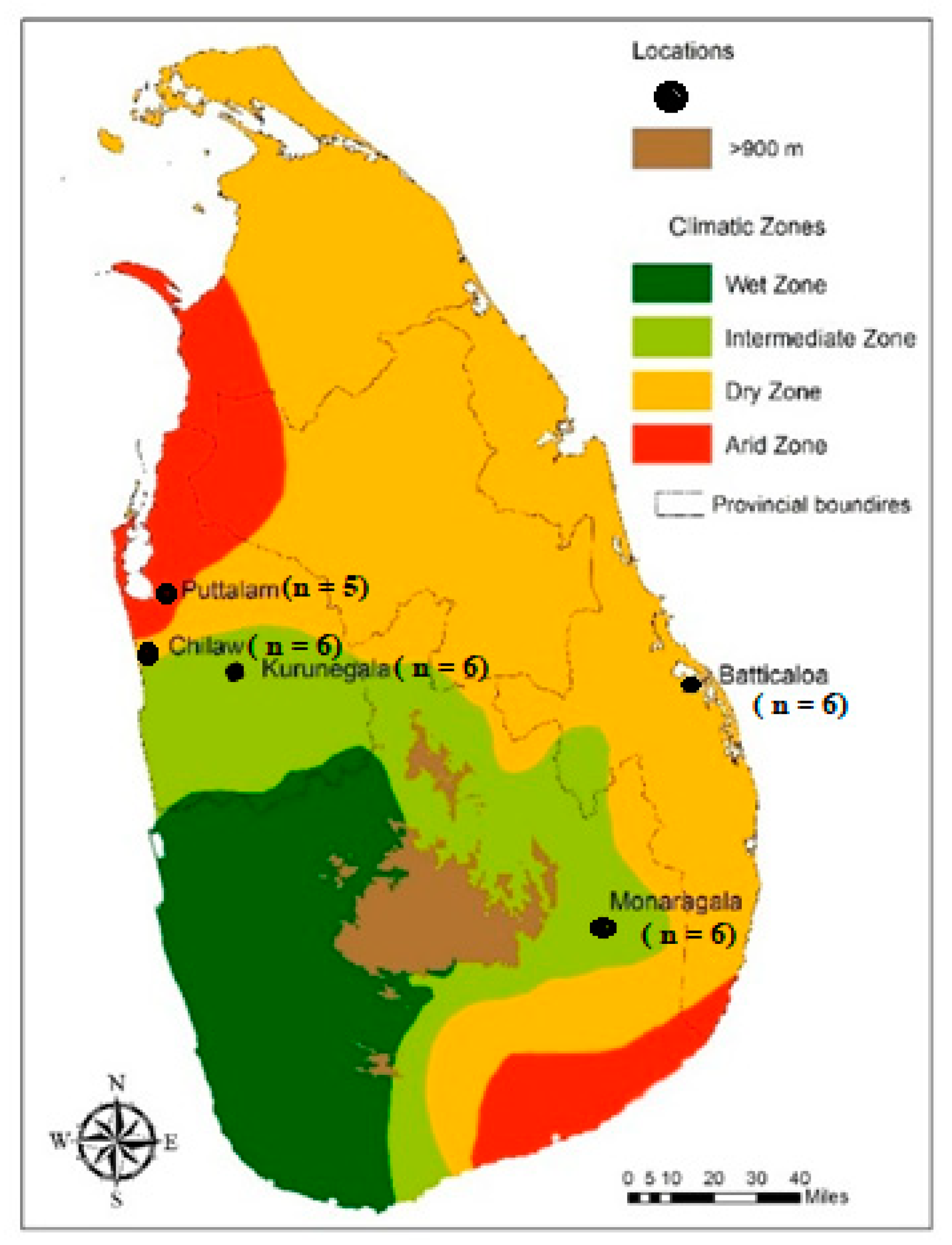
2.1. Mosquito Sampling
2.2. DNA Extraction and PCR Amplifications
2.3. Determination of Genetic Divergence of ITS2
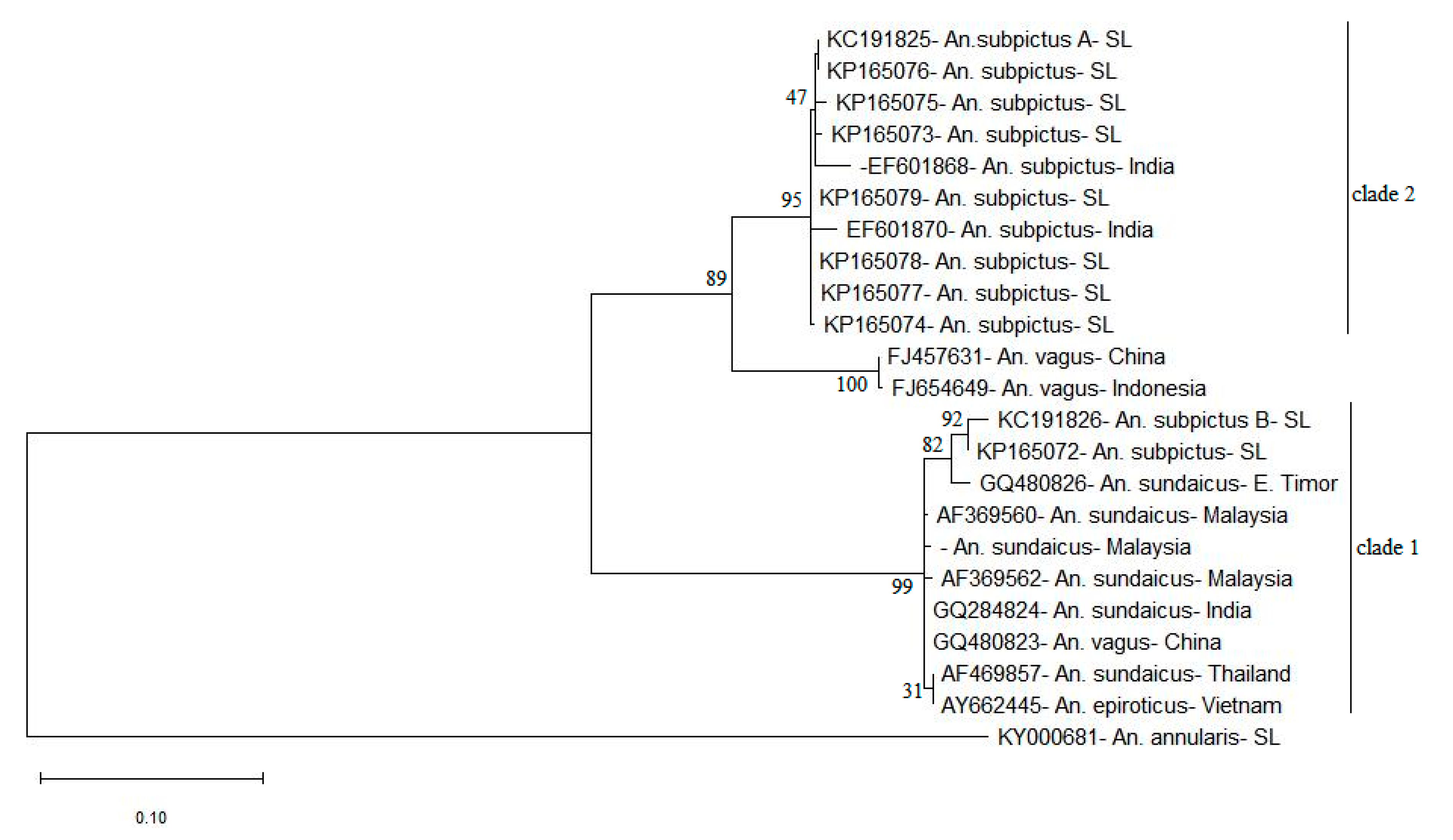
2.4. ITS2 Sequence Phylogeny
2.5. ITS2 Sequence-Structure Analysis
2.6. Analysis of Compensatory Base Changes (CBCs)
3. Results
3.1. Annotation of ITS2 Reveals Two Types of An. subpictus
3.2. ITS2 Secondary Structures Reveal Complimentary Base Changes
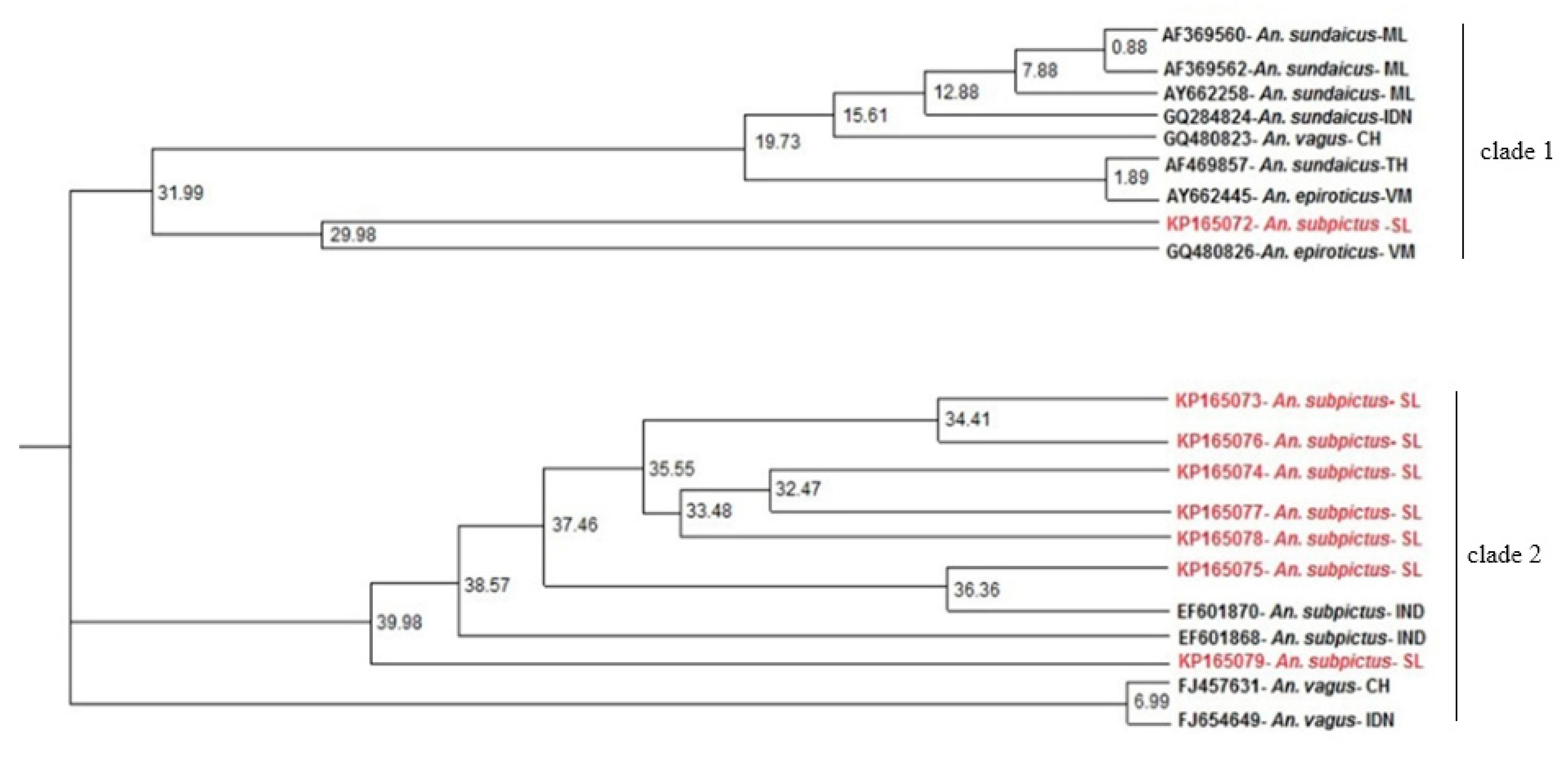
3.3. ITS2 Sequence-Structure Analysis and ITS2 Sequence Phylogeny Reveal Two Distinct Clades for An. subpictus s.l.
4. Discussion
Discrepancy on the Molecular Taxonomy of Anopheles subpictus, Anopheles sundaicus and Anopheles pseudosundaicus
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wickramasinghe, M.B. Malaria and its control in Sri Lanka. Ceylon Med. J. 1981, 26, 107–115. [Google Scholar] [PubMed]
- Abeyasinghe, R.R.; Galappaththy, G.N.L.; Gueye, C.S.; Kahn, F.G.; Feachem, R.G.A. Malaria Control and Elimination in Sri Lanka: Documenting Progress and Success Factors in a Conflict Setting. PLoS ONE 2012, 7. [Google Scholar] [CrossRef] [PubMed]
- Antimalarial Campaign, Sri Lanka. Available online: http://www.malariacampaign.gov.lk/index.php/en/ (accessed on 31 October 2018).
- Weeraratne, T.C.; Surendran, S.N.; Reimer, L.J.; Wondji, C.S.; Perera, M.D.B.; Walton, C.; Karunaratne, S.H.P.P. Molecular characterization of Anopheline (Diptera: Culicidae) mosquitoes from eight geographical locations of Sri Lanka. Malar J. 2017, 16. [Google Scholar] [CrossRef] [Green Version]
- Steyskal, G.C. The meaning of the term “sibling species”. Syst. Biol. 1972, 21. [Google Scholar] [CrossRef]
- Tripathy, A.; Samanta, L.; Das, S.; Parida1, S.K.; Marai, N.; Hazra, R.K.; Kar, S.K.; Mahapatra, N. Distribution of sibling species of Anopheles culicifacies s.l. and Anopheles fluviatilis s.l. and their vectorial capacity in eight different malaria endemic districts of Orissa, India. Mem. Inst. Oswaldo Cruz Rio de Janeiro 2010, 105, 981–987. [Google Scholar] [CrossRef] [Green Version]
- Karunaweera, N.D.; Galappaththy, G.N.L.; Wirth, G.F. On the road to eliminate malaria in Sri Lanka: Lessons from history, challenges, gaps in knowledge and research needs. Malar J. 2014, 13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amerasinghe, P.H.; Amerasinghe, F.P.; Wirtz, R.A.; Indrajith, N.G.; Somapala, W.; Pereira, L.R.; Rathnayake, A.M. Malaria transmission by Anopheles subpictus (Diptera: Culicidae) in a New Irrigation Project in Sri Lanka. J. Med. Entomol. 1992, 29, 577–581. [Google Scholar] [CrossRef]
- Jude P., J.; Ramasamy, R.; Surendran, S.N. Bionomic aspects of the Anopheles subpictus species complex in Sri Lanka. J. Insect Sci. 2014, 14. [Google Scholar] [CrossRef]
- Panicker, K.N.; GeethaBai, M.; Rao, B.U.S.; Wiswam, K.; Suryanarayanamurthy, U. Anopheles subpictus vector of malaria in coastal villages of South-East India. Curr. Sci. 1981, 50, 694–695. [Google Scholar]
- Reid, J.A. A Note on Anopheles subpictus Grassi and Anopheles indefinitus Ludlow (Diptera: Culicidae). J. Med. Entomol. 1966, 3, 327–331. [Google Scholar] [CrossRef]
- Reuben, R.; Suguna, S.G. Morphological differences between sibling species of the taxon Anopheles subpictus Grassi in India, with notes on relationships with known forms. Mosq. Syst. 1983, 15, 117–126. [Google Scholar]
- Suguna, S.G.; Rathinam, G.K.; Rajavel, A.R.; Dhanda, V. Morphological and chromosomal descriptions of new species in the Anopheles subpictus complex. Med. Vet. Entomol. 1994, 8, 88–94. [Google Scholar] [CrossRef]
- Abhayawardana, T.A.; Wijesuriya, S.R.G.; Dilrukshi, R.K.C. Anopheles subpictus Complex: Distribution of Sibling species in Sri Lanka. Indian J. Malariol 1996, 33, 53–60. [Google Scholar]
- Surendran, S.N.; Singh, O.P.; Jude, P.J.; Ramasamy, R. Genetic evidence for malaria vectors of the Anopheles sundaicus complex in Sri Lanka with morphological characteristics attributed to Anopheles subpictus species B. Malar J. 2010, 9. [Google Scholar] [CrossRef] [Green Version]
- Jayatunga, D.P.W.; Harischandra, I.N.; Chandrasekharan, N.V.; de Silva, B.G.D.N.K. Alterations and interchange of morphometric characteristics in different life cycle stages wit reference to the genomic variations of Anopheles subpictus (Diptera; Culicidae) sibling species complex in Sri Lanka. Insects 2018, 9, 89. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Surendan, S.N.; Sarma, D.K.; Jude, P.J.; Kemppainen, P.; Kanthakumaran, N.; Gajapathy, K.; Peiris, L.B.S.; Ramasamy, R.; Walton, C. Molecular characterization and identification of members of the Anopheles subpictus complex in Sri Lanka. Malar J. 2013, 12. [Google Scholar] [CrossRef] [Green Version]
- Wilai, P.; Ali, R.S.M.; Saingamsook, J.; Saeung, A.; Junkum, A.; Walton, C.; Harbach, R.E.; Somboon, P. Integrated systematics of Anopheles subpictus (Diptera: Culicidae) in the Oriental Region, with emphasis on forms in Thailand and Sulawesi, Indonesia. Acta Trop. 2020, 208, 1–8. [Google Scholar] [CrossRef]
- Sindhania, A.; Das, M.K.; Sharma, G.; Surendran, S.N.; Kaushal, B.R.; Lohani, H.P.; Singh, O.P. Molecular forms of Anopheles subpictus and Anopheles sundaicus in the Indian subcontinent. Malar J. 2020, 19. [Google Scholar] [CrossRef]
- Barraclough, T.G.; Nee, S. Phylogenetics and speciation. Trends Ecol Evol 2001, 16, 391–399. [Google Scholar] [CrossRef]
- Singh, B.N. Concepts of species and modes of speciation. Curr. Sci. 2012, 103, 784–790. [Google Scholar]
- Collins, F.H.; Paskewitz, M. A review of the use of ribosomal DNA (rDNA) to differentiate among cryptic Anopheles species. Insect Mol. Biol. 1996, 5, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Frankel, N.; Davis, G.K.; Vargas, D.; Wang, S.; Payre, F.; Stern, D.L. Phenotypic robustness conferred by apparently redundant transcriptional enhancers. Nature 2010, 466, 490–493. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nosil, P.; Feder, J.L. Widespread yet heterogeneous genomic divergence. Mol. Ecol. 2012, 21, 2829–2832. [Google Scholar] [CrossRef]
- Turner, T.L.; Hahn, M.W.; Nuzhdin, S.V. Genomic Islands of Speciation in Anopheles gambiae. PLoS Biol. 2005, 3, e285. [Google Scholar] [CrossRef]
- Clarkson, C.S.; Weetman, D.; Essandoh, J.; Yawson, A.E.; Maslen, G.; Manske, M.; Field, S.G.; Webster, M.; Antão, T.; MacInnis, B.; et al. Adaptive introgression between Anopheles sibling species eliminates a major genomic island but not reproductive isolation. Nat. Commun. 2014, 5. [Google Scholar] [CrossRef] [Green Version]
- Nei, M.; Rooney, A.P. Concerted and birth-and-death evolution of multigene families. Annu. Rev. Genet. 2005, 39, 121–152. [Google Scholar] [CrossRef] [Green Version]
- Dezfouli, S.R.N.; Oshaghi, M.A.; Vatandoost, H.; Assmar, M. rDNA-ITS2 based species-diagnostic Polymerase Chain Reaction assay for identification of sibling species of Anopheles fluviatilis in Iran. Southeast Asian J. Trop. Med. Public Health 2003, 34, 56–60. [Google Scholar]
- Li, C.; Wilkerson, R.C. Intragenomic rDNA ITS2 variation in the neotropical Anopheles (Nyssorhynchus) albitarsis complex (Diptera: Culicidae). J. Hered. 2007, 98, 51–59. [Google Scholar] [CrossRef] [Green Version]
- Paredes-Esquivel, C.; Donnelly, M.; Harbach, R.E.; Townson, H. A molecular phylogeny of mosquitoes in the Anopheles barbirostris Subgroup reveals cryptic species: Implications for identification of disease vectors. Mol. Phylogenet Evol. 2009, 50, 141–151. [Google Scholar] [CrossRef]
- Coleman, A.W.; Mai, J.C. Ribosomal DNA and ITS-2 sequences comparisons as a tool for predicting genetic relatedness. J. Mol. Evol. 1997, 45, 168–177. [Google Scholar] [CrossRef]
- Schultz, J.; Maisel, S.; Gerlach, D.; Muller, T.; Wolf, M. A common core of secondary structure of the internal transcribed spacer 2 (ITS2) throughout the Eukaryota. RNA 2005, 11, 164–361. [Google Scholar] [CrossRef] [Green Version]
- Gottschling, M.; Hilger, H.H.; Wolf, M.; Diane, N. Secondary structure of the ITS1 transcript and its application in a reconstruction of the phylogeny of Boraginales. Plant Biol. 2001, 3, 629–636. [Google Scholar] [CrossRef]
- Coleman, A.W. The Significance of a Coincidence between Evolutionary Landmarks Found in Mating Affinity and a DNA Sequence. Protist 2000, 151, 1–9. [Google Scholar] [CrossRef]
- Coleman, A.W.; Vacquier, V. Exploring the Phylogenetic Utility of ITS Sequences for Animals: A Test Case for Abalone (Haliotis). J. Mol. Evol. 2002, 54, 246–257. [Google Scholar] [CrossRef]
- Coleman, A.W. Pan-eukaryotic ITS2 homologies revealed by RNA secondary structure. Nucleic Acids Res. 2007, 35, 3322–3329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muller, T.; Philippi, N.; Dandekar, T.; Schultz, J.; Wolf, M. Distinguishing species. RNA 2007, 13, 1469–1472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coleman, A.W. Is there a molecular key to the level of “biological species” in eukaryotes? A DNA guide. Mol. Phylogenet. Evol. 2009, 50, 197–203. [Google Scholar] [CrossRef]
- Wolf, M.; Chen, S.; Song, J.; Ankenbrand, M.; Muller, T. Compensatory base changes in ITS2 secondary structures correlate with the biological species concept despite intragenomic variability in ITS2 sequences-A proof of concept. PLoS ONE 2013, 8. [Google Scholar] [CrossRef]
- Wolf, M.; Selig, C.; Müller, T.; Philippi, N.; Dandekar, T.; Schultz, J. Placozoa: At least two. Biologia 2007, 62, 641–645. [Google Scholar] [CrossRef]
- Schill, R.O.; Forster, F.; Dandekar, T.; Wolf, M. Using compensatory base change analysis of internal transcribed spacer 2 secondary structures to identify three new species in Paramacrobiotus (Tardigrada). Org. Divers. Evol. 2010, 10, 287–296. [Google Scholar] [CrossRef]
- Ruhl, M.W.; Wolf, M.; Jenkins, J.M. Compensatory base changes illuminate morphologically difficult taxonomy. Mol. Phylogenet Evol. 2010, 54, 664–669. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Zhao, H.; Zhao, F.; Jiang, L.; Peng, H.; Zhang, W.; Simmons, M.P. Alternative analyses of compensatory base changes in an ITS2 phylogeny of Corydalis (Papaveraceae). Ann. Bot. 2019, 124, 233–243. [Google Scholar] [CrossRef] [PubMed]
- Cai, R.; Kayal, E.; Alves-de-Souza, C.; Bigeard, E.; Corre, E.; Jeanthon, C.; Marie, D.; Porcel, B.M.; Siano, R.; Szymczak, J.; et al. Cryptic species in the parasitic Amoebophrya species complex revealed by a polyphasic approach. Sci. Rep. 2020, 10, 2531. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amato, A.; Kooistra, W.S.C.F.; Ghiron, J.H.L.; Mann, D.G.; Proschold, T.; Montresor, M. Reproductive Isolation among Sympatric Cryptic Species in Marine Diatoms. Protist 2007, 158, 193–207. [Google Scholar] [CrossRef] [PubMed]
- Bertrand, C.; Janzen, D.H.; Hallwachs, W.; Burns, J.M.; Gibson, J.F.; Shokralla, S.; Hajibabaei, M. Mitochondrial and nuclear phylogenetic analysis with Sanger and next-generation sequencing shows that, in Área de Conservación Guanacaste, northwestern Costa Rica, the skipper butterfly named Urbanus belli (family Hesperiidae) comprises three morphologically cryptic species. BMC Evol. Biol. 2014, 14. [Google Scholar] [CrossRef] [Green Version]
- Alquezar, D.E.; Hemmerter, S.; Cooper, R.D.; Beebe, N. Incomplete concerted evolution and reproductive isolation at the rDNA locus uncovers nine cryptic species within Anopheles longirostris from Papua New Guinea. BMC Evol. Biol. 2010, 10. [Google Scholar] [CrossRef] [Green Version]
- Amerasinghe, F.P. A Guide to the Identification of the Anopheline Mosquitoes (Diptera: Culicidae) of Sri Lanka, I. Adult Females. Cey J. Sci. (Bio Sci.) 1990, 21, 1–16. [Google Scholar]
- Ballinger-Crabtree, M.E.; Black IV, W.C.; Miller, B.R. Use of genetic polymorphisms detected by the Random-Amplified Polymorphic DNA Polymerase Chain Reaction (RAPD PCR) for differentiation and identification of Aedes aegypti subspecies and populations. Am. J. Trop. Med. Hyg. 1992, 47, 893–901. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. In Nucleic Acid Symptoms Serials; Information Retrieval Ltd.: London, UK, 1999; Volume 41, pp. 95–98. [Google Scholar]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformation 2009, 25, 1451–1452. [Google Scholar] [CrossRef] [Green Version]
- Sharma, D.; Issac, B.; Raghava, G.P.; Ramaswamy, R. Spectral Repeat Finder (SRF): Identification of repetitive sequences using Fourier transformation. Bioinformation 2004, 20, 1405–1412. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Posada, D. jModelTest: Phylogenetic model averaging. Mol. Biol. Evol. 2008, 25, 1253–1256. [Google Scholar] [CrossRef]
- Keller, A.; Schleicher, T.; Schultz, J.; Muller, T.; Dandekar, T.; Wolf, M. 5.8S-28S rRNA interaction and HMM-based ITS2 annotation. Gene 2009, 430, 50–57. [Google Scholar] [CrossRef]
- Reuter, J.S.; Mathews, D.H. RNAstructure: Software for RNA secondary structure prediction and analysis. BMC Bioinform. 2010, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seibel, P.N.; Müller, T.; Dandekar, T.; Schultz, J.; Wolf, M. 4SALE-A tool for synchronous RNA sequence and secondary structure alignment and editing. BMC Bioinform. 2006, 7. [Google Scholar] [CrossRef] [Green Version]
- Seibel, P.N.; Müller, T.; Dandekar, T.; Wolf, M. Synchronous visual analysis and editing of RNA sequence and secondary structure alignments using 4SALE. BMC Res. Notes 2008, 1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Byun, Y.; Han, K. PseudoViewer: Web application and web service for visualizing RNA pseudoknots and secondary structures. Nucleic Acids Res. 2006, 34, W416–W422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wolf, M.; Ruderisch, B.; Dandekar, T.; Schultz, J.; Müller, T. ProfDistS: (profile-) distance-based phylogeny on sequence—structure alignments. Bioinformation 2008, 24, 2401–2402. [Google Scholar] [CrossRef] [Green Version]
- Weimers, M.; Keller, A.; Wolf, M. ITS2 secondary structure improves phylogeny estimation in a radiation of blue butterflies of the subgenus Agrodiaetus (Lepidoptera: Lycaenidae: Polyommatus). BMC Evol. Biol. 2009, 26. [Google Scholar] [CrossRef] [Green Version]
- Chhilar, J.S.; Chaudhry, S. Phylogenetic Analysis of Anopheles (Cellia) subpictus Grassi using rDNA-ITS2 Sequence. Proc. Zool Soc. 2012, 65, 1–10. [Google Scholar] [CrossRef]
- Dassanayake, R.S.; Gunawardene, Y.I.N.S.; de Silva, B.G.D.N.K. ITS-2 secondary structures and phylogeny of Anopheles culicifacies species. Bioinformation 2008, 2, 456–460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tyagi, B.K.; Hiriyan, J.; Tewari, S.C.; Ayanar, K.; Samuel, P.P.; Arunachalam, N.; Paramasivan, R.; Krishnamoorthy, R.; Dhananjetan, K.J.; Leo, V.; et al. Description of a new species, Anopheles pseudosundaicus (Diptera: Culicidae) from Kerala, India. Zootaxa 2009, 2219, 49–60. [Google Scholar] [CrossRef] [Green Version]
- Linton, Y.-M.; Harbach, R.E.; Seng, C.M.; Anthony, T.G.; Matusop, A. Morphological and molecular identity of Anopheles (Cellia) sundaicus (Diptera: Culicidae), the nominotypical member of a malaria vector species complex in Southeast Asia. Syst. Entomol. 2001, 26, 357–366. [Google Scholar] [CrossRef]


| ITS2 | ||
| Size | 480 bp | 575 bp |
| No. of sequences (n) | 15 | 14 |
| No. of segregating sites, (S) | 0 | 1 |
| No. of haplotypes (h) | 1 | 2 |
| Haplotype diversity (Hd) | 0 | 0.263 |
| Nucleotide diversity (π) | 0 | 0.00059 |
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | |
| KP165072 | 2 | 2 | 2 | 2 | 2 | 2 | |
| KP165073 | 0.100 | 0 | 0 | 0 | 0 | 0 | |
| KP165074 | 0.100 | 0.004 | 0 | 0 | 0 | 0 | |
| KP165075 | 0.098 | 0.004 | 0.004 | 0 | 0 | 0 | |
| KP165076 | 0.098 | 0.003 | 0.004 | 0.002 | 0 | 0 | |
| KP165077 | 0.100 | 0.005 | 0.002 | 0.004 | 0.002 | 0 | |
| KP165078 | 0.098 | 0.005 | 0.002 | 0.004 | 0.002 | 0.000 | |
| KP165079 | 0.098 | 0.002 | 0.002 | 0.002 | 0.000 | 0.000 | 0.000 |
| An. subpictus | EF601868-An. subpictus-India | EF601870-An.. subpictus India | FJ457631-An. vagus China | FJ654649-An. vagus-Indonesia | AY662258-An. sundaicus-Malaysia | GQ480826-An. sundaicus-E. Timor | AF469857-An. sundaicus-Thailand | AY662445-An. epiroticus-Vietnam | GQ284824- An. sundaicus- Indonesia | GQ480823- An. vagus- China | AF369562- An. sundaicus- Malaysia | AF369560- An. sundaicus- Malaysia | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ITS2 575 bp | ITS2 480 bp | ||||||||||||||
| An. subpictus (Present study) | ITS2 575 bp (A) | - | 2 | 0 | 0 | 1 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| ITS2 480 bp (B) | 2 | - | 2 | 2 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| EF601868-An. subpictus-India | 0 | 2 | - | 0 | 1 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | |
| EF601870-An. subpictus-India | 0 | 2 | 0 | - | 1 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | |
| FJ457631-An. vagus- China | 1 | 1 | 1 | 1 | - | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| FJ654649-An. vagus-Indonesia | 1 | 1 | 1 | 1 | 0 | - | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| AY662258-An. sundaicus- Malaysia | 2 | 0 | 2 | 2 | 1 | 1 | - | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| GQ480826-An. Sundaicus-E. Timor | 2 | 0 | 2 | 2 | 1 | 1 | 0 | - | 0 | 0 | 0 | 0 | 0 | 0 | |
| AF469857-An. sundaicus-Thailand | 2 | 0 | 2 | 2 | 1 | 1 | 0 | 0 | - | 0 | 0 | 0 | 0 | 0 | |
| AY662445-An. epiroticus-Vietnam | 2 | 0 | 2 | 2 | 1 | 1 | 0 | 0 | 0 | - | 0 | 0 | 0 | 0 | |
| GQ284824- An. sundaicus- Indonesia | 2 | 0 | 2 | 2 | 1 | 1 | 0 | 0 | 0 | 0 | - | 0 | 0 | 0 | |
| GQ480823- An. vagus- China | 2 | 0 | 2 | 2 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | - | 0 | 0 | |
| AF369562- An. sundaicus- Malaysia | 2 | 0 | 2 | 2 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | - | 0 | |
| AF369560- An. sundaicus- Malaysia | 2 | 0 | 2 | 2 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | - | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jayatunga, D.P.W.; Harischandra, I.N.; Chandrasekharan, N.V.; de Silva, B.G.D.N.K. Compensatory Base Changes Reveal Sexual Incompatibility among Members of the Anopheles subpictus Sensu Lato (Diptera: Culicidae) Species Complex in Sri Lanka. Life 2021, 11, 211. https://doi.org/10.3390/life11030211
Jayatunga DPW, Harischandra IN, Chandrasekharan NV, de Silva BGDNK. Compensatory Base Changes Reveal Sexual Incompatibility among Members of the Anopheles subpictus Sensu Lato (Diptera: Culicidae) Species Complex in Sri Lanka. Life. 2021; 11(3):211. https://doi.org/10.3390/life11030211
Chicago/Turabian StyleJayatunga, D. P. W., I. N. Harischandra, N. V. Chandrasekharan, and B. G. D. N. K. de Silva. 2021. "Compensatory Base Changes Reveal Sexual Incompatibility among Members of the Anopheles subpictus Sensu Lato (Diptera: Culicidae) Species Complex in Sri Lanka" Life 11, no. 3: 211. https://doi.org/10.3390/life11030211
APA StyleJayatunga, D. P. W., Harischandra, I. N., Chandrasekharan, N. V., & de Silva, B. G. D. N. K. (2021). Compensatory Base Changes Reveal Sexual Incompatibility among Members of the Anopheles subpictus Sensu Lato (Diptera: Culicidae) Species Complex in Sri Lanka. Life, 11(3), 211. https://doi.org/10.3390/life11030211






