Structure, Activity and Function of the NSD3 Protein Lysine Methyltransferase
Abstract
:1. Introduction
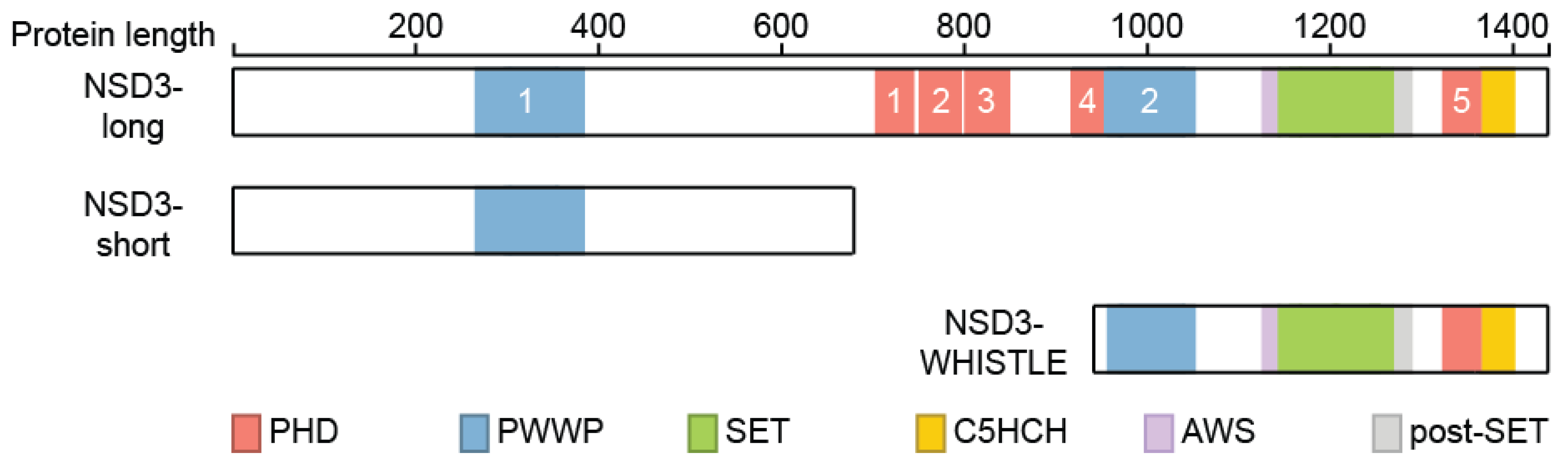
2. Structural Features
3. NSD3 Structure
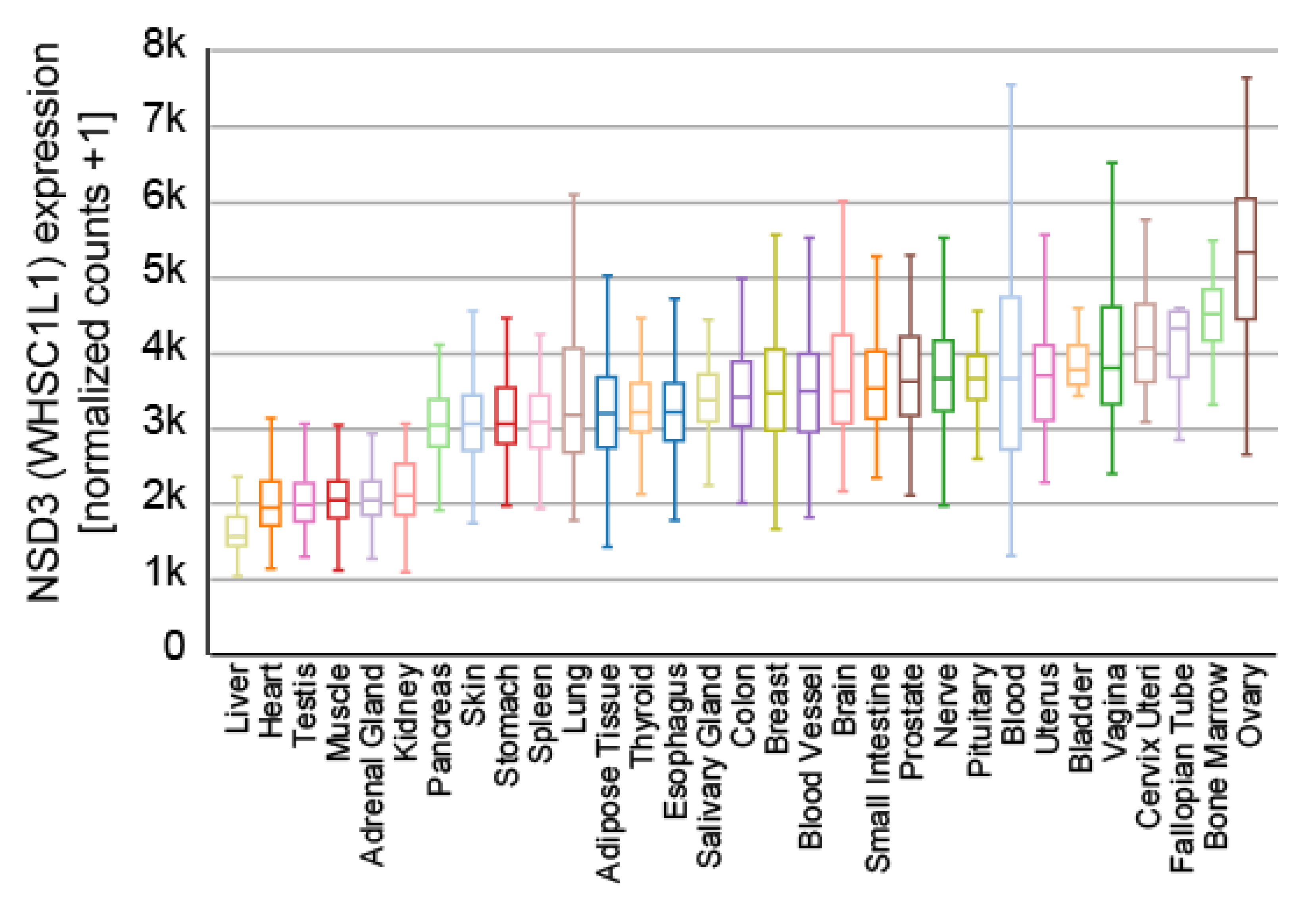
4. Biochemical Features
5. Cellular Features
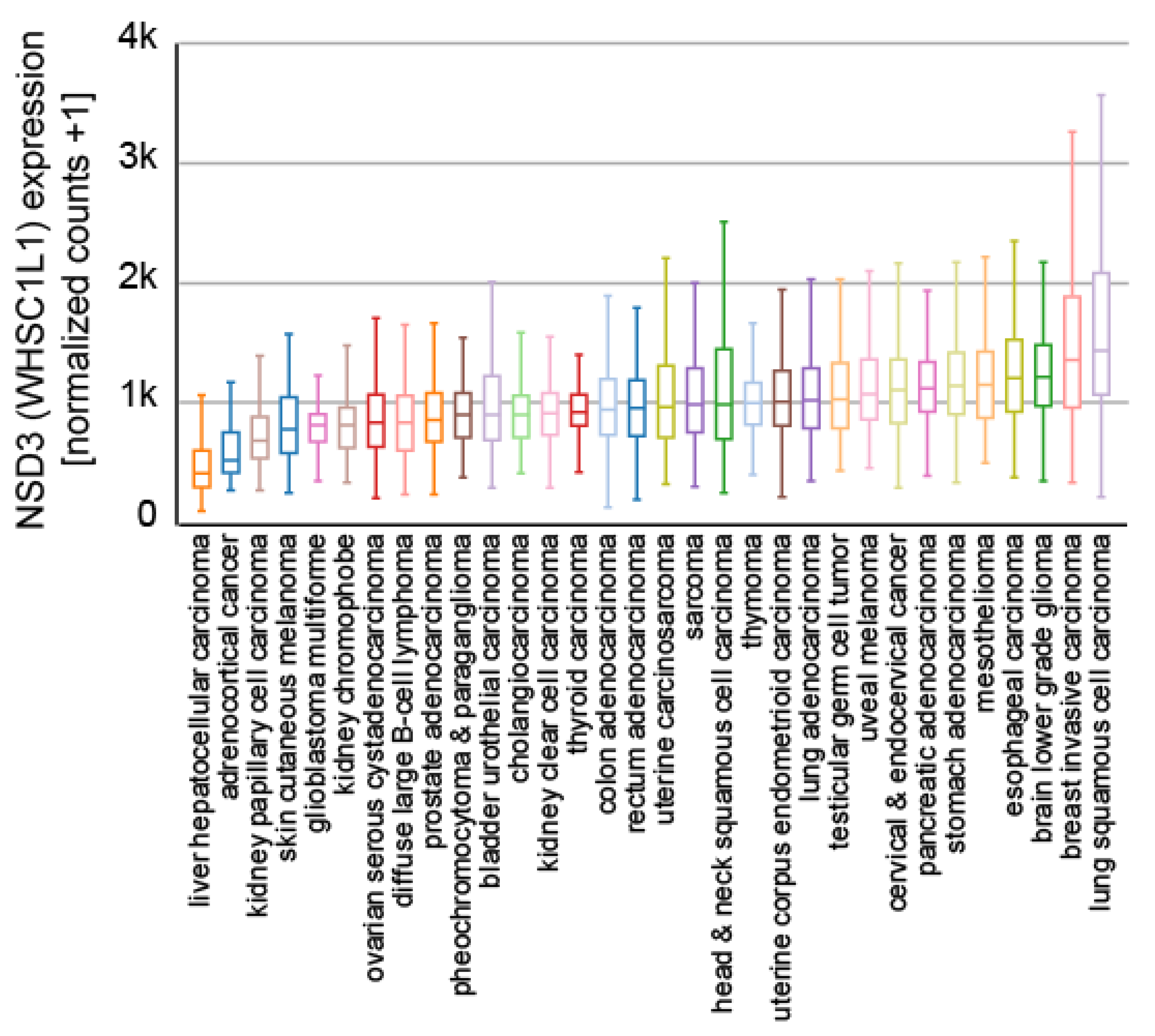
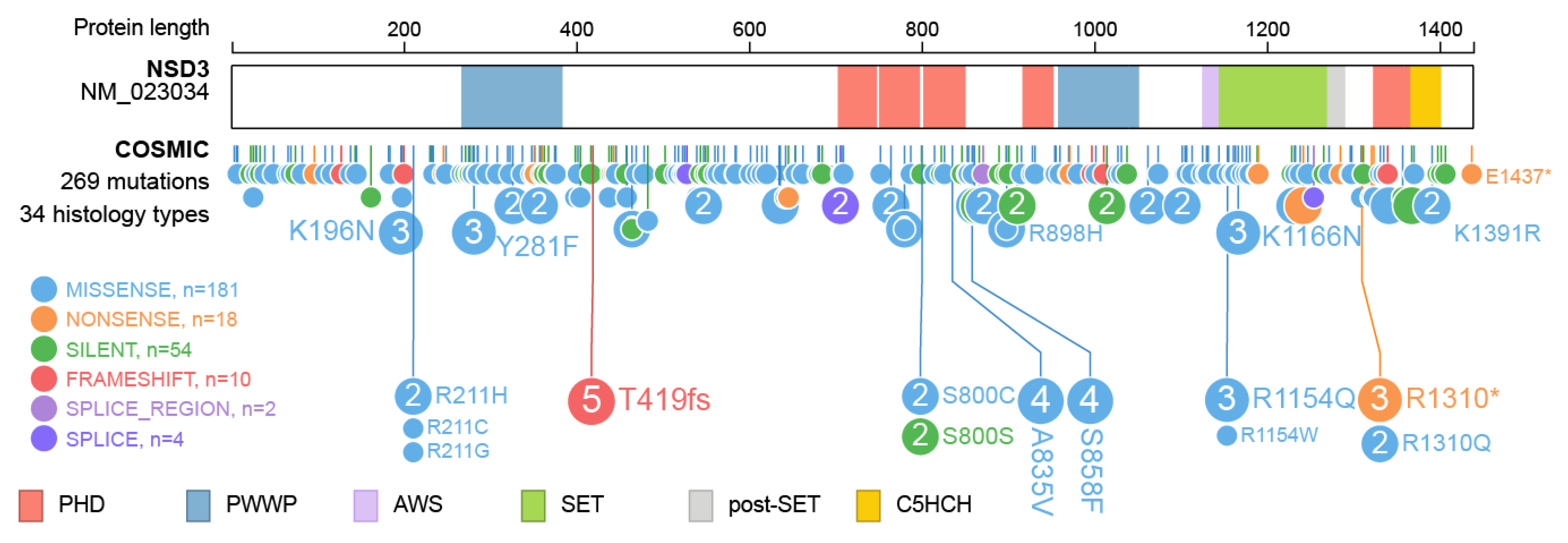
6. The Role of NSD3 in Cancer
7. Outlook
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kornberg, R.D.; Lorch, Y.L. Twenty-Five Years of the Nucleosome, Fundamental Particle of the Eukaryote Chromosome. Cell 1999, 98, 285–294. [Google Scholar] [CrossRef] [Green Version]
- Davis, L.; Onn, I.; Elliott, E. The emerging roles for the chromatin structure regulators CTCF and cohesin in neurodevelopment and behavior. Cell. Mol. Life Sci. 2018, 75, 1205–1214. [Google Scholar] [CrossRef]
- Dekker, J.; Mirny, L. The 3D Genome as Moderator of Chromosomal Communication. Cell 2016, 164, 1110–1121. [Google Scholar] [CrossRef] [Green Version]
- Atchison, M.L. Function of YY1 in Long-Distance DNA Interactions. Front. Immunol. 2014, 5, 45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hildebrand, E.M.; Dekker, J. Mechanisms and Functions of Chromosome Compartmentalization. Trends Biochem. Sci. 2020, 45, 385–396. [Google Scholar] [CrossRef] [PubMed]
- Turner, B.M. Histone acetylation and an epigenetic code. Bioessays 2000, 22, 836–845. [Google Scholar] [CrossRef]
- Strahl, B.D.; Allis, C.D. The language of covalent histone modifications. Nature 2000, 403, 41–45. [Google Scholar] [CrossRef] [PubMed]
- Fischle, W.; Wang, Y.M.; Allis, C.D. Histone and chromatin cross-talk. Curr. Opin. Cell Biol. 2003, 15, 172–183. [Google Scholar] [CrossRef] [Green Version]
- Jenuwein, T.; Allis, C.D. Translating the Histone Code. Science 2001, 293, 1074–1080. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cosgrove, M.S.; Wolberger, C. How does the histone code work? Biochem. Cell Biol. 2005, 83, 468–476. [Google Scholar] [CrossRef]
- Greer, E.L.; Shi, Y. Histone methylation: A dynamic mark in health, disease and inheritance. Nat. Rev. Genet. 2012, 13, 343–357. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Ahn, J.H.; Wang, G.G. Understanding histone H3 lysine 36 methylation and its deregulation in disease. Cell. Mol. Life Sci. 2019, 76, 2899–2916. [Google Scholar] [CrossRef]
- Bennett, R.L.; Swaroop, A.; Troche, C.; Licht, J.D. The Role of Nuclear Receptor–Binding SET Domain Family Histone Lysine Methyltransferases in Cancer. Cold Spring Harb. Perspect. Med. 2017, 7, a026708. [Google Scholar] [CrossRef] [Green Version]
- Angrand, P.-O.; Apiou, F.; Stewart, A.F.; Dutrillaux, B.; Losson, R.; Chambon, P. NSD3, a New SET Domain-Containing Gene, Maps to 8p12 and Is Amplified in Human Breast Cancer Cell Lines. Genomics 2001, 74, 79–88. [Google Scholar] [CrossRef]
- Stec, I.; Van Ommen, G.J.B.; den Dunnen, J.T. WHSC1L1, on Human Chromosome 8p11.2, Closely Resembles WHSC1 and Maps to a Duplicated Region Shared with 4p16.3. Genomics 2001, 76, 5–8. [Google Scholar] [CrossRef] [PubMed]
- Wood, A.; Shilatifard, A. Posttranslational Modifications of Histones by Methylation. Adv. Protein Chem. 2004, 67, 201–222. [Google Scholar] [CrossRef]
- Alvarez-Venegas, R.; Avramova, Z. SET-domain proteins of the Su(var)3-9, E(z) and Trithorax families. Gene 2002, 285, 25–37. [Google Scholar] [CrossRef]
- He, C.; Li, F.D.; Zhang, J.H.; Wu, J.H.; Shi, Y.Y. The Methyltransferase NSD3 Has Chromatin-binding Motifs, PHD5-C5HCH, That Are Distinct from Other NSD (Nuclear Receptor SET Domain) Family Members in Their Histone H3 Recognition. J. Biol. Chem. 2013, 288, 4692–4703. [Google Scholar] [CrossRef] [Green Version]
- Berardi, A.; Quilici, G.; Spiliotopoulos, D.; Corral-Rodriguez, M.A.; Martin-Garcia, F.; Degano, M.; Tonon, G.; Ghitti, M.; Musco, G. Structural basis for PHD(V)C5HCH(NSD1)-C2HR(Nizp1) interaction: Implications for Sotos syndrome. Nucleic Acids Res. 2016, 44, 3448–3463. [Google Scholar] [CrossRef] [Green Version]
- Nielsen, A.L.; Jorgensen, P.; Lerouge, T.; Cerviño, M.; Chambon, P.; Losson, R. Nizp1, a Novel Multitype Zinc Finger Protein That Interacts with the NSD1 Histone Lysine Methyltransferase through a Unique C2HR Motif. Mol. Cell. Biol. 2004, 24, 5184–5196. [Google Scholar] [CrossRef] [Green Version]
- Pasillas, M.P.; Shah, M.; Kamps, M.P. NSD1 PHD domains bind methylated H3K4 and H3K9 using interactions disrupted by point mutations in human sotos syndrome. Hum. Mutat. 2011, 32, 292–298. [Google Scholar] [CrossRef]
- Vermeulen, M.; Eberl, H.C.; Matarese, F.; Marks, H.; Denissov, S.; Butter, F.; Lee, K.K.; Olsen, J.V.; Hyman, A.A.; Stunnenberg, H.G.; et al. Quantitative Interaction Proteomics and Genome-wide Profiling of Epigenetic Histone Marks and Their Readers. Cell 2010, 142, 967–980. [Google Scholar] [CrossRef] [Green Version]
- Wu, H.; Zeng, H.; Lam, R.; Tempel, W.; Amaya, M.F.; Xu, C.; Dombrovski, L.; Qiu, W.; Wang, Y.; Min, J. Structural and Histone Binding Ability Characterizations of Human PWWP Domains. PLoS ONE 2011, 6, e18919. [Google Scholar] [CrossRef] [Green Version]
- Kuo, A.J.; Cheung, P.; Chen, K.F.; Zee, B.M.; Kioi, M.; Lauring, J.; Xi, Y.X.; Park, B.H.; Shi, X.B.; Garcia, B.A.; et al. NSD2 Links Dimethylation of Histone H3 at Lysine 36 to Oncogenic Programming. Mol. Cell 2011, 44, 609–620. [Google Scholar] [CrossRef] [Green Version]
- Morishita, M.; Mevius, D.; Di Luccio, E. In vitro histone lysine methylation by NSD1, NSD2/MMSET/WHSC1 and NSD3/WHSC1L. BMC Struct. Biol. 2014, 14, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Sankaran, S.M.; Wilkinson, A.W.; Elias, J.E.; Gozani, O. A PWWP Domain of Histone-Lysine N-Methyltransferase NSD2 Binds to Dimethylated Lys-36 of Histone H3 and Regulates NSD2 Function at Chromatin. J. Biol. Chem. 2016, 291, 8465–8474. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martinez-Garcia, E.; Popovic, R.; Min, D.-J.; Sweet, S.M.M.; Thomas, P.M.; Zamdborg, L.; Heffner, A.; Will, C.; Lamy, L.; Staudt, L.M.; et al. The MMSET histone methyl transferase switches global histone methylation and alters gene expression in t(4;14) multiple myeloma cells. Blood 2011, 117, 211–220. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Wu, H.P.; Chuai, S.; Xu, F.N.; Yan, F.; Englund, N.; Wang, Z.F.; Zhang, H.L.; Fang, M.; Wang, Y.Z.; et al. NSD2 Is Recruited through Its PHD Domain to Oncogenic Gene Loci to Drive Multiple Myeloma. Cancer Res. 2013, 73, 6277–6288. [Google Scholar] [CrossRef] [Green Version]
- Sims, R.J.; Reinberg, D. Histone H3 Lys 4 methylation: Caught in a bind? Genes Dev. 2006, 20, 2779–2786. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.L.; Chen, X.; Lu, C. The interplay between DNA and histone methylation: Molecular mechanisms and disease implications. EMBO Rep. 2021, 22, e51803. [Google Scholar] [CrossRef] [PubMed]
- Lienert, F.; Mohn, F.; Tiwari, V.K.; Baubec, T.; Roloff, T.C.; Gaidatzis, D.; Stadler, M.B.; Schubeler, D. Genomic Prevalence of Heterochromatic H3K9me2 and Transcription Do Not Discriminate Pluripotent from Terminally Differentiated Cells. PLoS Genet. 2011, 7, e1002090. [Google Scholar] [CrossRef]
- Kang, H.B.; Choi, Y.; Lee, J.M.; Choi, K.C.; Kim, H.C.; Yoo, J.Y.; Lee, Y.H.; Yoon, H.G. The histone methyltransferase, NSD2, enhances androgen receptor-mediated transcription. FEBS Lett. 2009, 583, 1880–1886. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, X.; Edmonson, M.N.; Wilkinson, M.R.; Patel, A.; Wu, G.; Liu, Y.; Li, Y.J.; Zhang, Z.J.; Rusch, M.C.; Parker, M.; et al. Exploring genomic alteration in pediatric cancer using ProteinPaint. Nat. Genet. 2016, 48, 4–6. [Google Scholar] [CrossRef] [Green Version]
- Morrison, M.J.; Boriack-Sjodin, P.A.; Swinger, K.K.; Wigle, T.J.; Sadalge, D.; Kuntz, K.W.; Scott, M.P.; Janzen, W.P.; Chesworth, R.; Duncan, K.W.; et al. Identification of a peptide inhibitor for the histone methyltransferase WHSC1. PLoS ONE 2018, 13, e0197082. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Tian, W.; Yuan, G.; Deng, P.; Sengupta, D.; Cheng, Z.; Cao, Y.; Ren, J.; Qin, Y.; Zhou, Y.; et al. Molecular basis of nucleosomal H3K36 methylation by NSD methyltransferases. Nature 2021, 590, 498–503. [Google Scholar] [CrossRef]
- Qiao, Q.; Li, Y.; Chen, Z.; Wang, M.Z.; Reinberg, D.; Xu, R.-M. The Structure of NSD1 Reveals an Autoregulatory Mechanism Underlying Histone H3K36 Methylation. J. Biol. Chem. 2011, 286, 8361–8368. [Google Scholar] [CrossRef] [Green Version]
- Böttcher, J.; Dilworth, D.; Reiser, U.; Neumüller, R.A.; Schleicher, M.; Petronczki, M.; Zeeb, M.; Mischerikow, N.; Allali-Hassani, A.; Szewczyk, M.M.; et al. Fragment-based discovery of a chemical probe for the PWWP1 domain of NSD3. Nat. Chem. Biol. 2019, 15, 822–829. [Google Scholar] [CrossRef]
- Qin, S.; Min, J.R. Structure and function of the nucleosome-binding PWWP domain. Trends Biochem. Sci. 2014, 39, 536–547. [Google Scholar] [CrossRef]
- Vezzoli, A.; Bonadies, N.; Allen, M.D.; Freund, S.M.V.; Santiveri, C.M.; Kvinlaug, B.T.; Huntly, B.J.P.; Gottgens, B.; Bycroft, M. Molecular basis of histone H3K36me3 recognition by the PWWP domain of Brpf1. Nat. Struct. Mol. Biol. 2010, 17, 617–619. [Google Scholar] [CrossRef] [PubMed]
- McDaniel, S.L.; Strahl, B.D. Shaping the cellular landscape with Set2/SETD2 methylation. Cell. Mol. Life Sci. 2017, 74, 3317–3334. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.M.; Kee, H.J.; Eom, G.H.; Choe, N.W.; Kim, J.Y.; Kim, Y.S.; Kim, S.K.; Kook, H.; Kook, H.; Seo, S.B. Characterization of a novel WHSC1-associated SET domain protein with H3K4 and H3K27 methyltransferase activity. Biochem. Biophys. Res. Commun. 2006, 345, 318–323. [Google Scholar] [CrossRef]
- Li, Y.; Trojer, P.; Xu, C.F.; Cheung, P.; Kuo, A.; Drury, W.J.; Qiao, Q.; Neubert, T.A.; Xu, R.M.; Gozani, O.; et al. The Target of the NSD Family of Histone Lysine Methyltransferases Depends on the Nature of the Substrate. J. Biol. Chem. 2009, 284, 34283–34295. [Google Scholar] [CrossRef] [Green Version]
- Yuan, G.; Ma, B.; Yuan, W.; Zhang, Z.Q.; Chen, P.; Ding, X.J.; Feng, L.; Shen, X.H.; Chen, S.; Li, G.H.; et al. Histone H2A Ubiquitination Inhibits the Enzymatic Activity of H3 Lysine 36 Methyltransferases. J. Biol. Chem. 2013, 288, 30832–30842. [Google Scholar] [CrossRef] [Green Version]
- Yuan, W.; Xu, M.; Huang, C.; Liu, N.; Chen, S.; Zhu, B. H3K36 Methylation Antagonizes PRC2-mediated H3K27 Methylation. J. Biol. Chem. 2011, 286, 7983–7989. [Google Scholar] [CrossRef] [Green Version]
- Streubel, G.; Watson, A.; Jammula, S.G.; Scelfo, A.; Fitzpatrick, D.J.; Oliviero, G.; McCole, R.; Conway, E.; Glancy, E.; Negri, G.L.; et al. The H3K36me2 Methyltransferase Nsd1 Demarcates PRC2-Mediated H3K27me2 and H3K27me3 Domains in Embryonic Stem Cells. Mol. Cell 2018, 70, 371–379.e5. [Google Scholar] [CrossRef] [Green Version]
- Goldman, M.J.; Craft, B.; Hastie, M.; Repecka, K.; McDade, F.; Kamath, A.; Banerjee, A.; Luo, Y.H.; Rogers, D.; Brooks, A.N.; et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat. Biotechnol. 2020, 38, 675–678. [Google Scholar] [CrossRef] [PubMed]
- Rahman, S.; Sowa, M.E.; Ottinger, M.; Smith, J.A.; Shi, Y.; Harper, J.W.; Howley, P.M. The Brd4 Extraterminal Domain Confers Transcription Activation Independent of pTEFb by Recruiting Multiple Proteins, Including NSD3. Mol. Cell. Biol. 2011, 31, 2641–2652. [Google Scholar] [CrossRef] [Green Version]
- Shen, C.; Ipsaro, J.J.; Shi, J.; Milazzo, J.P.; Wang, E.; Roe, J.S.; Suzuki, Y.; Pappin, D.J.; Joshua-Tor, L.; Vakoc, C.R. NSD3-Short Is an Adaptor Protein that Couples BRD4 to the CHD8 Chromatin Remodeler. Mol. Cell 2015, 60, 847–859. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.; Zeng, L.; Shen, C.; Ju, Y.; Konuma, T.; Zhao, C.; Vakoc, C.R.; Zhou, M.M. Structural Mechanism of Transcriptional Regulator NSD3 Recognition by the ET Domain of BRD4. Structure 2016, 24, 1201–1208. [Google Scholar] [CrossRef] [Green Version]
- Spriano, F.; Stathis, A.; Bertoni, F. Targeting BET bromodomain proteins in cancer: The example of lymphomas. Pharmacol. Ther. 2020, 215, 107631. [Google Scholar] [CrossRef]
- Zuber, J.; Shi, J.W.; Wang, E.; Rappaport, A.R.; Herrmann, H.; Sison, E.A.; Magoon, D.; Qi, J.; Blatt, K.; Wunderlich, M.; et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature 2011, 478, 524–528. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dawson, M.A.; Prinjha, R.; Dittman, A.; Giotopoulos, G.; Bantscheff, M.; Chan, W.-I.; Robson, S.; Chung, C.-W.; Hopf, C.; Savitski, M.; et al. Inhibition of BET Recruitment to Chromatin As An Effective Treatment for MLL-Fusion Leukaemia. Blood 2011, 118, 55. [Google Scholar] [CrossRef]
- Xu, Y.L.; Vakoc, C.R. Targeting Cancer Cells with BET Bromodomain Inhibitors. Cold Spring Harb. Perspect. Med. 2017, 7, a026674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.M.; Kee, H.J.; Choe, N.; Kim, J.Y.; Kook, H.; Kook, H.; Seo, S.B. The histone methyltransferase activity of WHISTLE is important for the induction of apoptosis and HDAC1-mediated transcriptional repression. Exp. Cell Res. 2007, 313, 975–983. [Google Scholar] [CrossRef]
- Bannister, A.J.; Schneider, R.; Myers, F.A.; Thorne, A.W.; Crane-Robinson, C.; Kouzarides, T. Spatial Distribution of Di- and Tri-methyl Lysine 36 of Histone H3 at Active Genes. J. Biol. Chem. 2005, 280, 17732–17736. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, M.S.; Deng, L.W.; Lacoste, V.; Park, H.U.; Pumfery, A.; Kashanchi, F.; Brady, J.N.; Kumar, A. Coordination of Transcription Factor Phosphorylation and Histone Methylation by the P-TEFb Kinase during Human Immunodeficiency Virus Type 1 Transcription. J. Virol. 2004, 78, 13522–13533. [Google Scholar] [CrossRef] [Green Version]
- Popovic, R.; Martine-Garcia, E.; Giannopoulou, E.G.; Zhang, Q.W.; Zhang, Q.Y.; Ezponda, T.; Shah, M.Y.; Zheng, Y.P.; Will, C.M.; Small, E.C.; et al. Histone Methyltransferase MMSET/NSD2 Alters EZH2 Binding and Reprograms the Myeloma Epigenome through Global and Focal Changes in H3K36 and H3K27 Methylation. PLoS Genet. 2014, 10, e1004566. [Google Scholar] [CrossRef]
- Weinberg, D.N.; Papillon-Cavanagh, S.; Chen, H.F.; Yue, Y.; Chen, X.; Rajagopalan, K.N.; Horth, C.; McGuire, J.T.; Xu, X.J.; Nikbakht, H.; et al. The histone mark H3K36me2 recruits DNMT3A and shapes the intergenic DNA methylation landscape. Nature 2019, 573, 281–286. [Google Scholar] [CrossRef]
- Piunti, A.; Shilatifard, A. The roles of Polycomb repressive complexes in mammalian development and cancer. Nat. Rev. Mol. Cell Biol. 2021, 22, 326–345. [Google Scholar] [CrossRef]
- Yuan, S.; Natesan, R.; Sanchez-Rivera, F.J.; Li, J.Y.; Bhanu, N.V.; Yamazoe, T.; Lin, J.H.; Merrell, A.J.; Sela, Y.; Thomas, S.K.; et al. Global Regulation of the Histone Mark H3K36me2 Underlies Epithelial Plasticity and Metastatic Progression. Cancer Discov. 2020, 10, 854–871. [Google Scholar] [CrossRef] [Green Version]
- Dhayalan, A.; Rajavelu, A.; Rathert, P.; Tamas, R.; Jurkowska, R.Z.; Ragozin, S.; Jeltsch, A. The Dnmt3a PWWP Domain Reads Histone 3 Lysine 36 Trimethylation and Guides DNA Methylation. J. Biol. Chem. 2010, 285, 26114–26120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bock, I.; Dhayalan, A.; Kudithipudi, S.; Brandt, O.; Rathert, P.; Jeltsch, A. Detailed specificity analysis of antibodies binding to modified histone tails with peptide arrays. Epigenetics 2011, 6, 256–263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carlson, S.M.; Gozani, O. Nonhistone Lysine Methylation in the Regulation of Cancer Pathways. Cold Spring Harb. Perspect. Med. 2016, 6, a026435. [Google Scholar] [CrossRef] [Green Version]
- Saloura, V.; Vougiouklakis, T.; Zewde, M.; Deng, X.; Kiyotani, K.; Park, J.-H.; Matsuo, Y.; Lingen, M.; Suzuki, T.; Dohmae, N.; et al. WHSC1L1-mediated EGFR mono-methylation enhances the cytoplasmic and nuclear oncogenic activity of EGFR in head and neck cancer. Sci. Rep. 2017, 7, 40664. [Google Scholar] [CrossRef] [PubMed]
- Lu, T.; Jackson, M.W.; Wang, B.L.; Yang, M.J.; Chance, M.R.; Miyagi, M.; Gudkov, A.V.; Stark, G.R. Regulation of NF-kappa B by NSD1/FBXL11-dependent reversible lysine methylation of p65. Proc. Natl. Acad. Sci. USA 2010, 107, 46–51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kudithipudi, S.; Lungu, C.; Rathert, P.; Happel, N.; Jeltsch, A. Substrate Specificity Analysis and Novel Substrates of the Protein Lysine Methyltransferase NSD1. Chem. Biol. 2014, 21, 226–237. [Google Scholar] [CrossRef] [Green Version]
- Voutsadakis, I.A. Amplification of 8p11.23 in cancers and the role of amplicon genes. Life Sci. 2021, 264, 118729. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Piao, L.; Zhuang, Q.; Yuan, X.; Liu, Z.; He, X. The role of histone lysine methyltransferase NSD3 in cancer. OncoTargets Ther. 2018, 11, 3847–3852. [Google Scholar] [CrossRef] [Green Version]
- Taketani, T.; Taki, T.; Nakamura, H.; Taniwaki, M.; Masuda, J.; Hayashi, Y. NUP98–NSD3 fusion gene in radiation-associated myelodysplastic syndrome with t(8;11)(p11;p15) and expression pattern of NSD family genes. Cancer Genet. Cytogenet. 2009, 190, 108–112. [Google Scholar] [CrossRef]
- Rosati, R.; La Starza, R.; Veronese, A.; Aventin, A.; Schwienbacher, C.; Vallespi, T.; Negrini, M.; Martelli, M.F.; Mecucci, C. NUP98 is fused to the NSD3 gene in acute myeloid leukemia associated with t(8;11)(p11.2;p15). Blood 2002, 99, 3857–3860. [Google Scholar] [CrossRef] [Green Version]
- Wang, G.G.; Cai, L.; Pasillas, M.P.; Kamps, M.P. NUP98–NSD1 links H3K36 methylation to Hox-A gene activation and leukaemogenesis. Nat. Cell Biol. 2007, 9, 804–812. [Google Scholar] [CrossRef] [PubMed]
- Khan, J.; Whaley, R.; Cheng, L. Primary Pulmonary NUT Carcinoma with NSD3-NUTM1 Fusion. Am. J. Clin. Pathol. 2020, 154, S83–S84. [Google Scholar] [CrossRef]
- Suzuki, S.; Kurabe, N.; Ohnishi, I.; Yasuda, K.; Aoshima, Y.; Naito, M.; Tanioka, F.; Sugimura, H. NSD3-NUT-expressing midline carcinoma of the lung: First characterization of primary cancer tissue. Pathol. Res. Pract. 2015, 211, 404–408. [Google Scholar] [CrossRef] [PubMed]
- Kuroda, S.; Suzuki, S.; Kurita, A.; Muraki, M.; Aoshima, Y.; Tanioka, F.; Sugimura, H. Cytological Features of a Variant NUT Midline Carcinoma of the Lung Harboring theNSD3-NUTFusion Gene: A Case Report and Literature Review. Case Rep. Pathol. 2015, 2015, 1–5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- French, C.A.; Rahman, S.; Walsh, E.M.; Kuhnle, S.; Grayson, A.R.; Lemieux, M.E.; Grunfeld, N.; Rubin, B.P.; Antonescu, C.R.; Zhang, S.L.; et al. NSD3–NUT Fusion Oncoprotein in NUT Midline Carcinoma: Implications for a Novel Oncogenic Mechanism. Cancer Discov. 2014, 4, 928–941. [Google Scholar] [CrossRef] [Green Version]
- Rathert, P.; Roth, M.; Neumann, T.; Muerdter, F.; Roe, J.-S.; Muhar, M.; Deswal, S.; Cerny-Reiterer, S.; Peter, B.; Jude, J.; et al. Transcriptional plasticity promotes primary and acquired resistance to BET inhibition. Nature 2015, 525, 543–547. [Google Scholar] [CrossRef] [PubMed]
- Mahmood, S.F.; Gruel, N.; Nicolle, R.; Chapeaublanc, E.; Delattre, O.; Radvanyi, F.; Bernard-Pierrot, I. PPAPDC1B and WHSC1L1 Are Common Drivers of the 8p11-12 Amplicon, Not Only in Breast Tumors But Also in Pancreatic Adenocarcinomas and Lung Tumors. Am. J. Pathol. 2013, 183, 1634–1644. [Google Scholar] [CrossRef]
- Kang, D.; Cho, H.S.; Toyokawa, G.; Kogure, M.; Yamane, Y.; Iwai, Y.; Hayami, S.; Tsunoda, T.; Field, H.I.; Matsuda, K.; et al. The histone methyltransferase Wolf–Hirschhorn syndrome candidate 1-like 1 (WHSC1L1) is involved in human carcinogenesis. Genes Chromosomes Cancer 2013, 52, 126–139. [Google Scholar] [CrossRef]
- Yuan, G.; Flores, N.M.; Hausmann, S.; Lofgren, S.M.; Kharchenko, V.; Angulo-Ibanez, M.; Sengupta, D.; Lu, X.; Czaban, I.; Azhibek, D.; et al. Elevated NSD3 histone methylation activity drives squamous cell lung cancer. Nature 2021, 590, 504–508. [Google Scholar] [CrossRef]
- Irish, J.C.; Mills, J.N.; Turner-Ivey, B.; Wilson, R.C.; Guest, S.T.; Rutkovsky, A.; Dombkowski, A.; Kappler, C.S.; Hardiman, G.; Ethier, S.P. Amplification of WHSC1L1 regulates expression and estrogen-independent activation of ERα in SUM-44 breast cancer cells and is associated with ERα over-expression in breast cancer. Mol. Oncol. 2016, 10, 850–865. [Google Scholar] [CrossRef]
- Rutkovsky, A.C.; Turner-Ivey, B.; Smith, E.L.; Spruill, L.S.; Mills, J.N.; Ethier, S.P. Development of mammary hyperplasia, dysplasia, and invasive ductal carcinoma in transgenic mice expressing the 8p11 amplicon oncogene NSD3 (WHSC1L1). Cancer Res. 2017, 77, 1835. [Google Scholar] [CrossRef]
- Liu, Z.; Piao, L.; Zhuang, M.; Qiu, X.; Xu, X.; Zhang, D.; Liu, M.; Ren, D. Silencing of histone methyltransferase NSD3 reduces cell viability in osteosarcoma with induction of apoptosis. Oncol. Rep. 2017, 38, 2796–2802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yi, L.; Yi, L.; Liu, Q.; Li, C. Downregulation of NSD3 (WHSC1L1) inhibits cell proliferation and migration via ERK1/2 deactivation and decreasing CAPG expression in colorectal cancer cells. OncoTargets Ther. 2019, 12, 3933–3943. [Google Scholar] [CrossRef] [Green Version]
- Brumbaugh, J.; Kim, I.S.; Ji, F.; Huebner, A.J.; Di Stefano, B.; Schwarz, B.A.; Charlton, J.; Coffey, A.; Choi, J.; Walsh, R.M.; et al. Inducible histone K-to-M mutations are dynamic tools to probe the physiological role of site-specific histone methylation in vitro and in vivo. Nat. Cell Biol. 2019, 21, 1449–1461. [Google Scholar] [CrossRef] [Green Version]
- Morgan, M.; Herz, H.M.; Gao, X.; Jackson, J.; Rickels, R.; Swanson, S.K.; Florens, L.; Washburn, M.P.; Eissenberg, J.C.; Shilatifard, A. Histone H3 lysine-to-methionine mutants as a paradigm to study chromatin signaling. FEBS J. 2015, 282, 406. [Google Scholar]
- Fang, D.; Gan, H.Y.; Lee, J.H.; Han, J.; Wang, Z.Q.; Riester, S.M.; Jin, L.; Chen, J.J.; Zhou, H.; Wang, J.L.; et al. The histone H3.3K36M mutation reprograms the epigenome of chondroblastomas. Science 2016, 352, 1344–1348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, C.; Jain, S.U.; Hoelper, D.; Bechet, D.; Molden, R.C.; Ran, L.L.; Murphy, D.; Venneti, S.; Hameed, M.; Pawel, B.R.; et al. Histone H3K36 mutations promote sarcomagenesis through altered histone methylation landscape. Science 2016, 352, 844–849. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Fang, D. The incorporation loci of H3.3K36M determine its preferential prevalence in chondroblastomas. Cell Death Dis. 2021, 12, 1–16. [Google Scholar] [CrossRef]
- Zhang, Y.L.; Shan, C.M.; Wang, J.Y.; Bao, K.; Tong, L.; Jia, S.T. Molecular basis for the role of oncogenic histone mutations in modulating H3K36 methylation. Sci. Rep. 2017, 7, 43906. [Google Scholar] [CrossRef] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rathert, P. Structure, Activity and Function of the NSD3 Protein Lysine Methyltransferase. Life 2021, 11, 726. https://doi.org/10.3390/life11080726
Rathert P. Structure, Activity and Function of the NSD3 Protein Lysine Methyltransferase. Life. 2021; 11(8):726. https://doi.org/10.3390/life11080726
Chicago/Turabian StyleRathert, Philipp. 2021. "Structure, Activity and Function of the NSD3 Protein Lysine Methyltransferase" Life 11, no. 8: 726. https://doi.org/10.3390/life11080726
APA StyleRathert, P. (2021). Structure, Activity and Function of the NSD3 Protein Lysine Methyltransferase. Life, 11(8), 726. https://doi.org/10.3390/life11080726





