Color-Specific Recovery to Extreme High-Light Stress in Plants
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Variety
2.2. Plant Growth and Sampling
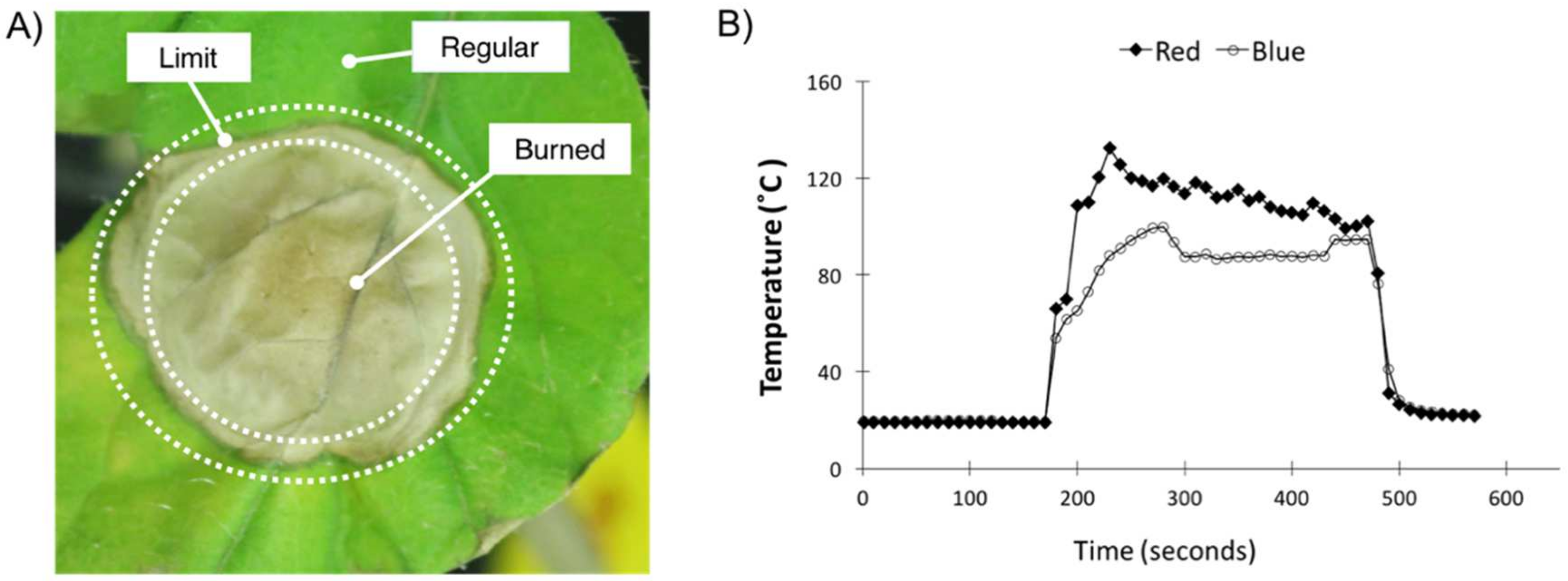
2.3. Light Treatment
2.4. Fluorescence and Analysis of Measurements
2.5. Net Photosynthesis Rate (Pn)
2.6. Protein Extraction and Digestion
2.7. Liquid Chromatography–Mass Spectrometry (LC−MS/MS)
2.8. Database Searching and Statistical Analysis
2.9. Bioinformatics
2.10. Quantitative Reverse Transcription PCR (RT-qPCR)
2.11. Statistical Analysis
3. Results
3.1. Plant Physiological Stress Measurements
3.2. Impact of High-Light Induced Heat Stress in Photosynthesis Efficiency
3.3. Photosystem II-Related Protein Abundance Comparisons
3.4. Correlation of Gene Expression with Protein Abundance
3.5. Other Proteins Identified in the BLT Dataset
4. Discussion
4.1. Implications in the Photosynthetic Responses to High-Light Stress under Different Wavelengths
4.2. Spatial Disparities in the Response of Leaves to High-Light Stress
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Demmig-Adams, B.; Adams, W.W. Xanthophyll cycle and light stress in nature: Uniform response to excess direct sunlight among higher plant species. Planta 1996, 198, 460–470. [Google Scholar] [CrossRef]
- Zavafer, A.; Chow, W.S.; Cheah, M.H. The action spectrum of Photosystem II photoinactivation in visible light. J. Photochem. Photobiol. B Biol. 2015, 152, 247–260. [Google Scholar] [CrossRef] [PubMed]
- Parrine, D.; Wu, B.-S.; Muhammad, B.; Rivera, K.; Pappin, D.; Zhao, X.; Lefsrud, M. Proteome modifications on tomato under extreme high light induced-stress. Proteome Sci. 2018, 16, 1–15. [Google Scholar] [CrossRef]
- Ruban, A.V.; Murchie, E.H. Assessing the photoprotective effectiveness of non-photochemical chlorophyll fluorescence quenching: A new approach. Biochim. Biophys. Acta BBA—Bioenergy 2012, 1817, 977–982. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olvera-González, E.; Alaniz-Lumbreras, D.; Ivanov-Tsonchev, R.; Villa-Hernández, J.; de la Rosa-Vargas, I.; López-Cruz, I.; Silos-Espino, H.; Lara-Herrera, A. Chlorophyll fluorescence emission of tomato plants as a response to pulsed light based LEDs. Plant Growth Regul. 2013, 69, 117–123. [Google Scholar] [CrossRef] [Green Version]
- Moon, Y.R.; Lee, M.H.; Tovuu, A.; Lee, C.-H.; Chung, B.Y.; Park, Y.-I.; Kim, J.-H. Acute exposure to UV-B sensitizes cucumber, tomato, and Arabidopsis plants to photooxidative stress by inhibiting thermal energy dissipation and antioxidant defense. J. Radiat. Res. 2011, 52, 238–248. [Google Scholar] [CrossRef] [Green Version]
- Raja, V.; Qadir, S.U.; Alyemeni, M.N.; Ahmad, P. Impact of drought and heat stress individually and in combination on physio-biochemical parameters, antioxidant responses, and gene expression in Solanum lycopersicum. 3 Biotech 2020, 10, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Ruban, A.V.; Johnson, M.P.; Duffy, C.D. The photoprotective molecular switch in the photosystem II antenna. Biochim. Biophys. Acta BBA—Bioenergy 2012, 1817, 167–181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Croce, R. PsbS is the plants’ pick for sun protection. Nat. Struct. Mol. Biol. 2015, 22, 650–652. [Google Scholar] [CrossRef] [PubMed]
- Fan, M.; Li, M.; Liu, Z.; Cao, P.; Pan, X.; Zhang, H.; Zhao, X.; Zhang, J.; Chang, W. Crystal structures of the PsbS protein essential for photoprotection in plants. Nat. Struct. Mol. Biol. 2015, 22, 729–735. [Google Scholar] [CrossRef]
- Kromdijk, J.; Głowacka, K.; Leonelli, L.; Gabilly, S.T.; Iwai, M.; Niyogi, K.K.; Long, S.P. Improving photosynthesis and crop productivity by accelerating recovery from photoprotection. Science 2016, 354, 857–861. [Google Scholar] [CrossRef] [Green Version]
- Suorsa, M.; Sirpiö, S.; Allahverdiyeva, Y.; Paakkarinen, V.; Mamedov, F.; Styring, S.; Aro, E.-M. PsbR, a missing link in the assembly of the oxygen-evolving complex of plant photosystem II. J. Biol. Chem. 2006, 281, 145–150. [Google Scholar] [CrossRef] [Green Version]
- Allahverdiyeva, Y.; Mamedov, F.; Suorsa, M.; Styring, S.; Vass, I.; Aro, E.-M. Insights into the function of PsbR protein in Arabidopsis thaliana. Biochim. Biophys. Acta BBA—Bioenergy 2007, 1767, 677–685. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakata, S.; Mizusawa, N.; Kubota-Kawai, H.; Sakurai, I.; Wada, H. Psb28 is involved in recovery of photosystem II at high temperature in Synechocystis sp. PCC 6803. Biochim. Biophys. Acta BBA—Bioenergy 2013, 1827, 50–59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weisz, D.A.; Liu, H.; Zhang, H.; Thangapandian, S.; Tajkhorshid, E.; Gross, M.L.; Pakrasi, H.B. Mass spectrometry-based cross-linking study shows that the Psb28 protein binds to cytochrome b559 in Photosystem II. Proc. Natl. Acad. Sci. USA 2017, 114, 2224–2229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pagliano, C.; Saracco, G.; Barber, J. Structural, functional and auxiliary proteins of photosystem II. Photosynth. Res. 2013, 116, 167–188. [Google Scholar] [CrossRef]
- Bečková, M.; Gardian, Z.; Yu, J.; Konik, P.; Nixon, P.J.; Komenda, J. Association of Psb28 and Psb27 proteins with PSII-PSI supercomplexes upon exposure of Synechocystis sp. PCC 6803 to high light. Mol. Plant 2017, 10, 62–72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tomato Genome Consortium. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 2012, 485, 635–641. [Google Scholar] [CrossRef] [Green Version]
- Hoagland, D.R.; Arnon, D.I. The water-culture method for growing plants without soil. Circ. Calif. Agric. Exp. Stn. 1950, 347, 1–32. [Google Scholar]
- Wu, B.-S.; Lefsrud, M.G. Photobiology eye safety for horticultural LED lighting: Transmittance performance of eyewear protection using high-irradiant monochromatic LEDs. J. Occup. Environ. Hyg. 2018, 15, 133–142. [Google Scholar] [CrossRef]
- Dixon, M.; Grace, J. Natural convection from leaves at realistic Grashof numbers. Plant Cell Environ. 1983, 6, 665–670. [Google Scholar]
- Murchie, E.H.; Lawson, T. Chlorophyll fluorescence analysis: A guide to good practice and understanding some new applications. J. Exp. Bot. 2013, 64, 3983–3998. [Google Scholar] [CrossRef] [Green Version]
- Abraham, P.; Adams, R.; Giannone, R.J.; Kalluri, U.; Ranjan, P.; Erickson, B.; Shah, M.; Tuskan, G.A.; Hettich, R.L. Defining the boundaries and characterizing the landscape of functional genome expression in vascular tissues of Populus using shotgun proteomics. J. Proteome Res. 2012, 11, 449–460. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Xiong, E.; Wang, W.; Scali, M.; Cresti, M. Universal sample preparation method integrating trichloroacetic acid/acetone precipitation with phenol extraction for crop proteomic analysis. Nat. Protoc. 2014, 9, 362–374. [Google Scholar] [CrossRef]
- Kislinger, T.; Gramolini, A.O.; MacLennan, D.H.; Emili, A. Multidimensional protein identification technology (MudPIT): Technical overview of a profiling method optimized for the comprehensive proteomic investigation of normal and diseased heart tissue. J. Am. Soc. Mass Spectrom. 2005, 16, 1207–1220. [Google Scholar] [CrossRef] [Green Version]
- Patel, A.K.; Huang, E.L.; Low-Décarie, E.; Lefsrud, M.G. Comparative shotgun proteomic analysis of wastewater-cultured microalgae: Nitrogen sensing and carbon fixation for growth and nutrient removal in Chlamydomonas reinhardtii. J. Proteome Res. 2015, 14, 3051–3067. [Google Scholar] [CrossRef]
- McDonald, W.H.; Ohi, R.; Miyamoto, D.T.; Mitchison, T.J.; Yates, J.R., III. Comparison of three directly coupled HPLC MS/MS strategies for identification of proteins from complex mixtures: Single-dimension LC-MS/MS, 2-phase MudPIT, and 3-phase MudPIT. Int. J. Mass Spectrom. 2002, 219, 245–251. [Google Scholar] [CrossRef]
- Wilmes, P.; Andersson, A.F.; Lefsrud, M.G.; Wexler, M.; Shah, M.; Zhang, B.; Hettich, R.L.; Bond, P.L.; VerBerkmoes, N.C.; Banfield, J.F. Community proteogenomics highlights microbial strain-variant protein expression within activated sludge performing enhanced biological phosphorus removal. ISME J. 2008, 2, 853–864. [Google Scholar] [CrossRef]
- Dorfer, V.; Pichler, P.; Stranzl, T.; Stadlmann, J.; Taus, T.; Winkler, S.; Mechtler, K. MS Amanda, a universal identification algorithm optimized for high accuracy tandem mass spectra. J. Proteome Res. 2014, 13, 3679–3684. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus computational platform for comprehensive analysis of (prote) omics data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef]
- Zybailov, B.; Mosley, A.L.; Sardiu, M.E.; Coleman, M.K.; Florens, L.; Washburn, M.P. Statistical analysis of membrane proteome expression changes in Saccharomyces cerevisiae. J. Proteome Res. 2006, 5, 2339–2347. [Google Scholar] [CrossRef] [PubMed]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.-H.; Pagès, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef] [Green Version]
- Huang, D.W.; Sherman, B.T.; Tan, Q.; Collins, J.R.; Alvord, W.G.; Roayaei, J.; Stephens, R.; Baseler, M.W.; Lane, H.C.; Lempicki, R.A. The DAVID Gene Functional Classification Tool: A novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007, 8, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Løvdal, T.; Lillo, C. Reference gene selection for quantitative real-time PCR normalization in tomato subjected to nitrogen, cold, and light stress. Anal. Biochem. 2009, 387, 238–242. [Google Scholar] [CrossRef]
- Dekkers, B.J.; Willems, L.; Bassel, G.W.; van Bolderen-Veldkamp, R.; Ligterink, W.; Hilhorst, H.W.; Bentsink, L.J.P. Identification of reference genes for RT–qPCR expression analysis in Arabidopsis and tomato seeds. Plant Cell Physiol. 2012, 53, 28–37. [Google Scholar] [CrossRef] [Green Version]
- Petroutsos, D.; Tokutsu, R.; Maruyama, S.; Flori, S.; Greiner, A.; Magneschi, L.; Cusant, L.; Kottke, T.; Mittag, M.; Hegemann, P. A blue-light photoreceptor mediates the feedback regulation of photosynthesis. Nature 2016, 537, 563–566. [Google Scholar] [CrossRef] [PubMed]
- Walters, R.G. Towards an understanding of photosynthetic acclimation. J. Exp. Bot. 2005, 56, 435–447. [Google Scholar] [CrossRef]
- Walters, R.G.; Horton, P. Acclimation of Arabidopsis thaliana to the light environment: Regulation of chloroplast composition. Planta 1995, 197, 475–481. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, S. Simulation of the canopy photosynthesis model of greenhouse tomato. Procedia Eng. 2011, 16, 632–639. [Google Scholar] [CrossRef] [Green Version]
- Yousef, A.F.; Ali, M.M.; Rizwan, H.M.; Tadda, S.A.; Kalaji, H.M.; Yang, H.; Ahmed, M.A.; Wróbel, J.; Xu, Y.; Chen, F. Photosynthetic apparatus performance of tomato seedlings grown under various combinations of LED illumination. PLoS ONE 2021, 16, e0249373. [Google Scholar]
- Bednarczyk, D.; Aviv-Sharon, E.; Savidor, A.; Levin, Y.; Charuvi, D. Influence of short-term exposure to high light on photosynthesis and proteins involved in photo-protective processes in tomato leaves. Environ. Exp. Bot. 2020, 179, 104198. [Google Scholar] [CrossRef]
- Arena, C.; Tsonev, T.; Doneva, D.; De Micco, V.; Michelozzi, M.; Brunetti, C.; Centritto, M.; Fineschi, S.; Velikova, V.; Loreto, F. The effect of light quality on growth, photosynthesis, leaf anatomy and volatile isoprenoids of a monoterpene-emitting herbaceous species (Solanum lycopersicum L.) and an isoprene-emitting tree (Platanus orientalis L.). Environ. Exp. Bot. 2016, 130, 122–132. [Google Scholar] [CrossRef]
- He, J.; Qin, L.; Chong, E.L.; Choong, T.-W.; Lee, S.K. Plant growth and photosynthetic characteristics of Mesembryanthemum crystallinum grown aeroponically under different blue-and red-LEDs. Front. Plant Sci. 2017, 8, 361. [Google Scholar] [CrossRef] [Green Version]
- Hakala, M.; Tuominen, I.; Keränen, M.; Tyystjärvi, T.; Tyystjärvi, E. Evidence for the role of the oxygen-evolving manganese complex in photoinhibition of photosystem II. Biochim. Biophys. Acta BBA—Bioenergy 2005, 1706, 68–80. [Google Scholar] [CrossRef] [Green Version]
- Hajihashemi, S.; Brestic, M.; Kalaji, H.; Skalicky, M.; Noedoost, F. Environmental pollution is reflected in the activity of the photosynthetic apparatus. Photosynthetica 2020, 58, 529–539. [Google Scholar] [CrossRef] [Green Version]
- Kalaji, H.M.; Jajoo, A.; Oukarroum, A.; Brestic, M.; Zivcak, M.; Samborska, I.A.; Cetner, M.D.; Łukasik, I.; Goltsev, V.; Ladle, R.J. Chlorophyll a fluorescence as a tool to monitor physiological status of plants under abiotic stress conditions. Acta Physiol. Plant. 2016, 38, 102. [Google Scholar] [CrossRef] [Green Version]
- Hao, Y.; Feng, Y.; Cai, L.; Wu, Q.; Song, L. Effect of ABA on Photosynthesis and Chlorophyll Fluorescence in Emmenopterys henri Oliv. under High Light. Russ. J. Plant Physiol. 2021, 68, 510–518. [Google Scholar] [CrossRef]
- Allen, J.F.; de Paula, W.B.; Puthiyaveetil, S.; Nield, J. A structural phylogenetic map for chloroplast photosynthesis. Trends Plant Sci. 2011, 16, 645–655. [Google Scholar] [CrossRef]
- Allen, J.F. The CoRR hypothesis for genes in organelles. J. Theor. Biol. 2017, 434, 50–57. [Google Scholar] [CrossRef]
- Li, D.; Wang, M.; Zhang, T.; Chen, X.; Li, C.; Liu, Y.; Brestic, M.; Chen, T.H.; Yang, X. Glycinebetaine mitigated the photoinhibition of photosystem II at high temperature in transgenic tomato plants. Photosynth. Res. 2021, 147, 301–315. [Google Scholar] [CrossRef]
- Górecka, M.; Lewandowska, M.; Dąbrowska-Bronk, J.; Białasek, M.; Barczak-Brzyżek, A.; Kulasek, M.; Mielecki, J.; Kozłowska-Makulska, A.; Gawroński, P.; Karpiński, S. Photosystem II 22 kDa protein level-a prerequisite for excess light-inducible memory, cross-tolerance to UV-C and regulation of electrical signalling. Plant Cell Environ. 2020, 43, 649–661. [Google Scholar] [CrossRef] [PubMed]
- Dobáková, M.; Sobotka, R.; Tichý, M.; Komenda, J. Psb28 protein is involved in the biogenesis of the photosystem II inner antenna CP47 (PsbB) in the Cyanobacterium synechocystis sp. PCC 6803. Plant Physiol. 2009, 149, 1076–1086. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maier, T.; Güell, M.; Serrano, L. Correlation of mRNA and protein in complex biological samples. FEBS Lett. 2009, 583, 3966–3973. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buccitelli, C.; Selbach, M. mRNAs, proteins and the emerging principles of gene expression control. Nat. Rev. Genet. 2020, 21, 630–644. [Google Scholar] [CrossRef] [PubMed]
- Shim, J.S.; Park, S.-H.; Lee, D.-K.; Kim, Y.S.; Park, S.-C.; Redillas, M.C.F.R.; Seo, J.S.; Kim, J.-K. The Rice GLYCINE-RICH PROTEIN 3 Confers Drought Tolerance by Regulating mRNA Stability of ROS Scavenging-Related Genes. Rice 2021, 14, 1–19. [Google Scholar] [CrossRef]
- Espinoza-Corral, R.; Schwenkert, S.; Lundquist, P.K. Molecular changes of Arabidopsis thaliana plastoglobules facilitate thylakoid membrane remodeling under high light stress. Plant J. 2021, 106, 1571–1587. [Google Scholar] [CrossRef]
- Gan, S.; Amasino, R.M. Making sense of senescence (molecular genetic regulation and manipulation of leaf senescence). Plant Physiol. 1997, 113, 313–319. [Google Scholar] [CrossRef] [Green Version]
- Bitrián, M.; Zarza, X.; Altabella, T.; Tiburcio, A.F.; Alcázar, R. Polyamines under abiotic stress: Metabolic crossroads and hormonal crosstalks in plants. Metabolites 2012, 2, 516–528. [Google Scholar] [CrossRef] [Green Version]




| Gene | Forward Primer | Reverse Primer | PCR Product Size (bp) | Source |
|---|---|---|---|---|
| psb28 | CCTCGCTCTCTTCTCGGAAT | GCAAAACGCGAACGGGATAG | 98 | This study |
| psbS | GGAATTGGCTTCACTAAGCA | AGTGGCTCTGCTTCATAGAT | 155 | This study |
| psbH | TCTGGTCCAAGACGAACTGC | CAAAGGGGTAGTTCCCCACC | 93 | This study |
| psbR | CAGGAAGCCCAAGGGAAAGG | GTCACCGCCCATATGGCTAA | 153 | This study |
| TPP2Acs | CGATGTGTGATCTCCTATGGTC | AAGCTGATGGGCTCTAGAAATC | 149 | Løvdal and Lillo [34] |
| Clat | ATGCAATCACACCAGCAC | ACTCAGCACAACAACAAAGG | 61 | Dekkers et al. [35] |
| BLT (log2FC) | RLT a (log2FC) | Location | Function | |||||
|---|---|---|---|---|---|---|---|---|
| Protein | Limit | Burned | Regular | Limit | Burned | Regular | ||
| Psb28 | 0.02 | −1.27 | 0.14 | 0.014 | 1.084 * | −0.044 | OEC, binds to cytochrome b559 | PSII assembly factor |
| PsbS | 1.45 | −0.34 | 0.15 | 0.516 | 1.322 * | 0.322 | LHCII | NPQ relaxation process |
| PsbH | 0.73 | −0.23 | 0.74 | −0.713 * | 1.646 * | 0.536 | PSII complex core | Electron transfer between QA and QB |
| PsbR | 1.24 | −0.12 | 1.52 | −0.168 | 1.111 * | 0.151 | OEC, binds to PsbQ and PsbP | OEC formation |
| Sample Name | Protein Accession | Description | Present in BLT and RLT |
|---|---|---|---|
| Regular | Q10712 | Leucine aminopeptidase 1, chloroplastic | X |
| P25306 | Threonine dehydratase biosynthetic, chloroplastic | X | |
| K4CVX0 | Uncharacterized protein | X | |
| Q5UNS1 | Arginase 2 | X | |
| K4CVX6 | Uncharacterized protein | X | |
| Burned | K4ATA4 | Uncharacterized protein | X |
| Limit | Q10712 | Leucine aminopeptidase 1, chloroplastic | X |
| K4CWC4 | PR10 protein | X | |
| K4CVX0 | Uncharacterized protein | X | |
| P25306 | Threonine dehydratase biosynthetic, chloroplastic | X | |
| Q01413 | Glucan endo-1,3-β-glucosidase B | X | |
| K4CVQ7 | Uncharacterized protein | X | |
| Q05539 | Acidic 26 kDa endochitinase | ||
| K4B0B4 | Uncharacterized protein | ||
| K4C3T2 | Uncharacterized protein | X | |
| A0RZD0 | Inducible plastid-lipid associated protein | X | |
| Q9LEG1 | Cathepsin D Inhibitor |
| Sample Name | Protein Accession | Description | Present in BLT and RLT |
|---|---|---|---|
| Regular | K4CAE2 | Uncharacterized protein | |
| K4ASV2 | ATP-dependent Clp protease proteolytic subunit | ||
| K4B7W7 | Uncharacterized protein | X | |
| K4CMI6 | Uncharacterized protein | X | |
| Burned | K4CVQ7 | Uncharacterized protein | X |
| K4BM57 | Uncharacterized protein | X | |
| K4BVE2 | 50S ribosomal protein L31 | X | |
| P37218 | Histone H1 | X | |
| K4AYJ8 | Uncharacterized protein | X | |
| K4B0G3 | Uncharacterized protein | X | |
| K4AX22 | Superoxide dismutase [Cu–Zn] | X | |
| K4C998 | Uncharacterized protein | X | |
| P04284 | Pathogenesis-related leaf protein 6 | X | |
| E5KBY0 | Snakin-2 | X | |
| Q2MI49 | Photosystem I iron-sulfur center | X | |
| K4C1V2 | Uncharacterized protein | ||
| K4CX44 | Uncharacterized protein | ||
| Q3I5C4 | Cytosolic ascorbate peroxidase 1 | ||
| K4BJY6 | Uncharacterized protein | ||
| P43282 | S-adenosylmethionine synthase 3 | ||
| C0KKU8 | Lipoxygenase | ||
| P10708 | Chlorophyll a-b binding protein 7, chloroplastic | ||
| Limit | K4BVE2 | 50S ribosomal protein L31 | |
| K4BX19 | Uncharacterized protein | X | |
| K4C1V2 | Uncharacterized protein | X | |
| K4BLU6 | Uncharacterized protein | ||
| K4D2D7 | Uncharacterized protein | X |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Parrine, D.; Greco, T.M.; Muhammad, B.; Wu, B.-S.; Zhao, X.; Lefsrud, M. Color-Specific Recovery to Extreme High-Light Stress in Plants. Life 2021, 11, 812. https://doi.org/10.3390/life11080812
Parrine D, Greco TM, Muhammad B, Wu B-S, Zhao X, Lefsrud M. Color-Specific Recovery to Extreme High-Light Stress in Plants. Life. 2021; 11(8):812. https://doi.org/10.3390/life11080812
Chicago/Turabian StyleParrine, Débora, Todd M. Greco, Bilal Muhammad, Bo-Sen Wu, Xin Zhao, and Mark Lefsrud. 2021. "Color-Specific Recovery to Extreme High-Light Stress in Plants" Life 11, no. 8: 812. https://doi.org/10.3390/life11080812
APA StyleParrine, D., Greco, T. M., Muhammad, B., Wu, B.-S., Zhao, X., & Lefsrud, M. (2021). Color-Specific Recovery to Extreme High-Light Stress in Plants. Life, 11(8), 812. https://doi.org/10.3390/life11080812







