A Systematic Comparison of Antiandrogens Identifies Androgen Receptor Protein Stability as an Indicator for Treatment Response
Abstract
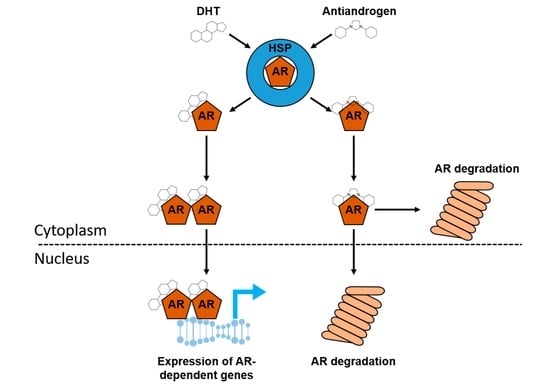
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Drug Treatment
2.3. Subcellular Fractionation
2.4. Western Blot Analysis
2.5. Ribonucleic Acid (RNA) Isolation and Quantitative Real-Time PCR (qPCR)
2.6. Measurement of Cell Viability
2.7. Statistical Analysis
3. Results
3.1. Antiandrogens Affect AR-Mediated Gene Transactivation in a Cell-Specific Manner
3.2. Cell Viability Is Affected in a Cell-Specific Manner by Antiandrogens
3.3. Bicalutamide, Enzalutamide, Apalutamide, and Darolutamide Reduce Nuclear AR Protein Levels after Treatment
3.4. Influence of Antiandrogen Treatment on AR mRNA and Protein Level
3.5. AR Protein Degradation Is an Early Event after Antiandrogen Treatment
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Fujita, K.; Nonomura, N. Role of androgen receptor in prostate cancer: A review. World J. Mens. Health 2019, 37, 288–295. [Google Scholar] [CrossRef]
- Crona, D.J.; Whang, Y.E. Androgen receptor-dependent and independent mechanisms involved in prostate cancer therapy resistance. Cancers 2017, 9, 67. [Google Scholar] [CrossRef]
- Santer, F.R.; Erb, H.H.; McNeill, R.V. Therapy escape mechanisms in the malignant prostate. Semin. Cancer Biol. 2015, 35, 133–144. [Google Scholar] [CrossRef] [Green Version]
- Estébanez-Perpiñá, E.; Bevan, C.L.; McEwan, I.J. Eighty years of targeting androgen receptor activity in prostate cancer: The fight goes on. Cancers 2021, 13, 509. [Google Scholar] [CrossRef]
- Cornford, P.; van den Bergh, R.C.N.; Briers, E.; Van den Broeck, T.; Cumberbatch, M.G.; De Santis, M.; Fanti, S.; Fossati, N.; Gandaglia, G.; Gillessen, S.; et al. EAU-EANM-ESTRO-ESUR-SIOG guidelines on prostate cancer. Part II-2020. Update: Treatment of relapsing and metastatic prostate cancer. Eur. Urol. 2021, 79, 263–282. [Google Scholar] [CrossRef]
- Mottet, N.; van den Bergh, R.C.N.; Briers, E.; Van den Broeck, T.; Cumberbatch, M.G.; De Santis, M.; Fanti, S.; Fossati, N.; Gandaglia, G.; Gillessen, S.; et al. EAU-EANM-ESTRO-ESUR-SIOG guidelines on prostate cancer-2020 Update. Part 1: Screening, diagnosis, and local treatment with curative intent. Eur. Urol. 2021, 79, 243–262. [Google Scholar] [CrossRef] [PubMed]
- Devlies, W.; Handle, F.; Devos, G.; Joniau, S.; Claessens, F. Preclinical models in prostate cancer: Resistance to AR targeting therapies in prostate cancer. Cancers 2021, 13, 915. [Google Scholar] [CrossRef] [PubMed]
- Maitland, N.J.; Frame, F.M.; Rane, J.K.; Erb, H.H.; Packer, J.R.; Archer, L.K.; Pellacani, D. Resolution of cellular heterogeneity in human prostate cancers: Implications for diagnosis and treatment. Adv. Exp. Med. Biol. 2019, 1164, 207–224. [Google Scholar] [CrossRef] [PubMed]
- Daoud, G.; Monzer, A.; Bahmad, H.; Chamaa, F.; Hamdar, L.; Mouhieddine, T.H.; Shayya, S.; Eid, A.; Kobeissy, F.; Liu, Y.N.; et al. Primary versus castration-resistant prostate cancer: Modeling through novel murine prostate cancer cell lines. Oncotarget 2016, 7, 28961–28975. [Google Scholar] [CrossRef] [PubMed]
- Maitland, N.J. Getting closer to prostate cancer in patients—What scientists should want from clinicians. J. Cancer Metastasis Treat. 2017, 3, 262–270. [Google Scholar] [CrossRef]
- Moilanen, A.M.; Riikonen, R.; Oksala, R.; Ravanti, L.; Aho, E.; Wohlfahrt, G.; Nykanen, P.S.; Tormakangas, O.P.; Palvimo, J.J.; Kallio, P.J. Discovery of ODM-201, a new-generation androgen receptor inhibitor targeting resistance mechanisms to androgen signaling-directed prostate cancer therapies. Sci. Rep. 2015, 5, 12007. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clegg, N.J.; Wongvipat, J.; Joseph, J.D.; Tran, C.; Ouk, S.; Dilhas, A.; Chen, Y.; Grillot, K.; Bischoff, E.D.; Cai, L.; et al. ARN-509: A novel antiandrogen for prostate cancer treatment. Cancer Res. 2012, 72, 1494–1503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Squillace, R.M.; Miller, D.; Wardwell, S.D.; Wang, F.; Clackson, T.; Rivera, V.M. Synergistic activity of the mTOR inhibitor ridaforolimus and the antiandrogen bicalutamide in prostate cancer models. Int. J. Oncol. 2012, 41, 425–432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jung, M.Y.D.; Sawyers, C.; Tran, C. Diarylthiohydantoin Compounds US2007254933 A1. Available online: https://patentimages.storage.googleapis.com/da/0c/bb/ab19dea2a7c04d/US20070254933A1.pdf (accessed on 20 August 2021).
- Tran, C.; Ouk, S.; Clegg, N.J.; Chen, Y.; Watson, P.A.; Arora, V.; Wongvipat, J.; Smith-Jones, P.M.; Yoo, D.; Kwon, A.; et al. Development of a second-generation antiandrogen for treatment of advanced prostate cancer. Science 2009, 324, 787–790. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thalmann, G.N.; Sikes, R.A.; Wu, T.T.; Degeorges, A.; Chang, S.M.; Ozen, M.; Pathak, S.; Chung, L.W. LNCaP progression model of human prostate cancer: Androgen-independence and osseous metastasis. Prostate 2000, 44, 91–103. [Google Scholar] [CrossRef]
- Erb, H.H.H.; Bodenbender, J.; Handle, F.; Diehl, T.; Donix, L.; Tsaur, I.; Gleave, M.; Haferkamp, A.; Huber, J.; Fuessel, S.; et al. Assessment of STAT5 as a potential therapy target in enzalutamide-resistant prostate cancer. PLoS ONE 2020, 15, e0237248. [Google Scholar] [CrossRef]
- Horoszewicz, J.S.; Leong, S.S.; Chu, T.M.; Wajsman, Z.L.; Friedman, M.; Papsidero, L.; Kim, U.; Chai, L.S.; Kakati, S.; Arya, S.K.; et al. The LNCaP cell line—A new model for studies on human prostatic carcinoma. Prog. Clin. Biol. Res. 1980, 37, 115–132. [Google Scholar]
- Klein, K.A.; Reiter, R.E.; Redula, J.; Moradi, H.; Zhu, X.L.; Brothman, A.R.; Lamb, D.J.; Marcelli, M.; Belldegrun, A.; Witte, O.N.; et al. Progression of metastatic human prostate cancer to androgen independence in immunodeficient SCID mice. Nat. Med. 1997, 3, 402–408. [Google Scholar] [CrossRef]
- Tai, S.; Sun, Y.; Squires, J.M.; Zhang, H.; Oh, W.K.; Liang, C.Z.; Huang, J. PC3 is a cell line characteristic of prostatic small cell carcinoma. Prostate 2011, 71, 1668–1679. [Google Scholar] [CrossRef] [Green Version]
- Kaighn, M.E.; Narayan, K.S.; Ohnuki, Y.; Lechner, J.F.; Jones, L.W. Establishment and characterization of a human prostatic carcinoma cell line (PC-3). Investig. Urol. 1979, 17, 16–23. [Google Scholar]
- Bonne, C.; Raynaud, J.P. Assay of androgen binding sites by exchange with methyltrienolone (R 1881). Steroids 1976, 27, 497–507. [Google Scholar] [CrossRef]
- Abazid, A.; Martin, B.; Choinowski, A.; McNeill, R.V.; Brandenburg, L.O.; Ziegler, P.; Zimmermann, U.; Burchardt, M.; Erb, H.; Stope, M.B. The androgen receptor antagonist enzalutamide induces apoptosis, dysregulates the heat shock protein system, and diminishes the androgen receptor and estrogen receptor beta1 expression in prostate cancer cells. J. Cell. Biochem. 2019, 120, 16711–16722. [Google Scholar] [CrossRef]
- Erb, H.H.H.; Oster, M.A.; Gelbrich, N.; Cammann, C.; Thomas, C.; Mustea, A.; Stope, M.B. Enzalutamide-induced proteolytic degradation of the androgen receptor in prostate cancer cells is mediated only to a limited extent by the proteasome system. Anticancer Res. 2021, 41, 3271–3279. [Google Scholar] [CrossRef] [PubMed]
- Massie, C.E.; Lynch, A.; Ramos-Montoya, A.; Boren, J.; Stark, R.; Fazli, L.; Warren, A.; Scott, H.; Madhu, B.; Sharma, N.; et al. The androgen receptor fuels prostate cancer by regulating central metabolism and biosynthesis. EMBO J. 2011, 30, 2719–2733. [Google Scholar] [CrossRef] [Green Version]
- Handle, F.; Puhr, M.; Schaefer, G.; Lorito, N.; Hoefer, J.; Gruber, M.; Guggenberger, F.; Santer, F.R.; Marques, R.B.; van Weerden, W.M.; et al. The STAT3 inhibitor galiellalactone reduces IL6-mediated AR activity in benign and malignant prostate models. Mol. Cancer Ther. 2018, 17, 2722–2731. [Google Scholar] [CrossRef] [Green Version]
- Jin, H.J.; Kim, J.; Yu, J. Androgen receptor genomic regulation. Transl. Androl. Urol. 2013, 2, 157–177. [Google Scholar] [CrossRef]
- Cleutjens, K.B.; van der Korput, H.A.; van Eekelen, C.C.; van Rooij, H.C.; Faber, P.W.; Trapman, J. An androgen response element in a far upstream enhancer region is essential for high, androgen-regulated activity of the prostate-specific antigen promoter. Mol. Endocrinol. 1997, 11, 148–161. [Google Scholar] [CrossRef]
- Xu, J.; Kalos, M.; Stolk, J.A.; Zasloff, E.J.; Zhang, X.; Houghton, R.L.; Filho, A.M.; Nolasco, M.; Badaro, R.; Reed, S.G. Identification and characterization of prostein, a novel prostate-specific protein. Cancer Res. 2001, 61, 1563–1568. [Google Scholar]
- Ng, K.; Smith, S.; Shamash, J. Metastatic hormone-sensitive prostate cancer (mHSPC): Advances and treatment strategies in the first-line setting. Oncol. Ther. 2020, 8, 209–230. [Google Scholar] [CrossRef]
- Verma, S.; Prajapati, K.S.; Kushwaha, P.P.; Shuaib, M.; Kumar Singh, A.; Kumar, S.; Gupta, S. Resistance to second generation antiandrogens in prostate cancer: Pathways and mechanisms. Cancer Drug Resist. 2020, 3, 742–761. [Google Scholar] [CrossRef]
- Davis, I.D.; Martin, A.J.; Stockler, M.R.; Begbie, S.; Chi, K.N.; Chowdhury, S.; Coskinas, X.; Frydenberg, M.; Hague, W.E.; Horvath, L.G.; et al. Enzalutamide with standard first-line therapy in metastatic prostate cancer. N. Engl. J. Med. 2019, 381, 121–131. [Google Scholar] [CrossRef]
- Chi, K.N.; Agarwal, N.; Bjartell, A.; Chung, B.H.; Pereira de Santana Gomes, A.J.; Given, R.; Juarez Soto, A.; Merseburger, A.S.; Ozguroglu, M.; Uemura, H.; et al. Apalutamide for Metastatic, Castration-Sensitive Prostate Cancer. N. Engl. J. Med. 2019, 381, 13–24. [Google Scholar] [CrossRef]
- Kwon, D.H.; Friedlander, T. A TITAN step forward: Apalutamide for metastatic castration-sensitive prostate cancer. Ann. Transl. Med. 2019, 7, S364. [Google Scholar] [CrossRef]
- Smith, M.R.; Saad, F.; Hussain, M.; Sternberg, C.N.; Fizazi, K.; Yamada, K.S.; Kappeler, C.; Kuss, I.; Tombal, B.F. ARASENS: A phase 3 trial of darolutamide in combination with docetaxel for men with metastatic hormone-sensitive prostate cancer (mHSPC). J. Clin. Oncol. 2018, 36, TPS383. [Google Scholar] [CrossRef]
- Heidenreich, A.; Chowdhury, S.; Klotz, L.; Siemens, D.R.; Villers, A.; Ivanescu, C.; Holmstrom, S.; Baron, B.; Wang, F.; Lin, P.; et al. Impact of enzalutamide compared with bicalutamide on quality of life in men with metastatic castration-resistant prostate cancer: Additional analyses from the TERRAIN randomised clinical trial. Eur. Urol. 2017, 71, 534–542. [Google Scholar] [CrossRef]
- Kuil, C.W.; Brinkmann, A.O. Androgens, antiandrogens and androgen receptor abnormalities. Eur. Urol. 1996, 29 (Suppl. 2), 78–82. [Google Scholar] [CrossRef]
- Fizazi, K.; Albiges, L.; Loriot, Y.; Massard, C. ODM-201: A new-generation androgen receptor inhibitor in castration-resistant prostate cancer. Expert Rev. Anticancer Ther. 2015, 15, 1007–1017. [Google Scholar] [CrossRef]
- Hay, C.W.; McEwan, I.J. The impact of point mutations in the human androgen receptor: Classification of mutations on the basis of transcriptional activity. PLoS ONE 2012, 7, e32514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lallous, N.; Volik, S.V.; Awrey, S.; Leblanc, E.; Tse, R.; Murillo, J.; Singh, K.; Azad, A.A.; Wyatt, A.W.; LeBihan, S.; et al. Functional analysis of androgen receptor mutations that confer anti-androgen resistance identified in circulating cell-free DNA from prostate cancer patients. Genome Biol. 2016, 17, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martignano, F.; Lolli, C.; Ravaglia, G.; Gallà, V.; Gurioli, G.; Salvi, S. Emerging mutations and functional changes of androgen receptor associated with treatment resistance in prostate cancer. Transl. Cancer Res. 2016, 5, S803–S808. [Google Scholar] [CrossRef]
- Veldscholte, J.; Berrevoets, C.A.; Ris-Stalpers, C.; Kuiper, G.G.; Jenster, G.; Trapman, J.; Brinkmann, A.O.; Mulder, E. The androgen receptor in LNCaP cells contains a mutation in the ligand binding domain which affects steroid binding characteristics and response to antiandrogens. J. Steroid Biochem. Mol. Biol. 1992, 41, 665–669. [Google Scholar] [CrossRef]
- Zhao, J.; Ning, S.; Lou, W.; Yang, J.C.; Armstrong, C.M.; Lombard, A.P.; D’Abronzo, L.S.; Evans, C.P.; Gao, A.C.; Liu, C. Cross-resistance among next-generation antiandrogen drugs through the AKR1C3/AR-V7 axis in advanced prostate cancer. Mol. Cancer Ther. 2020, 19, 1708–1718. [Google Scholar] [CrossRef]
- Wadosky, K.M.; Koochekpour, S. Androgen receptor splice variants and prostate cancer: From bench to bedside. Oncotarget 2017, 8, 18550–18576. [Google Scholar] [CrossRef] [Green Version]
- Puhr, M.; Hoefer, J.; Eigentler, A.; Ploner, C.; Handle, F.; Schaefer, G.; Kroon, J.; Leo, A.; Heidegger, I.; Eder, I.; et al. The glucocorticoid receptor is a key player for prostate cancer cell survival and a target for improved antiandrogen therapy. Clin. Cancer Res. 2018, 24, 927–938. [Google Scholar] [CrossRef] [Green Version]
- Ishikura, N.; Kawata, H.; Nishimoto, A.; Nakamura, R.; Tsunenari, T.; Watanabe, M.; Tachibana, K.; Shiraishi, T.; Yoshino, H.; Honma, A.; et al. CH5137291, an androgen receptor nuclear translocation-inhibiting compound, inhibits the growth of castration-resistant prostate cancer cells. Int. J. Oncol. 2015, 46, 1560–1572. [Google Scholar] [CrossRef]
- Lv, S.; Song, Q.; Chen, G.; Cheng, E.; Chen, W.; Cole, R.; Wu, Z.; Pascal, L.E.; Wang, K.; Wipf, P.; et al. Regulation and targeting of androgen receptor nuclear localization in castration-resistant prostate cancer. J. Clin. Investig. 2021, 131. [Google Scholar] [CrossRef]
- Hoefer, J.; Akbor, M.; Handle, F.; Ofer, P.; Puhr, M.; Parson, W.; Culig, Z.; Klocker, H.; Heidegger, I. Critical role of androgen receptor level in prostate cancer cell resistance to new generation antiandrogen enzalutamide. Oncotarget 2016, 7, 59781–59794. [Google Scholar] [CrossRef] [Green Version]
- Brooke, G.N.; Gamble, S.C.; Hough, M.A.; Begum, S.; Dart, D.A.; Odontiadis, M.; Powell, S.M.; Fioretti, F.M.; Bryan, R.A.; Waxman, J.; et al. Antiandrogens act as selective androgen receptor modulators at the proteome level in prostate cancer cells. Mol. Cell. Proteom. 2015, 14, 1201–1216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lakshmana, G.; Baniahmad, A. Interference with the androgen receptor protein stability in therapy-resistant prostate cancer. Int. J. Cancer 2019, 144, 1775–1779. [Google Scholar] [CrossRef]
- Liu, C.; Lou, W.; Yang, J.C.; Liu, L.; Armstrong, C.M.; Lombard, A.P.; Zhao, R.; Noel, O.D.V.; Tepper, C.G.; Chen, H.-W.; et al. Proteostasis by STUB1/HSP70 complex controls sensitivity to androgen receptor targeted therapy in advanced prostate cancer. Nat. Commun. 2018, 9, 4700. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhang, H.; Zhang, X.; Wang, P.; Wang, H.; Huang, F.; Zhou, C.; Zhou, J.; Li, S. PC-1 works in conjunction with E3 ligase CHIP to regulate androgen receptor stability and activity. Oncotarget 2016, 7, 81377–81388. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kregel, S.; Wang, C.; Han, X.; Xiao, L.; Fernandez-Salas, E.; Bawa, P.; McCollum, B.L.; Wilder-Romans, K.; Apel, I.J.; Cao, X.; et al. Androgen receptor degraders overcome common resistance mechanisms developed during prostate cancer treatment. Neoplasia 2020, 22, 111–119. [Google Scholar] [CrossRef]






| Cell Line | Characteristics | Origin | Patients Background | Reference |
|---|---|---|---|---|
| LNCaP | AR (T877A), Androgen dependent | Lymph node | 50-year-old Caucasian male | [19] |
| C4-2 | AR (T877A), Androgen independent, LNCaP sub-cell line | Lymph node | 50-year-old Caucasian male | [17] |
| LAPC4 | AR wt, Androgen dependent | Lymph node | non applicable | [20] |
| PC3 | Cells express no AR protein, Androgen independent, small cell neuroendocrine carcinoma | Bone metastasis | 62-year-old Caucasian male | [21,22] |
| Name | Company | Lot | Dilution |
|---|---|---|---|
| Androgen Receptor (D6F11) XP Rabbit mAb | Cell Signaling Technology, Cambridge, UK | 9 | 1:5000 |
| Lamin A/C (4C11) Mouse mAb | Cell Signaling Technology, Cambridge, UK | 5 | 1:2000 |
| Mouse Monoclonal anti-GAPDH (6C5) | Novus Biologicals, Littleton, CO, USA | 19/05-G4cc-C5cc | 1:10,000 |
| PSA/Kallikrein 3 (KLK3) (D6B1) XP Rabbit mAb | Cell Signaling Technology, Cambridge, UK | 4 | 1:1000 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Siciliano, T.; Simons, I.H.; Beier, A.-M.K.; Ebersbach, C.; Aksoy, C.; Seed, R.I.; Stope, M.B.; Thomas, C.; Erb, H.H.H. A Systematic Comparison of Antiandrogens Identifies Androgen Receptor Protein Stability as an Indicator for Treatment Response. Life 2021, 11, 874. https://doi.org/10.3390/life11090874
Siciliano T, Simons IH, Beier A-MK, Ebersbach C, Aksoy C, Seed RI, Stope MB, Thomas C, Erb HHH. A Systematic Comparison of Antiandrogens Identifies Androgen Receptor Protein Stability as an Indicator for Treatment Response. Life. 2021; 11(9):874. https://doi.org/10.3390/life11090874
Chicago/Turabian StyleSiciliano, Tiziana, Ingo H. Simons, Alicia-Marie K. Beier, Celina Ebersbach, Cem Aksoy, Robert I. Seed, Matthias B. Stope, Christian Thomas, and Holger H. H. Erb. 2021. "A Systematic Comparison of Antiandrogens Identifies Androgen Receptor Protein Stability as an Indicator for Treatment Response" Life 11, no. 9: 874. https://doi.org/10.3390/life11090874