MED16 Promotes Tumour Progression and Tamoxifen Sensitivity by Modulating Autophagy through the mTOR Signalling Pathway in ER-Positive Breast Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Samples
2.2. Bioinformatics Data Analysis
2.3. Cell Culture
2.4. Plasmid Construction and shRNA and siRNA Transfection
2.5. Cell Proliferation and Drug Sensitivity Assay
2.6. Colony Formation Assay
2.7. Immunohistochemistry (IHC) Analysis
2.8. RNA Isolation, Reverse-Transcription Reaction and Quantitative Real-Time PCR (qPCR)
2.9. Western Blot Analysis
2.10. 5-Ethynyl-2′-Deoxyuridine (Edu) Staining
2.11. Cell Cycle Assay and Flow Cytometry Analysis
2.12. Mammosphere Formation Assay (MSF)
2.13. Statistical Analysis
3. Results
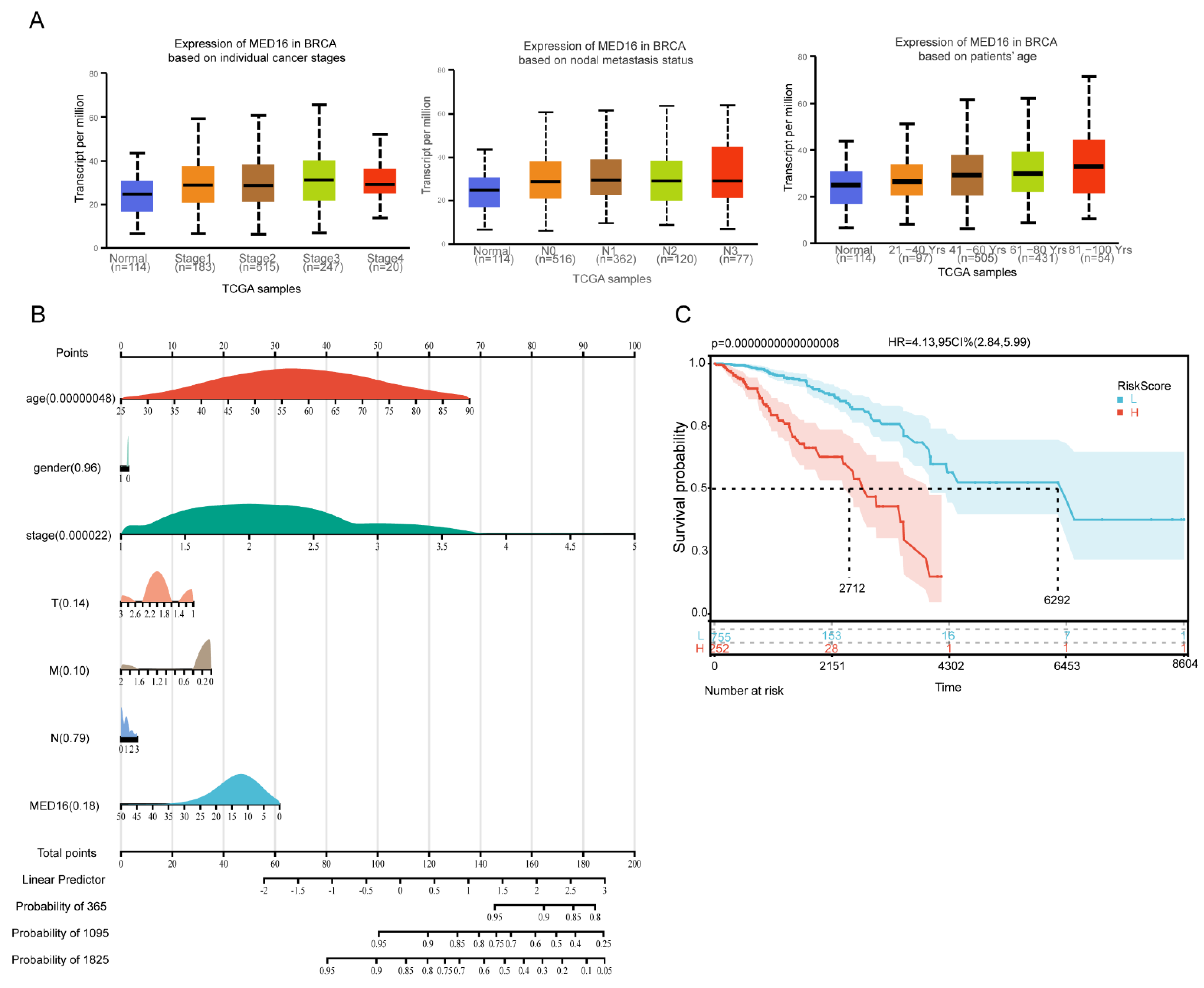
3.1. MED16 Expression Is Markedly Upregulated in BC
3.2. Diagnostic and Survival Value Analysis of MED16
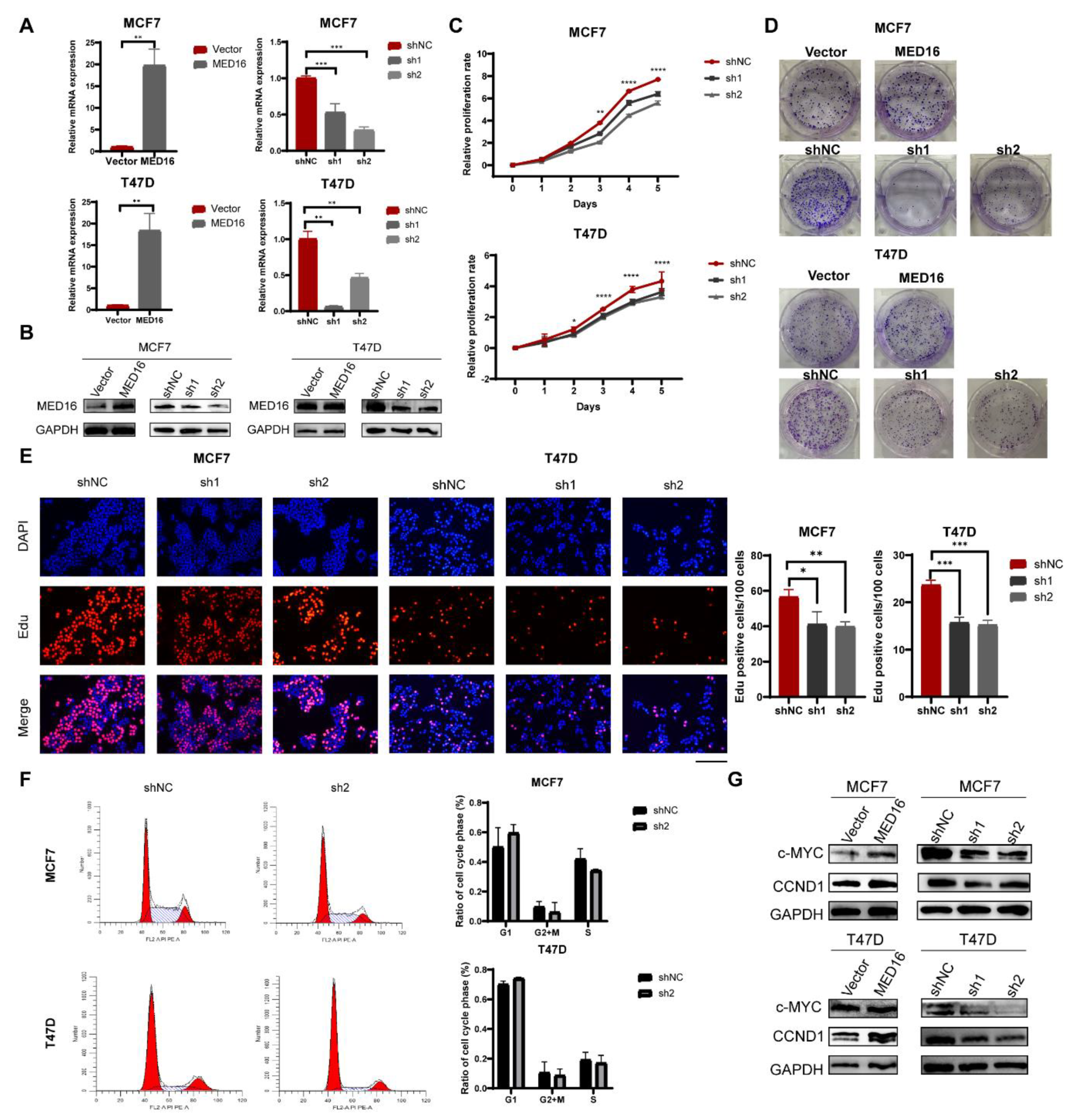
3.3. Knockdown of MED16 Inhibits ER+ BC Proliferation
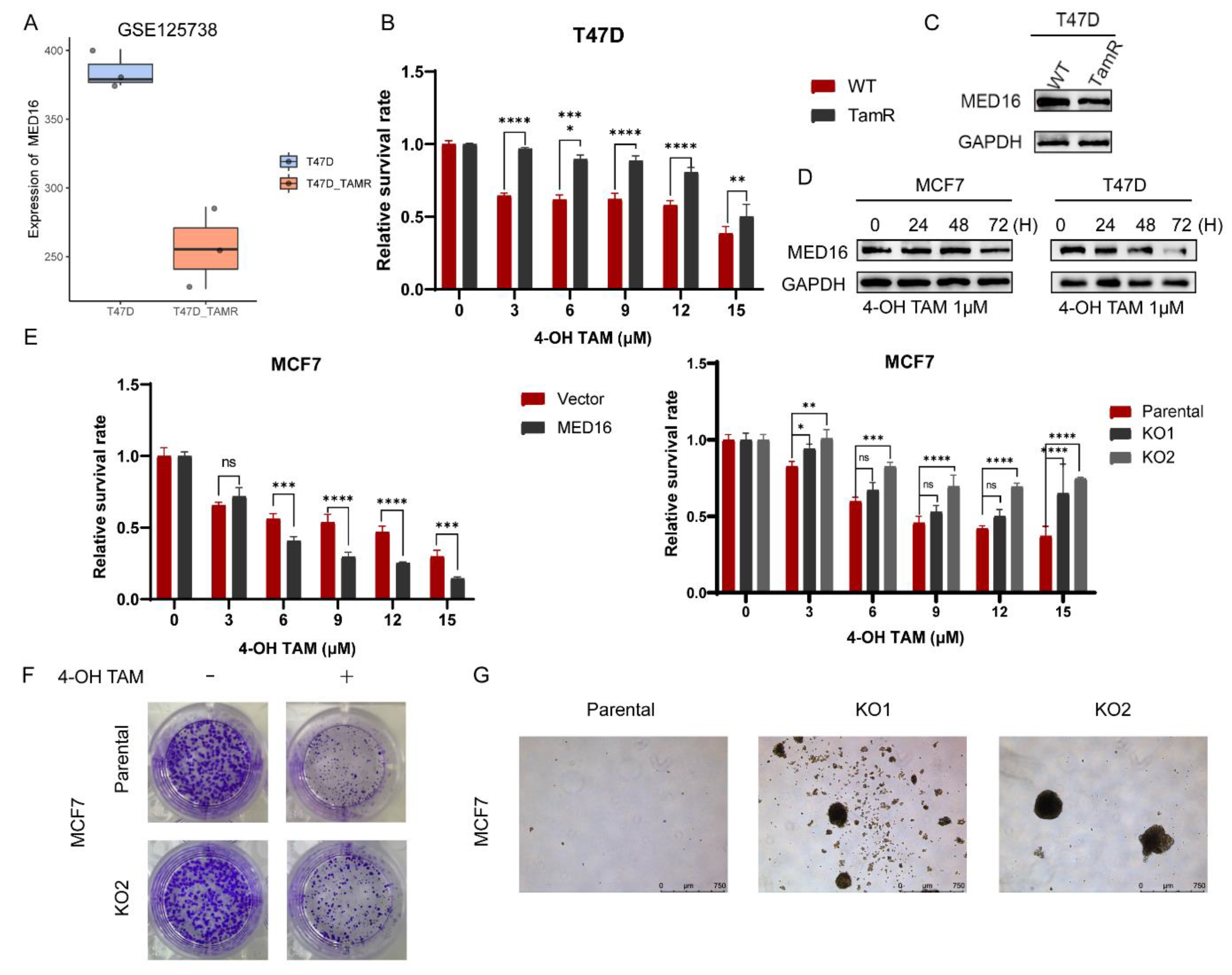
3.4. Knockdown of MED16 Reduces Tamoxifen Sensitivity
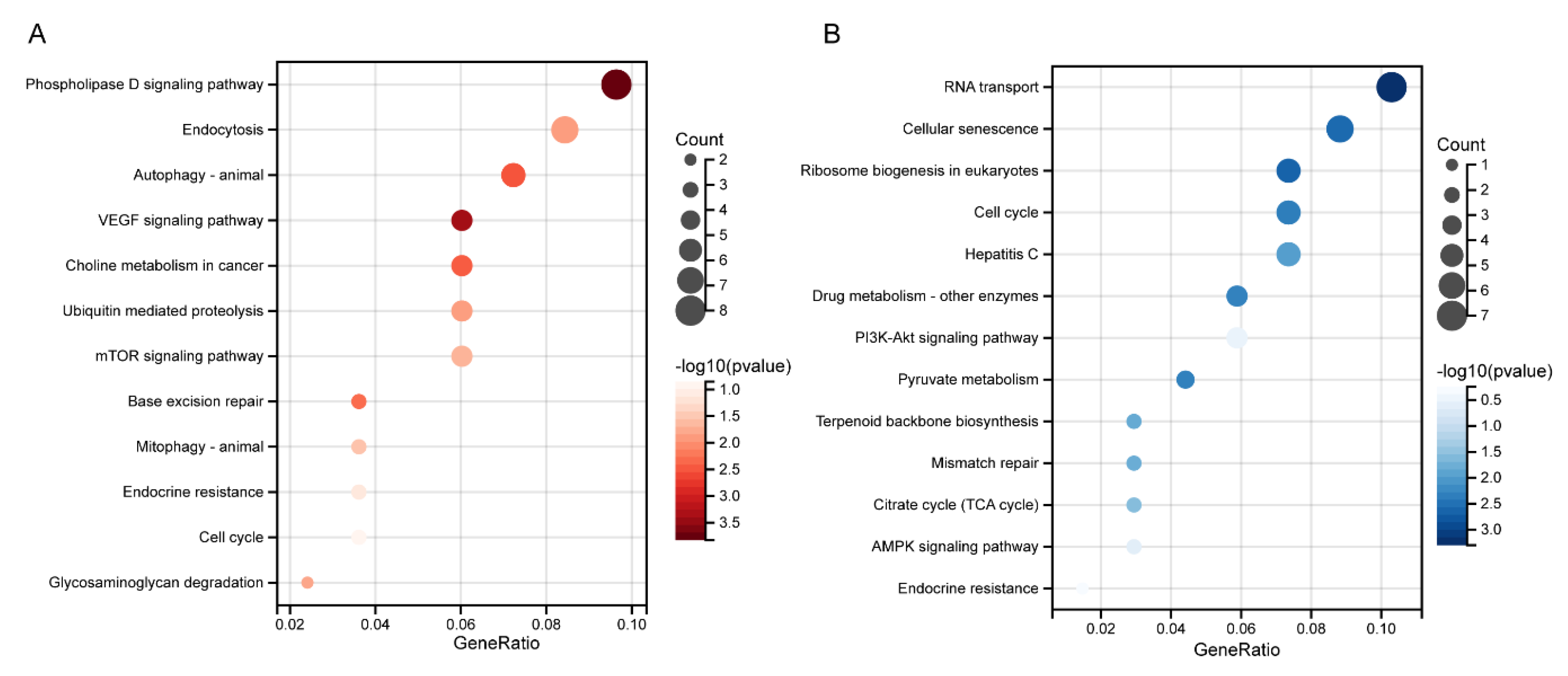
3.5. Biological Functional Analysis of MED16-Related Genes
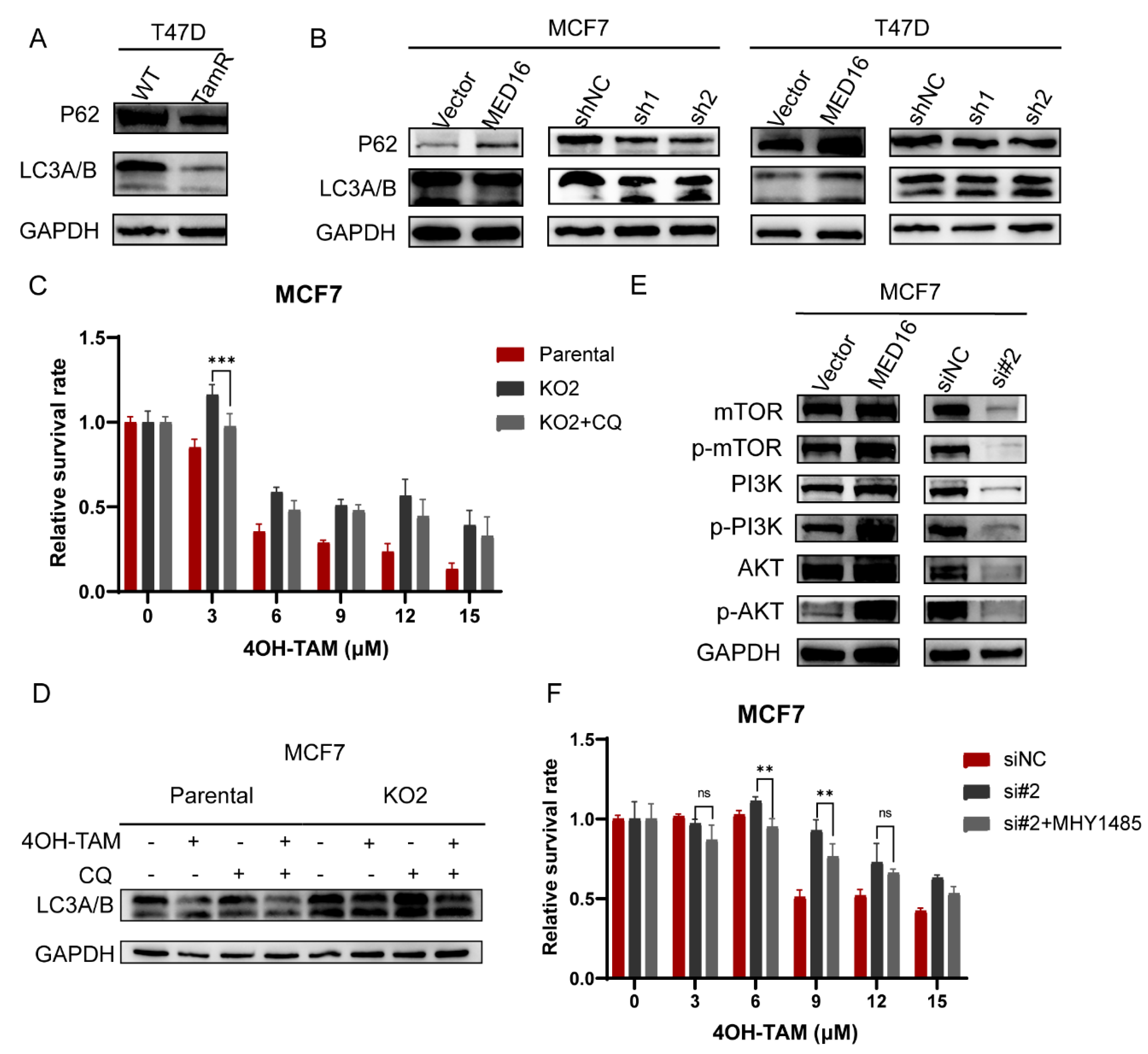
3.6. Knockdown of MED16 Induces Autophagy by Inhibiting the PI3K/AKT/mTOR Signalling Pathway
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Tseng, C.-W.; Kuo, W.-H.; Chan, S.-H.; Chan, H.-L.; Chang, K.-J.; Wang, L.-H. Transketolase Regulates the Metabolic Switch to Control Breast Cancer Cell Metastasis via the α-Ketoglutarate Signaling Pathway. Cancer Res. 2018, 78, 2799–2812. [Google Scholar] [CrossRef]
- Zhu, X.; Qiu, J.; Zhang, T.; Yang, Y.; Guo, S.; Li, T.; Jiang, K.; Zahoor, A.; Deng, G.; Qiu, C. MicroRNA-188-5p promotes apoptosis and inhibits cell proliferation of breast cancer cells via the MAPK signaling pathway by targeting Rap2c. J. Cell. Physiol. 2020, 235, 2389–2402. [Google Scholar] [CrossRef]
- Howlader, N.; Altekruse, S.F.; Li, C.I.; Chen, V.W.; Clarke, C.A.; Ries, L.A.; Cronin, K.A. US incidence of breast cancer subtypes defined by joint hormone receptor and HER2 status. J. Natl. Cancer Inst. 2014, 106, dju055. [Google Scholar] [CrossRef]
- Jeselsohn, R.; Buchwalter, G.; DE Angelis, C.; Brown, M.; Schiff, R. ESR1 mutations—A mechanism for acquired endocrine resistance in breast cancer. Nat. Rev. Clin. Oncol. 2015, 12, 573–583. [Google Scholar] [CrossRef]
- Szostakowska, M.; Trębińska-Stryjewska, A.; Grzybowska, E.A.; Fabisiewicz, A. Resistance to endocrine therapy in breast cancer: Molecular mechanisms and future goals. Breast Cancer Res. Treat. 2019, 173, 489–497. [Google Scholar] [CrossRef]
- Yao, J.; Deng, K.; Huang, J.; Zeng, R.; Zuo, J. Progress in the Understanding of the Mechanism of Tamoxifen Resistance in Breast Cancer. Front. Pharmacol. 2020, 11, 592912. [Google Scholar] [CrossRef]
- Qadir, M.A.; Kwok, B.; Dragowska, W.H.; To, K.H.; Le, D.; Bally, M.B.; Gorski, S.M. Macroautophagy inhibition sensitizes tamoxifen-resistant breast cancer cells and enhances mitochondrial depolarization. Breast Cancer Res. Treat. 2008, 112, 389–403. [Google Scholar] [CrossRef]
- Rudraraju, B.; Droog, M.; Abdel-Fatah, T.M.A.; Zwart, W.; Giannoudis, A.; Malki, M.I.; Moore, D.; Patel, H.; Shaw, J.; Ellis, I.O.; et al. Phosphorylation of activating transcription factor-2 (ATF-2) within the activation domain is a key determinant of sensitivity to tamoxifen in breast cancer. Breast Cancer Res. Treat. 2014, 147, 295–309. [Google Scholar] [CrossRef]
- Jeronimo, C.; Robert, F. The Mediator Complex: At the Nexus of RNA Polymerase II Transcription. Trends Cell Biol. 2017, 27, 765–783. [Google Scholar] [CrossRef]
- Kim, Y.-J.; Björklund, S.; Li, Y.; Sayre, M.H.; Kornberg, R.D. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell 1994, 77, 599–608. [Google Scholar] [CrossRef]
- Soutourina, J. Transcription regulation by the Mediator complex. Nat. Rev. Mol. Cell Biol. 2018, 19, 262–274. [Google Scholar] [CrossRef] [PubMed]
- Maldonado, E.; Morales-Pison, S.; Urbina, F.; Jara, L.; Solari, A. Role of the mediator complex and microRNAs in breast cancer etiology. Genes 2022, 13, 234. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Bai, P.; Xiao, L.; Shen, M.; Yu, Q.; Lei, Y.; Huang, W.; Lin, X.; Zheng, X.; Wei, T.; et al. Mediator complex subunit 16 is down-regulated in papillary thyroid cancer, leading to increased transforming growth factor-β signaling and radioiodine resistance. J. Biol. Chem. 2020, 295, 10726–10740. [Google Scholar] [CrossRef]
- Binkhorst, L.; van Gelder, T.; Mathijssen, R.H. Individualization of tamoxifen treatment for breast carcinoma. Clin. Pharmacol. Ther. 2012, 92, 431–433. [Google Scholar] [CrossRef]
- Takebe, N.; Miele, L.; Harris, P.J.; Jeong, W.; Bando, H.; Kahn, M.G.; Yang, S.X.; Ivy, S.P. Targeting Notch, Hedgehog, and Wnt pathways in cancer stem cells: Clinical update. Nat. Rev. Clin. Oncol. 2015, 12, 445–464. [Google Scholar] [CrossRef]
- Samaddar, J.S.; Gaddy, V.T.; Duplantier, J.; Thandavan, S.P.; Shah, M.; Smith, M.J.; Browning, D.; Rawson, J.; Smith, S.B.; Barrett, J.T.; et al. A role for macroautophagy in protection against 4-hydroxytamoxifen-induced cell death and the development of antiestrogen resistance. Mol. Cancer Ther. 2008, 7, 2977–2987. [Google Scholar] [CrossRef]
- Post, A.E.M.; Bussink, J.; Sweep, F.C.G.J.; Span, P.N. Changes in DNA Damage Repair Gene Expression and Cell Cycle Gene Expression Do Not Explain Radioresistance in Tamoxifen-Resistant Breast Cancer. Oncol. Res. 2020, 28, 33–40. [Google Scholar] [CrossRef]
- Bi, M.; Zhang, Z.; Jiang, Y.-Z.; Xue, P.; Wang, H.; Lai, Z.; Fu, X.; De Angelis, C.; Gong, Y.; Gao, Z.; et al. Enhancer reprogramming driven by high-order assemblies of transcription factors promotes phenotypic plasticity and breast cancer endocrine resistance. Nat. Cell Biol. 2020, 22, 701–715. [Google Scholar] [CrossRef]
- Firestein, R.; Bass, A.J.; Kim, S.Y.; Dunn, I.F.; Silver, S.J.; Guney, I.; Freed, E.; Ligon, A.H.; Vena, N.; Ogino, S.; et al. CDK8 is a colorectal cancer oncogene that regulates beta-catenin activity. Nature 2008, 455, 547–551. [Google Scholar] [CrossRef]
- Antonioli, M.; Di Rienzo, M.; Piacentini, M.; Fimia, G.M. Emerging Mechanisms in Initiating and Terminating Autophagy. Trends Biochem. Sci. 2017, 42, 28–41. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.H.; Koh, D.; Na, H.; Ka, N.L.; Kim, S.; Kim, H.J.; Hong, S.; Shin, Y.K.; Seong, J.K.; Lee, M.O. MTA1 is a novel regulator of autophagy that induces tamoxifen resistance in breast cancer cells. Autophagy 2018, 14, 812–824. [Google Scholar] [CrossRef] [PubMed]
- Cook, K.L.; Shajahan, A.N.; Clarke, R. Autophagy and endocrine resistance in breast cancer. Expert Rev. Anticancer Ther. 2011, 11, 1283–1294. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Wang, Q.; Lv, X.; Sha, L.; Qin, H.; Wang, L.; Li, L. Expression and localization of estrogen receptor in human breast cancer and its clinical significance. Cell Biochem. Biophys. 2015, 71, 63–68. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Xie, S.; Yang, J.; Xiong, H.; Jia, Y.; Zhou, Y.; Chen, Y.; Ying, X.; Chen, C.; Ye, C.; et al. The long noncoding RNA H19 promotes tamoxifen resistance in breast cancer via autophagy. J. Hematol. Oncol. 2019, 12, 81. [Google Scholar] [CrossRef]
- Sun, M.; Zhao, S.; Duan, Y.; Ma, Y.; Wang, Y.; Ji, H.; Zhang, Q. GLUT1 participates in tamoxifen resistance in breast cancer cells through autophagy regulation. Naunyn-Schmiedeberg’s Arch. Pharmacol. 2021, 394, 205–216. [Google Scholar] [CrossRef]
- Mansouri, S.; Farahmand, L.; Teymourzadeh, A.; Majidzadeh-A, K. Clinical Evidence on the Magnitude of Change in Growth Pathway Activity in Relation to Tamoxifen Resistance is Required. Curr. Cancer Drug Targets 2018, 18, 668–676. [Google Scholar] [CrossRef]
- Cheng, X.; Tan, S.; Duan, F.; Yuan, Q.; Li, Q.; Deng, G. Icariin induces apoptosis by suppressing autophagy in tamoxifen-resistant breast cancer cell line MCF-7/TAM. Breast Cancer 2019, 26, 766–775. [Google Scholar] [CrossRef]
- Li, G.; Zhang, J.; Xu, Z.; Li, Z. ERα36 as a Potential Therapeutic Target for Tamoxifen-Resistant Breast Cancer Cell Line Through EGFR/ERK Signaling Pathway. Cancer Manag. Res. 2020, 12, 265–275. [Google Scholar] [CrossRef]
- Iksen; Pothongsrisit, S.; Pongrakhananon, V. Targeting the PI3K/AKT/mTOR signaling pathway in lung cancer: An update regarding potential drugs and natural products. Molecules 2021, 26, 4100. [Google Scholar] [CrossRef]
- Abukhdeir, A.M.; Park, B.H. P21 and p27: Roles in carcinogenesis and drug resistance. Expert Rev. Mol. Med. 2008, 10, e19. [Google Scholar] [CrossRef] [PubMed]
- Bristol, M.L.; Di, X.; Beckman, M.J.; Wilson, E.N.; Henderson, S.C.; Maiti, A.; Fan, Z.; Gewirtz, D.A. Dual functions of autophagy in the response of breast tumor cells to radiation: Cytoprotective autophagy with radiation alone and cytotoxic autophagy in radiosensitization by vitamin D 3. Autophagy 2012, 8, 739–753. [Google Scholar] [CrossRef] [PubMed]
- Scott, R.C.; Juhasz, G.; Neufeld, T.P. Direct induction of autophagy by Atg1 inhibits cell growth and induces apoptotic cell death. Curr. Biol. 2007, 17, 1–11. [Google Scholar] [CrossRef] [PubMed]

| Characteristics | HR Positive (N = 18) | HR Negative (N = 7) | Total (N = 25) | p Value | FDR |
|---|---|---|---|---|---|
| Age (years) | 0.01 | 0.09 | |||
| >50 | 6 | 7 | 13 | ||
| ≤50 | 12 | 0 | 12 | ||
| Tumour size (cm) | 0.79 | 1 | |||
| >2 | 10 | 5 | 15 | ||
| ≤2 | 8 | 2 | 10 | ||
| Histological grade | 0.52 | 1 | |||
| II | 12 | 3 | 15 | ||
| III | 6 | 4 | 10 | ||
| Lymph node metastasis | 0.37 | 1 | |||
| Negative | 5 | 4 | 9 | ||
| Positive | 10 | 2 | 12 | ||
| Unknown | 3 | 1 | 4 | ||
| ER status | 0.000047 | 0.00047 | |||
| Negative | 1 | 7 | 8 | ||
| Positive | 17 | 0 | 17 | ||
| PR status | 0.01 | 0.09 | |||
| Negative | 6 | 7 | 13 | ||
| Positive | 12 | 0 | 12 | ||
| HER2 status | 0.34 | 1 | |||
| Negative | 10 | 6 | 16 | ||
| Positive | 8 | 1 | 9 | ||
| Ki-67 (%) | 0.56 | 1 | |||
| >30 | 5 | 3 | 8 | ||
| Positive | 2 | 0 | 2 | ||
| ≤30 | 11 | 4 | 15 | ||
| Molecular subtypes | 0.00005 | 0.00047 | |||
| HER2 positive | 0 | 1 | 1 | ||
| Luminal A | 10 | 0 | 10 | ||
| Luminal B | 8 | 0 | 8 | ||
| Triple Negative | 0 | 6 | 6 | ||
| Adjuvant therapy | 0.31 | 1 | |||
| No | 11 | 2 | 13 | ||
| Yes | 7 | 5 | 12 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, H.; Li, K.; Shu, D.; Shen, M.; Tan, Z.; Zhang, W.; Pu, D.; Tan, W.; Tang, Z.; Jin, A.; et al. MED16 Promotes Tumour Progression and Tamoxifen Sensitivity by Modulating Autophagy through the mTOR Signalling Pathway in ER-Positive Breast Cancer. Life 2022, 12, 1461. https://doi.org/10.3390/life12101461
Li H, Li K, Shu D, Shen M, Tan Z, Zhang W, Pu D, Tan W, Tang Z, Jin A, et al. MED16 Promotes Tumour Progression and Tamoxifen Sensitivity by Modulating Autophagy through the mTOR Signalling Pathway in ER-Positive Breast Cancer. Life. 2022; 12(10):1461. https://doi.org/10.3390/life12101461
Chicago/Turabian StyleLi, Han, Kang Li, Dan Shu, Meiying Shen, Zhaofu Tan, Wenjie Zhang, Dongyao Pu, Wenhao Tan, Zhenrong Tang, Aishun Jin, and et al. 2022. "MED16 Promotes Tumour Progression and Tamoxifen Sensitivity by Modulating Autophagy through the mTOR Signalling Pathway in ER-Positive Breast Cancer" Life 12, no. 10: 1461. https://doi.org/10.3390/life12101461
APA StyleLi, H., Li, K., Shu, D., Shen, M., Tan, Z., Zhang, W., Pu, D., Tan, W., Tang, Z., Jin, A., & Liu, S. (2022). MED16 Promotes Tumour Progression and Tamoxifen Sensitivity by Modulating Autophagy through the mTOR Signalling Pathway in ER-Positive Breast Cancer. Life, 12(10), 1461. https://doi.org/10.3390/life12101461






