Optimized Metabotype Definition Based on a Limited Number of Standard Clinical Parameters in the Population-Based KORA Study
Abstract
1. Introduction
2. Materials and Methods
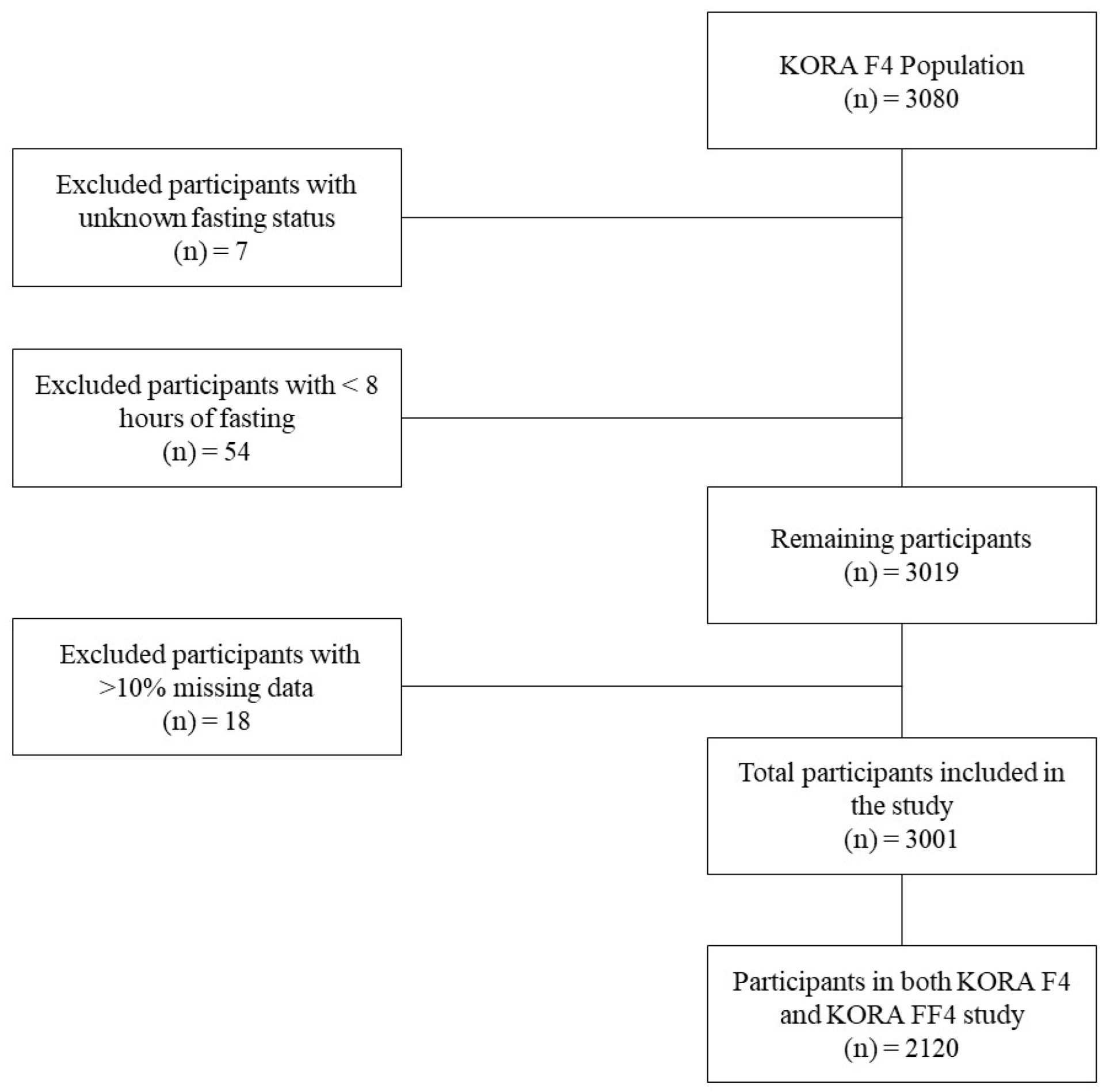
2.1. Study Population
2.2. Biochemical and Anthropometric Parameters
2.3. Socio-Demographic and Lifestyle Variables
2.4. Health Status
2.5. Data Preprocessing
2.6. Descriptive Statistics
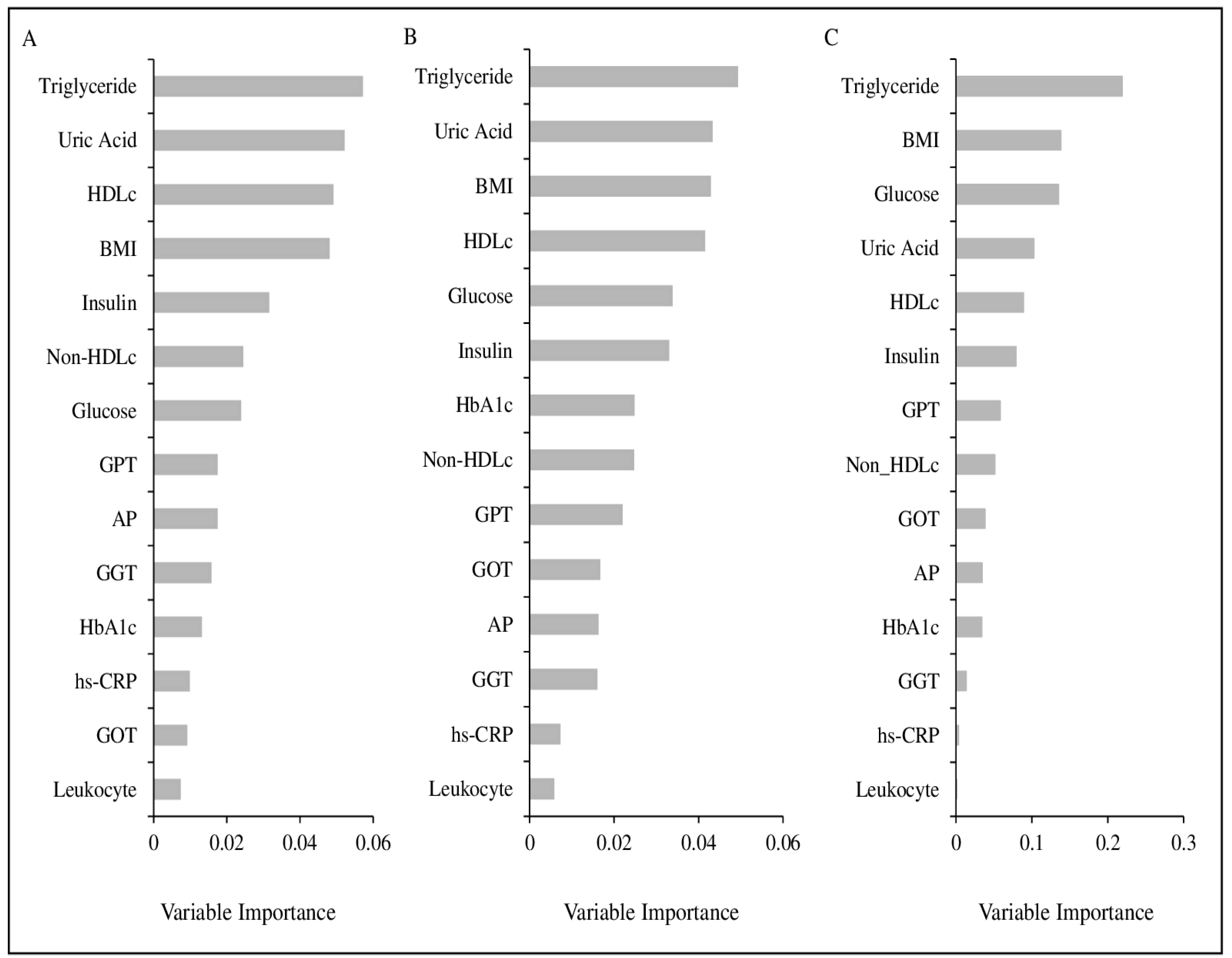
2.7. Parameter Selection
2.8. Metabotyping
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BMI | body mass index |
| HDL | high-density lipoprotein |
| TG | triglyceride |
| GGT | gamma-glutamyltransferase |
| GOT | glutamate-oxaloacetate transaminase |
| GPT | glutamate-pyruvate transaminase |
| HbA1c | glycated hemoglobin |
| hs-CRP | high-sensitive C-reactive protein |
| AP | alkaline phosphatase |
| KORA | Cooperative Health Research in the Region of Augsburg |
References
- Holmes, E.; Wilson, I.D.; Nicholson, J.K. Metabolic Phenotyping in Health and Disease. Cell 2008, 134, 714–717. [Google Scholar] [CrossRef] [PubMed]
- O’donovan, C.B.; Walsh, M.C.; Gibney, M.J.; Gibney, E.R.; Brennan, L. Can metabotyping help deliver the promise of personalised nutrition? Proc. Nutr. Soc. 2015, 106–114. [Google Scholar] [CrossRef] [PubMed]
- Riedl, A.; Gieger, C.; Hauner, H.; Daniel, H.; Linseisen, J. Metabotyping and its application in targeted nutrition: An overview. Br. J. Nutr. 2017, 117, 1631–1644. [Google Scholar] [CrossRef] [PubMed]
- Hillesheim, E.; Brennan, L. Metabotyping and its role in nutrition research. Nutr. Res. Rev. 2019, 33, 1–10. [Google Scholar] [CrossRef] [PubMed]
- O’Donovan, C.B.; Walsh, M.C.; Nugent, A.P.; McNulty, B.; Walton, J.; Flynn, A.; Gibney, M.J.; Gibney, E.R.; Brennan, L. Use of metabotyping for the delivery of personalised nutrition. Mol. Nutr. Food Res. 2015, 59, 377–385. [Google Scholar] [CrossRef]
- Brennan, L. Use of metabotyping for optimal nutrition. Curr. Opin. Biotechnol. 2017, 44, 35–38. [Google Scholar] [CrossRef]
- De Roos, B. Personalised nutrition: Ready for practice? In Proceedings of the Conference on ‘Future Food and Health’ Symposium II: Diet–Gene Interactions; Implications for Future Diets and Health, Aberdeen, UK, 26–27 March 2012. [Google Scholar] [CrossRef]
- Riedl, A.; Hillesheim, E.; Wawro, N.; Meisinger, C.; Peters, A.; Roden, M.; Kronenberg, F.; Herder, C.; Rathmann, W.; Völzke, H.; et al. Evaluation of the Metabotype Concept Identified in an Irish Population in the German KORA Cohort Study. Mol. Nutr. Food Res. 2020, 64, 1900918. [Google Scholar] [CrossRef]
- Zeevi, D.; Korem, T.; Zmora, N.; Israeli, D.; Rothschild, D.; Weinberger, A.; Ben-Yacov, O.; Lador, D.; Avnit-Sagi, T.; Lotan-Pompan, M.; et al. Personalized Nutrition by Prediction of Glycemic Responses. Cell 2015, 163, 1079–1094. [Google Scholar] [CrossRef]
- Celis-Morales, C.; Livingstone, K.M.; Marsaux, C.F.; Macready, A.L.; Fallaize, R.; O’donovan, C.B.; Woolhead, C.; Forster, H.; Walsh, M.C.; Navas-Carretero, S.; et al. Effect of personalized nutrition on health-related behaviour change: Evidence from the Food4Me European randomized controlled trial. Int. J. Epidemiol. 2017, 46, 578–588. [Google Scholar] [CrossRef]
- Livingstone, K.M.; Celis-Morales, C.; Navas-Carretero, S.; San-Cristobal, R.; Forster, H.; Woolhead, C.; O’Donovan, C.B.; Moschonis, G.; Manios, Y.; Traczyk, I.; et al. Personalized Nutrition Advice Reduces Intake of Discretionary Foods and Beverages: Findings From the Food4Me Randomized Controlled Trial. Curr. Dev. Nutr. 2021, 5, 152. [Google Scholar] [CrossRef]
- Palmnäs, M.; Brunius, C.; Shi, L.; Rostgaard-Hansen, A.; Torres, N.E.; González-Domínguez, R.; Zamora-Ros, R.; Ye, Y.L.; Halkjær, J.; Tjønneland, A.; et al. Perspective: Metabotyping—A Potential Personalized Nutrition Strategy for Precision Prevention of Cardiometabolic Disease. Adv. Nutr. 2019, 10, S308–S319. [Google Scholar] [CrossRef] [PubMed]
- Brennan, L. Symposium on “The challenge of translating nutrition research into public health nutrition” Session 2: Personalised nutrition Metabolomic applications in nutritional research. Proc. Nutr. Soc. 2020, 67, 404–408. [Google Scholar] [CrossRef] [PubMed]
- Bouwman, J.; Vogels, J.T.; Wopereis, S.; Rubingh, C.M.; Bijlsma, S.; van Ommen, B. Visualization and identification of health space, based on personalized molecular phenotype and treatment response to relevant underlying biological processes. BMC Med. Genomics. 2012, 5, 1. [Google Scholar] [CrossRef] [PubMed]
- Riedl, A.; Wawro, N.; Gieger, C.; Meisinger, C.; Peters, A.; Roden, M.; Kronenberg, F.; Herder, C.; Rathmann, W.; Völzke, H.; et al. Identification of Comprehensive Metabotypes Associated with Cardiometabolic Diseases in the Population-Based KORA Study. Mol. Nutr. Food Res. 2018, 62, 1–9. [Google Scholar] [CrossRef]
- Vázquez-Fresno, R.; Llorach, R.; Perera, A.; Mandal, R.; Tinahones, F.J.; Wishart, D.S.; Andrés-Lacueva, C. Clinical phenotype clustering in cardiovascular risk patients for the identification of responsive metabotypes after red wine polyphenol intake. J. Nutr. Biochem. 2016, 28, 114–120. [Google Scholar] [CrossRef]
- Moazzami, A.A.; Shrestha, A.; Morrison, D.A.; Poutanen, K.; Mykkänen, H. Metabolomics Reveals Differences in Postprandial Responses to Breads and Fasting Metabolic Characteristics Associated with Postprandial Insulin Demand in Postmenopausal Women. J. Nutr. 2014, 144, 807–814. [Google Scholar] [CrossRef]
- Mäkinen, V.; Soininen, P.; Forsblom, C.; Parkkonen, M.; Ingman, P.; Kaski, K.; Groop, P.-H.; Ala-Korpela, M. 1H NMR metabonomics approach to the disease continuum of diabetic complications and premature death. Mol. Syst. Biol. 2008, 4, 167. [Google Scholar] [CrossRef]
- Hillesheim, E.; Ryan, M.F.; Gibney, E.; Roche, H.M.; Brennan, L. Optimisation of a metabotype approach to deliver targeted dietary advice. Nutr. Metab. 2020, 17, 1–12. [Google Scholar] [CrossRef]
- Zubair, N.; Kuzawa, C.W.; McDade, T.W.; Adair, L.S. Cluster analysis reveals important determinants of cardiometabolic risk patterns in Filipino women. Asia Pac. J. Clin. Nutr. 2012, 21, 271–281. [Google Scholar] [CrossRef]
- Holle, R.; Happich, M.; Löwel, H.; Wichmann, H.E. KORA-A research platform for population based health research. Gesundheitswesen 2005, 67, 19–25. [Google Scholar] [CrossRef]
- Herder, C.; Bongaerts, B.W.C.; Rathmann, W.; Heier, M.; Kowall, B.; Koenig, W.; Thorand, B.; Roden, M.; Meisinger, C.; Ziegler, D. Association of subclinical inflammation with polyneuropathy in the older population: KORA F4 study. Diabetes Care 2013, 36, 3663–3670. [Google Scholar] [CrossRef]
- Stöckl, D.; Meisinger, C.; Peters, A.; Thorand, B.; Huth, C.; Heier, M.; Rathmann, W.; Kowall, B.; Stöckl, H.; Döring, A. Age at menarche and its association with the metabolic syndrome and its components: Results from the Kora F4 study. PLoS ONE 2011, 6, 26076. [Google Scholar] [CrossRef] [PubMed]
- Kowall, B.; Rathmann, W.; Stang, A.; Bongaerts, B.; Kuss, O.; Herder, C.; Roden, M.; Quante, A.; Holle, R.; Huth, C.; et al. Perceived risk of diabetes seriously underestimates actual diabetes risk: The KORA FF4 study. PLoS ONE 2017, 12, e0171152. [Google Scholar] [CrossRef] [PubMed]
- WHO/Europe|Nutrition-Body Mass Index-BMI. Available online: http://www.euro.who.int/en/health-topics/disease-prevention/nutrition/a-healthy-lifestyle/body-mass-index-bmi (accessed on 3 April 2020).
- van Buuren, S.; Groothuis-Oudshoorn, K. Mice:Multivariate Imputation by Chained Equations in R. J. Stat. Softw. 2011, 45, 1–67. [Google Scholar] [CrossRef]
- Riedl, A.; Wawro, N.; Gieger, C.; Meisinger, C.; Peters, A.; Rathmann, W.; Koenig, W.; Strauch, K.; Quante, A.S.; Thorand, B.; et al. Modifying effect of metabotype on diet–diabetes associations. Eur. J. Nutr. 2019, 59, 1357–1369. [Google Scholar] [CrossRef] [PubMed]
- Tanabe, N.; Iso, H.; Okada, K.; Nakamura, Y.; Harada, A.; Ohashi, Y.; Ando, T.; Ueshima, H. Serum total and non-high-density lipoprotein cholesterol and the risk prediction of cardiovascular events: -The JALS-ecc-. Circ. J. 2010, 74, 1347–1356. [Google Scholar] [CrossRef]
- Cui, Y.; Blumenthal, R.S.; Flaws, J.A.; Whiteman, M.K.; Langenberg, P.; Bachorik, P.S.; Bush, T.L. Non-high-density lipoprotein cholesterol level as a predictor of cardiovascular disease mortality. Arch. Intern. Med. 2001, 161, 1413–1419. [Google Scholar] [CrossRef]
- Angoorani, P.; Khademian, M.; Ejtahed, H.S.; Heshmat, R.; Motlagh, M.E.; Vafaeenia, M.; Shafiee, G.; Mahdivi-Gorabi, A.; Qorbani, M.; Kelishadi, R. Are non-high-density lipoprotein fractions associated with pediatric metabolic syndrome? The CASPIAN-V study. Lipids Health Dis. 2018, 17. [Google Scholar] [CrossRef]
- Brunner, F.J.; Waldeyer, C.; Ojeda, F.; Salomaa, V.; Kee, F.; Sans, S.; Thorand, B.; Giampaoli, S.; Brambilla, P.; Tunstall-Pedoe, H.; et al. Application of non-HDL cholesterol for population-based cardiovascular risk stratification: Results from the Multinational Cardiovascular Risk Consortium. Lancet 2019, 394, 2173–2183. [Google Scholar] [CrossRef]
- Putin, E.; Mamoshina, P.; Aliper, A.; Korzinkin, M.; Moskalev, A.; Kolosov, A.; Ostrovskiy, A.; Cantor, C.; Vijg, J.; Zhavoronkov, A. Deep biomarkers of human aging: Application of deep neural networks to biomarker development. Aging 2016, 8, 1021. [Google Scholar] [CrossRef]
- Huynh-Thu, V.A.; Saeys, Y.; Wehenkel, L.; Geurts, P. Statistical interpretation of machine learning-based feature importance scores for biomarker discovery. Bioinformatics 2012, 28, 1766–1774. [Google Scholar] [CrossRef] [PubMed]
- Prosperi, M.C.; Marinho, S.; Simpson, A.; Custovic, A.; Buchan, I.E. Predicting phenotypes of asthma and eczema with machine learning. BMC Med. Genomics. 2014, 7. [Google Scholar] [CrossRef] [PubMed]
- Altmann, A.; Toloşi, L.; Sander, O.; Lengauer, T. Permutation importance: A corrected feature importance measure. Bioinformatics 2010, 26, 1340–1347. [Google Scholar] [CrossRef] [PubMed]
- PIMP Function|R Documentation. Available online: https://www.rdocumentation.org/packages/vita/versions/1.0.0/topics/PIMP (accessed on 9 March 2020).
- Leo Breiman, A.C. Random Forests. Signal and Image Processing for Remote Sensing. 2003. Available online: http://www.stat.berkeley.edu/breiman/RandomForests/cchome.htm (accessed on 3 September 2020).
- Janitza, S.; Celik, E.; Boulesteix, A.L. A computationally fast variable importance test for random forests for high-dimensional data. Adv. Data Anal. Classif. 2018, 12, 885–915. [Google Scholar] [CrossRef]
- CVPVI Function|R Documentation. Available online: https://www.rdocumentation.org/packages/vita/versions/1.0.0/topics/CVPVI (accessed on 17 February 2021).
- Xu, Z.; Huang, G.; Weinberger, K.Q.; Zheng, A.X. Gradient boosted feature selection. In Proceedings of the 20th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, New York, NY, USA, 24–27 August 2014; pp. 522–531. [Google Scholar] [CrossRef]
- Hastie, T.; Tibshirani, R.; Friedman, J. Boosting and Additive Trees. In The Elements of Statistical Learning, 2nd ed.; Springer: New York, NY, USA, 2017; pp. 337–384. Available online: https://web.stanford.edu/~hastie/ElemStatLearn/ (accessed on 18 February 2021).
- Feature Importance and Feature Selection with XGBoost in Python. Available online: https://machinelearningmastery.com/feature-importance-and-feature-selection-with-xgboost-in-python/ (accessed on 18 February 2021).
- Chen, T.; Guestrin, C. XGBoost: A Scalable Tree Boosting System. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 785–794. [Google Scholar] [CrossRef]
- NbClust Function -RDocumentation. Available online: https://www.rdocumentation.org/packages/NbClust/versions/3.0.1/topics/NbClust (accessed on 5 September 2022).
- Barrera-Gómez, J.; Maintainer, X.B. Miclust:Multiple Imputation in Cluster Analysis. Available online: https://cran.r-project.org/web/packages/miclust/miclust.pdf (accessed on 9 March 2020).
- Wawro, N.; Pestoni, G.; Riedl, A.; Breuninger, T.A.; Peters, A.; Rathmann, W.; Koenig, W.; Huth, C.; Meisinger, C.; Rohrmann, S.; et al. Association of dietary patterns and type-2 diabetes mellitus in metabolically homogeneous subgroups in the KORA FF4 study. Nutrients 2020, 12, 1684. [Google Scholar] [CrossRef]
- van Bochove, K.; van Schalkwijk, D.B.; Parnell, L.D.; Lai, C.-Q.; Ordovás, J.M.; de Graaf, A.A.; van Ommen, B.; Arnett, D.K. Clustering by Plasma Lipoprotein Profile Reveals Two Distinct Subgroups with Positive Lipid Response to Fenofibrate Therapy. PLoS ONE 2012, 7, e38072. [Google Scholar] [CrossRef]
- O’Sullivan, A.; Gibney, M.J.; Connor, A.O.; Mion, B.; Kaluskar, S.; Cashman, K.D.; Flynn, A.; Shanahan, F.; Brennan, L. Biochemical and metabolomic phenotyping in the identification of a vitamin D responsive metabotype for markers of the metabolic syndrome. Mol. Nutr. Food Res. 2011, 55, 679–690. [Google Scholar] [CrossRef]
- Kamrath, C.; Hartmann, M.F.; Pons-Kühnemann, J.; Wudy, S.A. Urinary GC–MS steroid metabotyping in treated children with congenital adrenal hyperplasia. Metabolism 2020, 112, 154354. [Google Scholar] [CrossRef]
- Li, K.; Brennan, L.; McNulty, B.A.; Bloomfield, J.F.; Duff, D.J.; Devlin, N.F.C.; Gibney, M.J.; Flynn, A.; Walton, J.; Nugent, A.P. Plasma fatty acid patterns reflect dietary habits and metabolic health: A cross-sectional study. Mol. Nutr. Food Res. 2016, 60, 2043–2052. [Google Scholar] [CrossRef]
- Prendiville, O.; Walton, J.; Flynn, A.; Nugent, A.P.; Mcnulty, B.A.; Brennan, L. Classifying Individuals Into a Dietary Pattern Based on Metabolomic Data. Mol. Nutr. Food Res. 2021, 65, 2001183. [Google Scholar] [CrossRef]
- Sujana, C.; Seissler, J.; Jordan, J.; Rathmann, W.; Koenig, W. Associations of cardiac stress biomarkers with incident type 2 diabetes and changes in glucose metabolism: KORA F4/FF4 study. Cardiovasc. Diabetol. 2020, 19, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Fiamoncini, J.; Rundle, M.; Gibbons, H.; Thomas, E.L.; Geillinger-Kästle, K.; Bunzel, D.; Trezzi, J.-P.; Kiselova-Kaneva, Y.; Wopereis, S.; Wahrheit, J.; et al. Plasma metabolome analysis identifies distinct human metabotypes in the postprandial state with different susceptibility to weight loss-mediated metabolic improvements. FASEB J. 2018, 32, 5447–5458. [Google Scholar] [CrossRef] [PubMed]
- Dahal, C.; Wawro, N.; Meisinger, C.; Brandl, B.; Skurk, T.; Volkert, D.; Hauner, H.; Linseisen, J. Evaluation of the metabotype concept after intervention with oral glucose tolerance test and dietary fiber-enriched food: An enable study. Nutr. Metab. Cardiovasc. Dis. 2022. [Google Scholar] [CrossRef] [PubMed]
- O’donovan, C.B.; Walsh, M.C.; Woolhead, C.; Forster, H.; Celis-Morales, C.; Fallaize, R.; Macready, A.L.; Marsaux, C.F.M.; Navas-Carretero, S.; San-Cristobal, S.R.; et al. Metabotyping for the development of tailored dietary advice solutions in a European population: The Food4Me study. Br. J. Nutr. 2017, 118. [Google Scholar] [CrossRef] [PubMed]
- Rogers, H.L.; Fernández, S.N.; Hernando, S.P.; Sanchez, A.; Martos, C.; Moreno, M.; Gonzalo, G. “My Patients Asked Me if I Owned a Fruit Stand in Town or Something.” Barriers and Facilitators of Personalized Dietary Advice Implemented in a Primary Care Setting. J. Pers. Med. 2021, 11, 747. [Google Scholar] [CrossRef]
- Brotonsc, C.; Björkelund, C.; Bulc, M.; Ciurana, R.; Godycki-Cwirko, M.; Jurgova, E.; Kloppe, P.; Lionis, C.; Mierzecki, A.; Piñeiro, R.; et al. Prevention and health promotion in clinical practice: The views of general practitioners in Europe. Prev. Med. 2005, 40, 595–601. [Google Scholar] [CrossRef]
- Morris, C.; O’Grada, C.; Ryan, M.; Roche, H.M.; Gibney, M.J.; Gibney, E.R.; Brennan, L. Identification of Differential Responses to an Oral Glucose Tolerance Test in Healthy Adults. Federici M, editor. PLoS ONE 2013, 8, e72890. [Google Scholar] [CrossRef]
- Frazier-Wood, A.C.; Glasser, S.; Garvey, W.T.; Kabagambe, E.K.; Borecki, I.B.; Tiwari, H.K.; Tsai, M.Y.; Hopkins, P.N.; Ordovas, J.M.; Arnett, D.K. A clustering analysis of lipoprotein diameters in the metabolic syndrome. Lipids Health Dis. 2011, 10. [Google Scholar] [CrossRef]
- Chua, E.C.P.; Shui, G.; Lee, I.T.G.; Lau, P.; Tan, L.C.; Yeo, S.C.; Lam, B.D.; Bulchand, S.; Summers, S.A.; Puvanendran, K.; et al. Extensive diversity in circadian regulation of plasma lipids and evidence for different circadian metabolic phenotypes in humans. Proc. Natl. Acad. Sci. USA 2013, 110, 14468–14473. [Google Scholar] [CrossRef]
- Wilcox, M.A.; Wyszynski, D.F.; Panhuysen, C.I.; Ma, Q.; Yip, A.; Farrell, J.; Farrer, L.A. Empirically derived phenotypic subgroups-qualitative and quantitative trait analyses. BMC Genet. 2003, 4 (Suppl. 1). [Google Scholar] [CrossRef]
- Muniandy, M.; Velagapudi, V.; Hakkarainen, A.; Lundbom, J.; Lundbom, N.; Rissanen, A.; Kaprio, J.; Pietiläinen, K.H.; Ollikainen, M. Plasma metabolites reveal distinct profiles associating with different metabolic risk factors in monozygotic twin pairs. Int. J. Obes. 2019, 43, 487–502. [Google Scholar] [CrossRef] [PubMed]
- Urpi-Sarda, M.; Almanza-Aguilera, E.; Llorach, R.; Vázquez-Fresno, R.; Estruch, R.; Corella, D.; Sorli, J.V.; Carmona, F.; Sanchez-Pla, A.; Salas-Salvadó, J.; et al. Non-targeted metabolomic biomarkers and metabotypes of type 2 diabetes: A cross-sectional study of PREDIMED trial participants. Diabetes Metab. 2019, 45, 167–174. [Google Scholar] [CrossRef] [PubMed]
- Tzeng, C.R.; Chang, Y.C.I.; Chang, Y.C.; Wang, C.W.; Chen, C.H.; Hsu, M.I. Cluster analysis of cardiovascular and metabolic risk factors in women of reproductive age. Fertil Steril. 2014, 101, 1404–1414. [Google Scholar] [CrossRef] [PubMed]
| Total | Men | Women | p-Value | |
|---|---|---|---|---|
| n = 3001 | n = 1450 | n = 1551 | ||
| Socio-demographic characteristics | ||||
| Age (years) | 0.03 | |||
| Median (IQR) | 56.0 (22.0) | 57.0 (23.0) | 55.0 (22.0) | |
| Education | <0.001 | |||
| <10 years | 261 (8.7%) | 58 (4.0%) | 203 (13.1%) | |
| 10–<12 years | 1499 (50.0%) | 661 (45.6%) | 838 (54.0%) | |
| ≥12 years | 1236 (41.2%) | 728 (50.2%) | 508 (32.8%) | |
| Missing | 5 (0.2%) | 3 (0.2%) | 2 (0.1%) | |
| BMI (kg/m2) | <0.001 | |||
| Median (IQR) | 27.0 (5.9) | 27.3 (5.1) | 26.3 (7.1) | |
| Normal weight (18.5–<25) | 941 (31.4%) | 345(23.8%) | 596 (38.4%) | |
| Overweight (25–<30) | 1253 (41.8%) | 726(50.1%) | 527 (34.0%) | |
| Obese (≥30) | 793 (26.4%) | 375(25.9%) | 418 (27.0%) | |
| Missing | 14 (0.5%) | 4 (0.3%) | 10 (0.6%) | |
| Physical Activity | 0.409 | |||
| Active | 1641 (54.7%) | 780 (53.8%) | 861 (55.5%) | |
| Inactive | 1356 (45.2%) | 666 (45.9%) | 690 (44.5%) | |
| Missing | 4 (0.1%) | 4 (0.3%) | 0 (0.0%) | |
| Smoking | <0.001 | |||
| Smoker | 524 (17.5%) | 281 (19.4%) | 243 (15.7%) | |
| Ex-Smoker | 1218 (40.6%) | 715 (49.3%) | 503 (32.4%) | |
| Never-Smoker | 1254 (41.8%) | 450 (31.0%) | 804 (51.8%) | |
| Missing | 5 (0.2%) | 4 (0.3%) | 1 (0.1%) | |
| Alcohol consumption | <0.001 | |||
| ≥40 g/day | 336 (11.2%) | 290 (20.0%) | 46 (3.0%) | |
| 20–<40 g/day | 540 (18.0%) | 351 (24.2%) | 189 (12.2%) | |
| 0–<20 g/day | 1221 (40.7%) | 508 (35.0%) | 713 (46.0%) | |
| 0 g/day | 900 (30.0%) | 297 (20.5%) | 603 (38.9%) | |
| Missing | 4 (0.1%) | 4 (0.3%) | 0 (0.0%) | |
| Prevalence of disease n (%) | ||||
| Type 2 diabetes mellitus | 242 (8.1%) | 144 (9.9%) | 98 (6.3%) | <0.001 |
| Hypertension | 1150 (38.3%) | 639 (44.1%) | 511 (32.9%) | <0.001 |
| Missing | 7 (0.2%) | 4 (0.3%) | 3 (0.2%) | |
| Hyperuricemia | 113 (3.8%) | 90 (6.2%) | 23 (1.5%) | <0.001 |
| Missing | 2 (0.1%) | 2 (0.1%) | 0 (0.0%) | |
| Dyslipidemia | 386 (12.9%) | 219 (15.1%) | 167 (10.8%) | <0.001 |
| Missing | 3 (0.1%) | 3 (0.2%) | 0 (0.0%) | |
| Any metabolic disease | 1309 (43.6%) | 730 (50.3%) | 579 (37.3%) | <0.001 |
| Missing | 8 (0.3%) | 4 (0.3%) | 3 (0.2%) | |
| Stroke | 71 (2.4%) | 46 (3.2%) | 25 (1.6%) | 0.007 |
| Myocardial infarction | 79 (2.6%) | 62 (4.3%) | 17 (1.1%) | <0.001 |
| Any cardiovascular disease | 142 (4.7%) | 100 (6.9%) | 42 (2.7%) | <0.001 |
| Total | Metabotype | p-Value | |||
|---|---|---|---|---|---|
| Cluster 1 | Cluster 2 | Cluster 3 | |||
| Model 7 | n = 3001 | n = 1189 | n = 1440 | n = 372 | |
| Age | 56.0 (22.0) | 51.0 (20.0) | 57.0 (22.0) | 65.0 (15.0) | <0.001 |
| Median (IQR) | |||||
| Sex | <0.001 | ||||
| Male | 1450 (48.3%) | 282 (23.7%) | 942 (65.4%) | 226 (60.8%) | |
| Female | 1551 (51.7%) | 907 (76.3%) | 498 (34.6%) | 146 (39.2%) | |
| Education | <0.001 | ||||
| <10 years | 261 (8.7%) | 86 (7.2%) | 124 (8.6%) | 51 (13.7%) | |
| 10–<12 years | 1499 (50.0%) | 584 (49.2%) | 721 (50.1%) | 194 (52.2%) | |
| ≥12 years | 1236 (41.3%) | 516 (43.5%) | 593 (41.2%) | 127 (34.1%) | |
| BMI | <0.001 <0.001 | ||||
| Median (IQR) | 27.0 (5.9) | 24.2 (3.9) | 28.2 (4.6) | 33.2 (6.5) | |
| Normal weight (18.5–<25) | 941 (31.5%) | 723 (61.2%) | 205 (14.3%) | 13 (3.5%) | |
| Overweight (25–<30) | 1253 (41.9%) | 387 (32.8%) | 793 (55.2%) | 73 (19.8%) | |
| Obese (≥30) | 793 (26.5%) | 71 (6.0%) | 439 (30.5%) | 283 (76.7%) | |
| Physical Activity | <0.001 | ||||
| Active | 1641 (54.8%) | 719 (60.5%) | 767 (53.4%) | 155 (41.7%) | |
| Inactive | 1356 (45.2%) | 469 (39.5%) | 670 (46.6%) | 217 (58.3%) | |
| Smoking | <0.001 | ||||
| Smoker | 524 (17.5%) | 210 (17.7%) | 282 (19.6%) | 32 (8.6%) | |
| Ex-smoker | 1218 (40.7%) | 419 (35.3%) | 594 (41.3%) | 205 (55.1%) | |
| Never smoker | 1254 (41.9%) | 558 (47.0%) | 561 (39.0%) | 135 (36.3%) | |
| Alcohol consumption | <0.001 | ||||
| ≥40 g/day | 336 (11.2%) | 97 (8.2%) | 181 (12.6%) | 58 (15.6%) | |
| 20–<40 g/day | 540 (18.0%) | 211 (17.8%) | 273 (19.0%) | 56 (15.1%) | |
| 0–<20 g/day | 1221 (40.7%) | 534 (44.9%) | 561 (39.0%) | 126 (33.9%) | |
| 0 g/day | 900 (30.0%) | 346 (29.1%) | 422 (29.4%) | 132 (35.5%) | |
| Model 17 | n = 3001 | n = 1253 | n = 1476 | n = 281 | |
| Age | <0.001 | ||||
| Median (IQR) | 56.0 (22.0) | 52.0 (22.0) | 57.0 (21.0) | 64.0 (16.0) | |
| Sex | <0.001 | ||||
| Male | 1450 (48.3%) | 384 (30.6%) | 868 (59.2%) | 198 (70.5%) | |
| Female | 1551 (51.7%) | 869 (69.4%) | 599 (40.8%) | 83 (29.5%) | |
| Education | 0.024 | ||||
| <10 years | 261 (8.7%) | 99 (7.9%) | 132 (9.0%) | 30 (10.7%) | |
| 10–<12 years | 1499 (50.0%) | 620 (49.6%) | 719 (49.1%) | 160 (56.9%) | |
| ≥12 years | 1236 (41.3%) | 532 (42.5%) | 613 (41.9%) | 91 (32.4%) | |
| BMI | <0.001 <0.001 | ||||
| Median (IQR) | 27.0 (5.9) | 25.0 (5.3) | 28.0 (5.2) | 30.5 (5.9) | |
| Normal weight (18.5–<25) | 941 (31.5%) | 632 (50.6%) | 287 (19.7%) | 22 (7.9%) | |
| Overweight (25–<30) | 1253 (41.9%) | 437 (35.0%) | 716 (49.0%) | 100 (36.0%) | |
| Obese (≥30) | 793 (26.5%) | 180 (14.4%) | 457 (31.3%) | 156 (56.1%) | |
| Physical Activity | <0.001 | ||||
| Active | 1641 (54.8%) | 763 (61.0%) | 763 (52.1%) | 115 (40.9%) | |
| Inactive | 1356 (45.2%) | 488 (39.0%) | 702 (47.9%) | 166 (59.1%) | |
| Smoking | <0.001 | ||||
| Smoker | 524 (17.5%) | 195 (15.6%) | 277 (18.9%) | 52 (18.5%) | |
| Ex-smoker | 1218 (40.7%) | 476 (38.1%) | 604 (41.2%) | 138 (49.1%) | |
| Never smoker | 1254 (41.9%) | 579 (46.3%) | 584 (39.9%) | 91 (32.4%) | |
| Alcohol consumption | <0.001 | ||||
| ≥40 g/day | 336 (11.2%) | 128 (10.2%) | 155 (10.6%) | 53 (18.9%) | |
| 20–<40 g/day | 540 (18.0%) | 212 (16.9%) | 282 (19.2%) | 46 (16.4%) | |
| 0–<20 g/day | 1221 (40.7%) | 568 (45.4%) | 560 (38.2%) | 93 (33.1%) | |
| 0 g/day | 900 (30.0%) | 343 (27.4%) | 468 (31.9%) | 89 (31.7%) |
| Total | Metabotype | p-Value | |||
|---|---|---|---|---|---|
| Cluster 1 | Cluster 2 | Cluster 3 | |||
| Model 7 | n = 3001 | n = 1189 | n = 1440 | n = 372 | |
| Parameters used for metabotyping | |||||
| BMI [kg/m2] | 26.90 (5.93) | 24.19 (4.31) a | 28.24 (4.85) b | 33.17 (6.47) c | <0.001 |
| Uric acid [µmol/L] | 299.41 (114.12) | 243.52 (79.29) a | 334.12 (93.88) b | 375.29 (113.76) c | <0.001 |
| Glucose [mg/dL] | 94.00 (14.00) | 89.00 (10.80) a | 96.00 (12.40) b | 122.00 (28.40) c | <0.001 |
| HDLc [mmol/L] | 1.39 (0.52) | 1.70 (0.45) a | 1.26 (0.37) b | 1.18 (0.39) c | <0.001 |
| Non-HDLc [mmol/L] | 4.05 (1.32) | 3.59 (1.21) a | 4.49 (1.28) b | 4.01(1.25) c | <0.001 |
| Other Parameters | |||||
| TG [mmol/L] | 1.19 (0.90) | 0.82 (0.56) a | 1.46 (0.87) b | 1.76 (1.22) c | <0.001 |
| AP [µmol/L] | 1.10 (0.42) | 1.00 (0.43) a | 1.14 (0.39) b | 1.21 (0.43) c | <0.001 |
| GPT [µkat/L] | 0.35 (0.23) | 0.28 (0.16) a | 0.40 (0.24) b | 0.48 (0.32) c | <0.001 |
| GOT [µkat/L] | 0.41 (0.14) | 0.38 (0.12) a | 0.43 (0.14) b | 0.45 (0.19) c | <0.001 |
| GGT [µkat/L] | 0.43 (0.4) | 0.31 (0.26) a | 0.50 (0.42) b | 0.63 (0.53) c | <0.001 |
| HbA1c [%] | 5.50 (0.5) | 5.30 (0.42) a | 5.50 (0.5) b | 6.10 (0.98) c | <0.001 |
| hs-CRP [mg/L] | 1.18 (2.03) | 0.76 (1.38) a | 1.38 (2.08) b | 2.49 (3.27) c | <0.001 |
| Leukocytes (n/L) | 5.70 (2) | 5.30 (1.84) a | 5.90 (1.93) b | 6.30 (2) c | <0.001 |
| Insulin [µU/mL] | 8.80 (6.70) | 6.60 (4.14) a | 10.00 (6.46) b | 18.00 (11.74) c | <0.001 |
| Model 17 | n = 3001 | n = 1253 | n = 1467 | n = 281 | |
| Parameters used for metabotyping | |||||
| TG [mmol/L] | 1.19 (0.90) | 1.18 (0.90) a | 1.47 (0.78) b | 2.71 (1.74) c | <0.001 |
| Glucose [mg/dL] | 94.00 (14.00) | 90.00 (11.60) a | 96.00 (13.20) b | 124.00 (38.00) c | <0.001 |
| HDLc [mmol/L] | 1.39 (0.52) | 1.70 (0.45) a | 1.24 (0.37) b | 1.08 (0.35) c | <0.001 |
| Non-HDLc [mmol/L] | 4.05 (1.32) | 3.48 (1.05) a | 4.49 (1.19) b | 4.49 (1.27) b | <0.001 |
| Other Parameters | |||||
| BMI [kg/m2] | 26.99 (5.93) | 24.95 (5.42) a | 27.95 (5.35) b | 30.54 (5.93) c | <0.001 |
| Uric acid [µmol/L] | 299.41 (114.12) | 260.5 (96.70) a | 321.17 (107.05) b | 378.23 (117.8) c | <0.001 |
| AP [µmol/L] | 1.10 (0.42) | 1.01 (0.42) a | 1.15 (0.41) b | 1.20 (0.44)b | <0.001 |
| GPT [µkat/L] | 0.35 (0.23) | 0.30 (0.18) a | 0.39 (0.24) b | 0.48 (0.31) c | <0.001 |
| GOT [µkat/L] | 0.41 (0.14) | 0.39 (0.13) a | 0.42 (0.15) b | 0.45 (0.19) c | <0.001 |
| GGT [µkat/L] | 0.43 (0.40) | 0.34 (0.28) a | 0.48 (0.42) b | 0.71 (0.59) c | <0.001 |
| HbA1c [%] | 5.50 (0.50) | 5.40 (0.50) a | 5.50 (0.50) b | 6.20 (1.22) c | <0.001 |
| hs-CRP [mg/L] | 1.18 (2.03) | 0.86 (1.55) a | 1.39 (2.23) b | 1.97 (2.57) | <0.001 |
| Leukocytes (n/L) | 5.70 (2.00) | 5.40 (1.84) a | 5.90 (2.00) b | 6.40 (2.16) | <0.001 |
| Insulin [µU/mL] | 8.80 (6.70) | 6.90 (4.62) a | 10.00 (6.54) b | 17.00 (10.94) | <0.001 |
| Total | Metabotype | p-Value | |||
|---|---|---|---|---|---|
| Cluster 1 | Cluster 2 | Cluster 3 | |||
| Model 7 | n = 3001 | n = 1189 | n = 1440 | n = 372 | |
| Prevalence of disease in KORA F4; n (%) | |||||
| Type 2 diabetes | 242 (8.06%) | 15 (1.26%) | 52 (3.6%) | 175 (47.0%) | <0.001 |
| Hypertension | 1150 (38.4%) | 249 (21.0%) | 616 (42.9%) | 285 (76.6%) | <0.001 |
| Hyperuricemia | 113 (3.8%) | 15 (1.3%) | 58 (4.0%) | 40 (10.8%) | <0.001 |
| Dyslipidemia | 386 (12.9%) | 101 (8.5%) | 173 (12.0%) | 112 (30.2%) | <0.001 |
| Any metabolic diseases | 1309 (43.7%) | 299 (25.2%) | 684 (47.6%) | 326 (87.9%) | <0.001 |
| Stroke | 71 (2.4%) | 19 (1.6%) | 32 (2.2%) | 20 (5.4%) | <0.001 |
| Myocardial infraction | 79 (2.6%) | 12 (1.0%) | 38 (2.6%) | 29 (7.8%) | <0.001 |
| Any cardiovascular disease | 142 (4.7%) | 27 (2.3%) | 68 (4.7%) | 47 (12.6%) | <0.001 |
| Incidence of disease in KORA FF4; n (%) | n = 2120 | n = 895 | n = 1003 | n = 222 | |
| Type 2 diabetes | 94 (4.7%) | 13 (1.5%) | 43 (4.4%) | 38 (30.2%) | <0.001 |
| Hypertension | 230 (10.9%) | 86 (9.6%) | 114 (11.4%) | 30 (13.6%) | 0.187 |
| Hyperuricemia | 45 (2.1%) | 0 (0.0%) | 24 (2.4%) | 21 (9.5%) | <0.001 |
| Dyslipidemia | 157 (7.4%) | 27 (3.0%) | 92 (9.2%) | 38 (17.2%) | <0.001 |
| Any metabolic diseases | 442 (21.9%) | 114 (13.0%) | 233 (23.7%) | 95 (62.0%) | <0.001 |
| Stroke | 35 (1.7%) | 10 (1.1%) | 15 (1.5%) | 10 (4.6%) | 0.001 |
| Myocardial infraction | 27 (1.3%) | 2 (0.2%) | 20 (2.0%) | 5 (2.4%) | <0.001 |
| Any cardiovascular disease | 60 (2.9%) | 12 (1.4%) | 33 (3.4%) | 15 (7.4%) | <0.001 |
| Model 17 | n = 3001 | n = 1235 | n = 1467 | n = 281 | |
| Prevalence of disease in KORA F4; n (%) | |||||
| Type 2 diabetes | 242 (8.1%) | 39 (3.1%) | 67 (4.6%) | 136 (48.4%) | <0.001 |
| Hypertension | 1150 (38.4%) | 332 (26.6%) | 620 (42.4%) | 198 (70.5%) | <0.001 |
| Hyperuricemia | 113 (3.8%) | 22 (1.8%) | 53 (3.6%) | 38 (13.6%) | <0.001 |
| Dyslipidemia | 386 (12.9%) | 140 (11.2%) | 168 (11.5%) | 78 (27.9%) | <0.001 |
| Any metabolic diseases | 1309 (43.7%) | 385 (31.0%) | 694 (47.4%) | 230 (82.1%) | <0.001 |
| Stroke | 71 (2.4%) | 26 (2.1%) | 35 (2.4%) | 10 (3.6%) | 0.334 |
| Myocardial infraction | 79 (2.6%) | 24 (1.9%) | 37 (2.5%) | 18 (6.4%) | <0.001 |
| Any cardiovascular disease | 142 (4.7%) | 46 (3.6%) | 70 (4.8%) | 26 (9.2%) | <0.001 |
| Incidence of disease in KORA FF4; n (%) | n = 2120 | n = 916 | n = 1040 | n = 164 | |
| Type 2 diabetes | 94 (4.7%) | 15 (1.7%) | 54 (5.4%) | 25 (26.9%) | <0.001 |
| Hypertension | 230 (10.9%) | 85 (9.3%) | 125 (12.0%) | 20 (12.3%) | 0.127 |
| Hyperuricemia | 45 (2.1%) | 9 (0.1%) | 29 (2.8%) | 7 (4.3%) | 0.002 |
| Dyslipidemia | 157 (7.4%) | 30 (3.3%) | 92 (8.8%) | 35 (21.5%) | <0.001 |
| Any metabolic diseases | 442 (21.9%) | 120 (13.5%) | 256 (25.3%) | 66 (58.4%) | <0.001 |
| Stroke | 35 (1.7%) | 11 (1.2%) | 14 (1.4%) | 10 (6.2%) | <0.001 |
| Myocardial infraction | 27 (1.3%) | 2 (0.2%) | 21 (2.0%) | 4 (2.6%) | <0.001 |
| Any cardiovascular disease | 60 (2.9%) | 13 (1.5%) | 33 (3.3%) | 14 (9.1%) | <0.001 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dahal, C.; Wawro, N.; Meisinger, C.; Breuninger, T.A.; Thorand, B.; Rathmann, W.; Koenig, W.; Hauner, H.; Peters, A.; Linseisen, J. Optimized Metabotype Definition Based on a Limited Number of Standard Clinical Parameters in the Population-Based KORA Study. Life 2022, 12, 1460. https://doi.org/10.3390/life12101460
Dahal C, Wawro N, Meisinger C, Breuninger TA, Thorand B, Rathmann W, Koenig W, Hauner H, Peters A, Linseisen J. Optimized Metabotype Definition Based on a Limited Number of Standard Clinical Parameters in the Population-Based KORA Study. Life. 2022; 12(10):1460. https://doi.org/10.3390/life12101460
Chicago/Turabian StyleDahal, Chetana, Nina Wawro, Christa Meisinger, Taylor A. Breuninger, Barbara Thorand, Wolfgang Rathmann, Wolfgang Koenig, Hans Hauner, Annette Peters, and Jakob Linseisen. 2022. "Optimized Metabotype Definition Based on a Limited Number of Standard Clinical Parameters in the Population-Based KORA Study" Life 12, no. 10: 1460. https://doi.org/10.3390/life12101460
APA StyleDahal, C., Wawro, N., Meisinger, C., Breuninger, T. A., Thorand, B., Rathmann, W., Koenig, W., Hauner, H., Peters, A., & Linseisen, J. (2022). Optimized Metabotype Definition Based on a Limited Number of Standard Clinical Parameters in the Population-Based KORA Study. Life, 12(10), 1460. https://doi.org/10.3390/life12101460






