Complement Regulation in Immortalized Fibroblast-like Synoviocytes and Primary Human Endothelial Cells in Response to SARS-CoV-2 Nucleocapsid Protein and Pro-Inflammatory Cytokine TNFα
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Isolation, Cell Culture and Cell Stimulation
2.2. Viability Assays
2.2.1. Vitality Staining of Synovial Cells
2.2.2. CellTiter96® Aqueous One Solution Cell Proliferation Assay
2.2.3. CyQUANT® NF Cell Proliferation Assay
2.3. Gene Expression Analysis
2.3.1. RNA Isolation and cDNA Synthesis
2.3.2. qPCR
2.4. Protein Expression Analysis
2.5. Statistical Analysis
3. Results
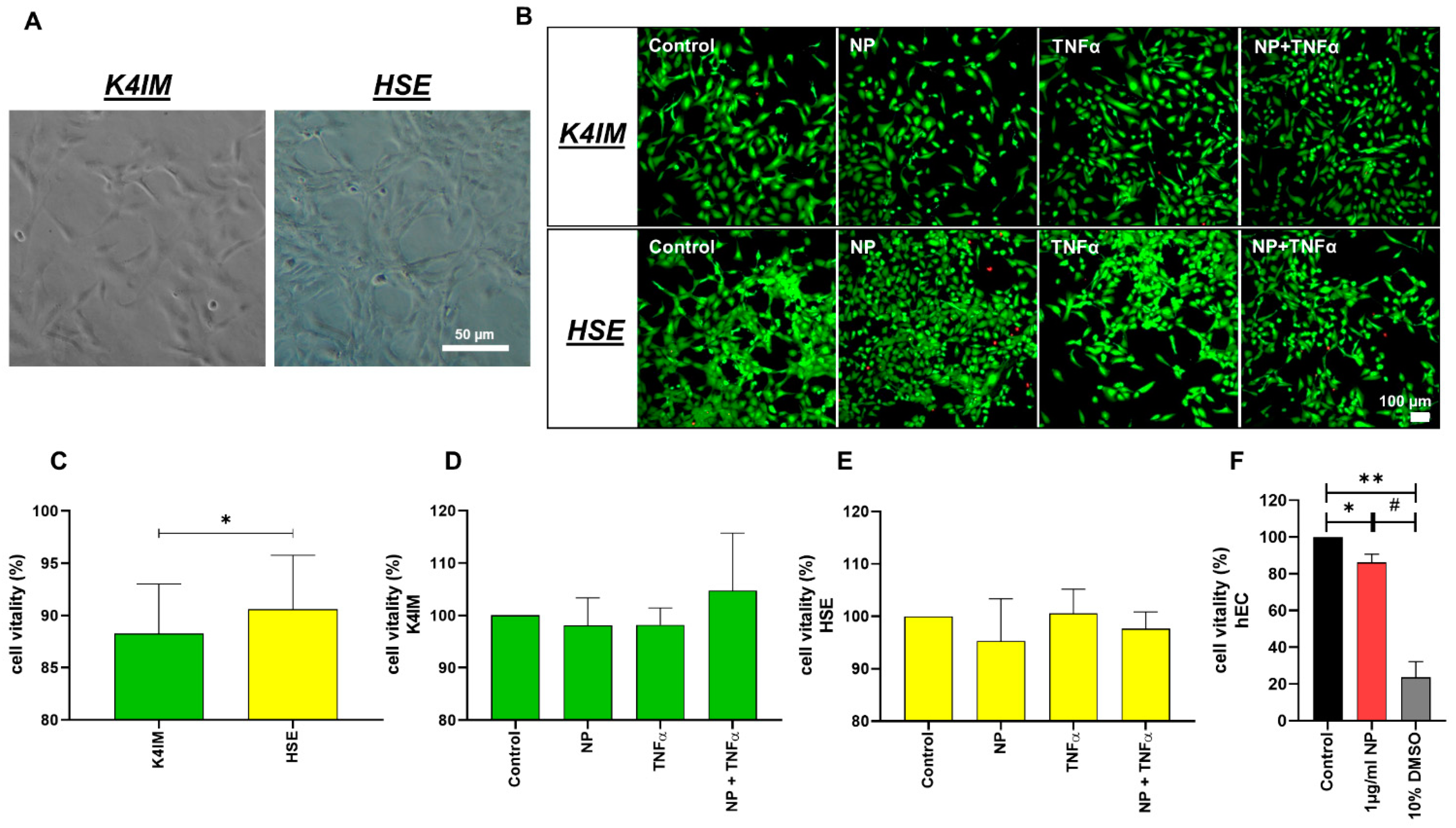
3.1. Vitality Assays
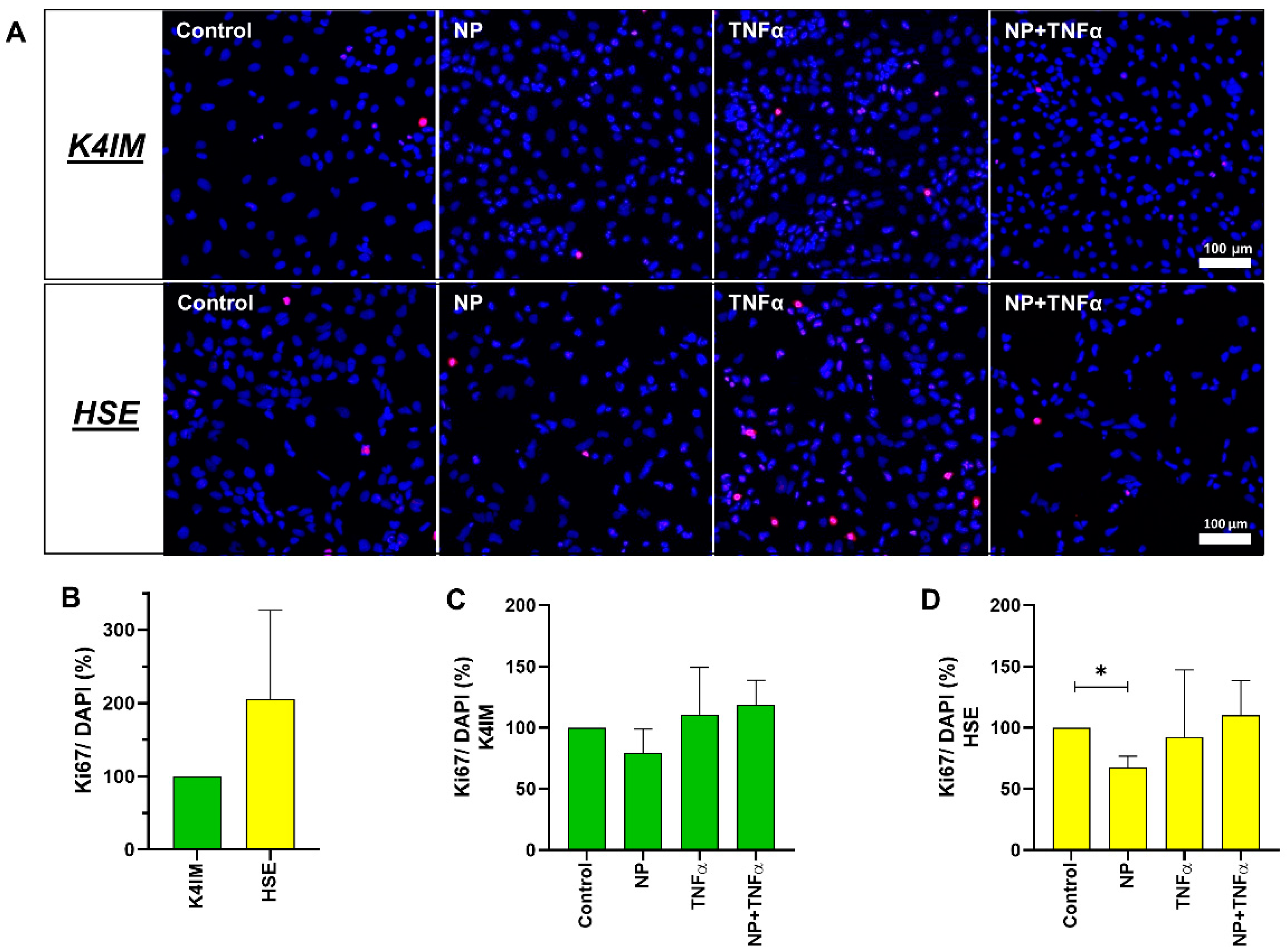
3.2. DNA Quantification and Cell Proliferation
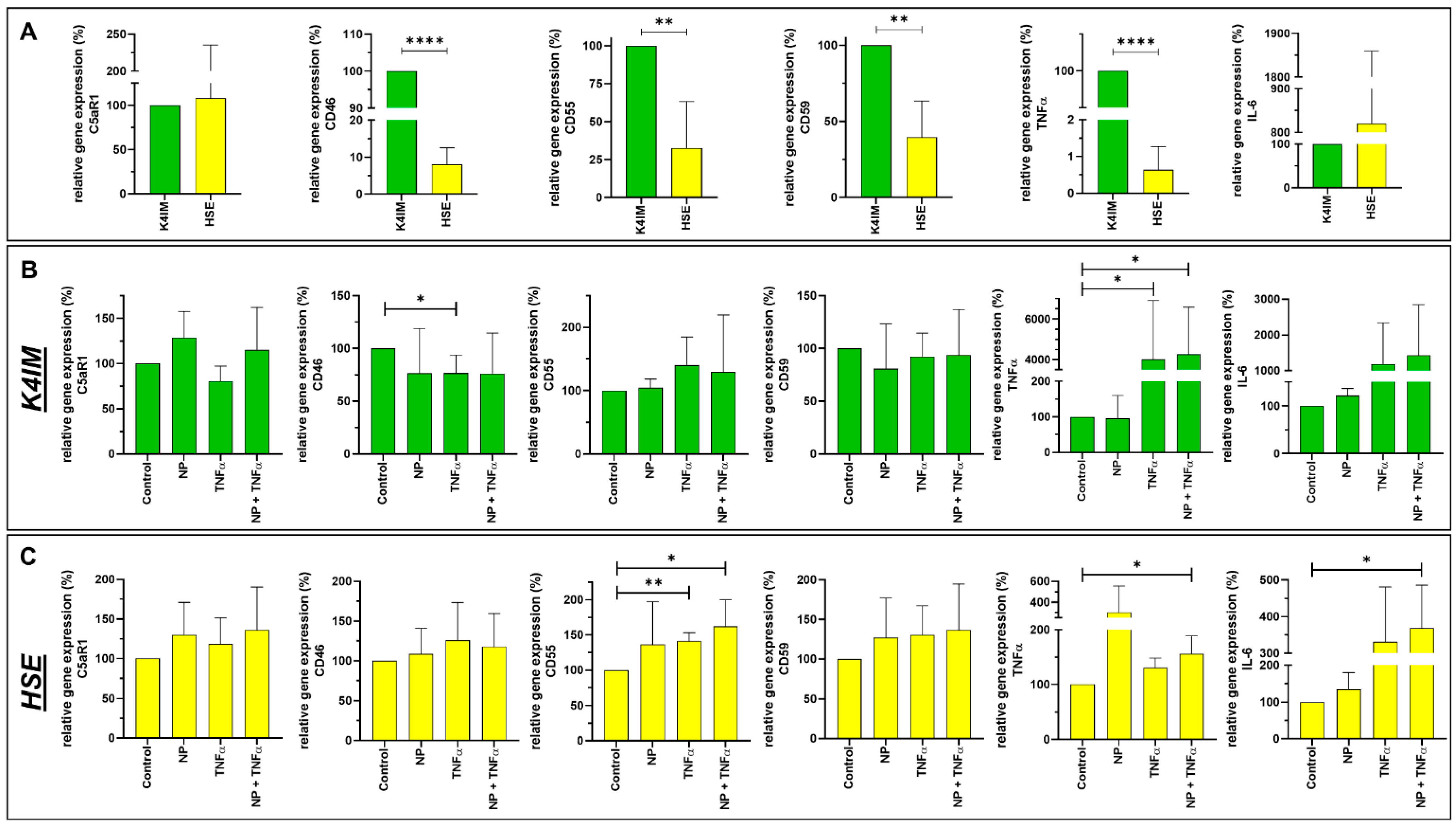
3.3. Gene Expression
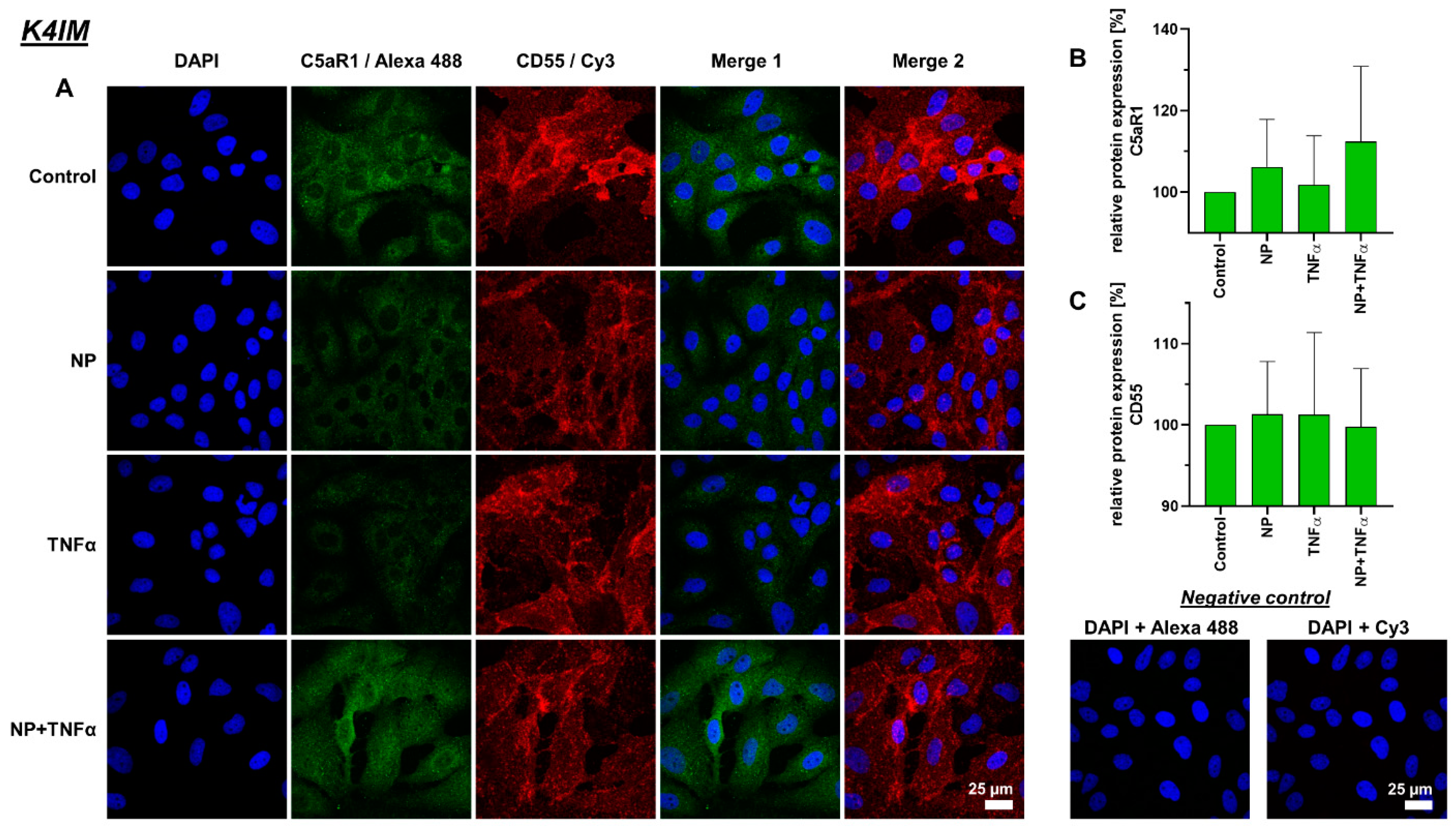
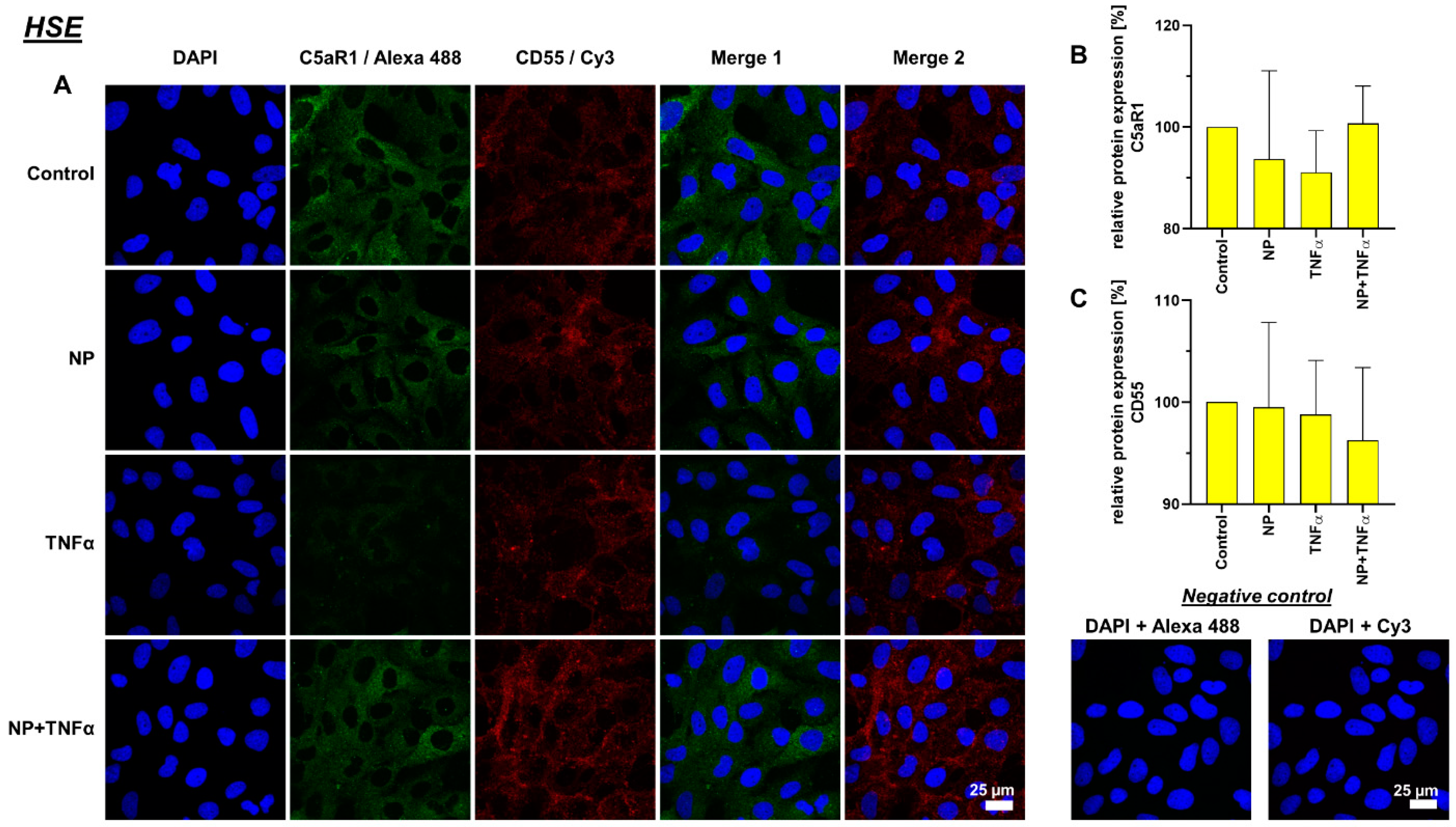
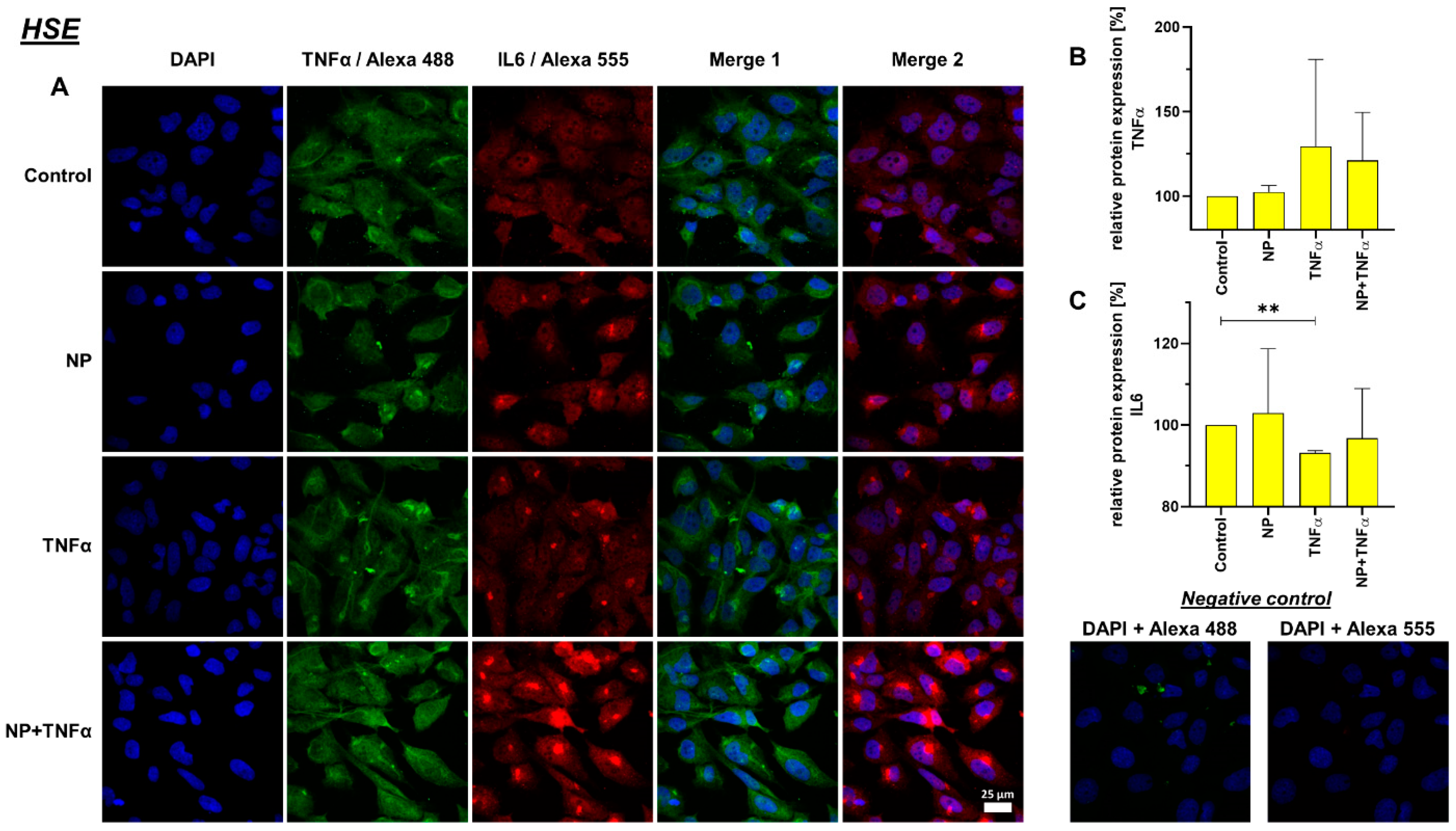
3.4. Protein Synthesis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ACPA | Anti-citrullinated peptide antibody |
| bFGF | Basic fibroblast growth factor |
| C3aR1 | C3a receptor 1 |
| C5aR1 | C5a receptor 1 |
| cDNA | Complementary DNA |
| CRP | Complement regulatory protein |
| CTCF | Corrected total cell fluorescence |
| DAPI | 4′,6-Diamidin-2-phenylindol |
| DMSO | Dimethyl sulfoxide |
| FBS | Fetal bovine serum |
| FDA | Fluorescein diacetate |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase |
| h | Hour |
| HBSS | Hanks’ balanced salt solution |
| hEC | Human endothelial cells |
| HSE | Immortalized human rheumatoid arthritic fibroblast-like synoviocytes |
| IL | Interleukin |
| K4IM | Immortalized human non-arthritic fibroblast-like synoviocytes |
| MAC | Membrane attack complex |
| MAPK | Mitogen activated protein kinase |
| MASP-2 | Mannose-binding protein-associated serine protease 2 |
| Min | Minutes |
| NF-κB | Nuclear factor ‘kappa-light-chain-enhancer’ of activated B-cells |
| NP | Nucleocapsid protein |
| PAMP | Pathogen-associated molecular patterns |
| PBS | Phosphate-buffered saline |
| PFA | Paraformaldehyde solution |
| qPCR | Real time detection polymerase chain reaction |
| RT | Room temperature |
| SARS-CoV-2 | Severe acute respiratory syndrome coronavirus type 2 |
| TBS | Tris buffered saline |
| TCC | Terminal complement complex |
| TLR | Toll-like receptors |
| TNFα | Tumor necrosis factor |
| VEGF | Vascular endothelial growth factor |
References
- Fehr, A.R.; Perlman, S. Coronaviruses: An overview of their replication and pathogenesis. Methods Mol Biol. 2015, 1282, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Satarker, S.; Nampoothiri, M. Structural Proteins in Severe Acute Respiratory Syndrome Coronavirus-2. Arch. Med. Res. 2020, 51, 482–491. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Grunewald, M.; Perlman, S. Coronaviruses: An Updated Overview of Their Replication and Pathogenesis. Methods Mol Biol. 2020, 2203, 1–29. [Google Scholar] [CrossRef] [PubMed]
- Bárcena, M.; Oostergetel, G.T.; Bartelink, W.; Faas, F.G.; Verkleij, A.; Rottier, P.J.; Koster, A.J.; Bosch, B.J. Cryo-electron tomography of mouse hepatitis virus: Insights into the structure of the coronavirion. Proc. Natl. Acad. Sci. USA 2009, 106, 582–587. [Google Scholar] [CrossRef] [PubMed]
- Smits, V.A.J.; Hernández-Carralero, E.; Paz-Cabrera, M.C.; Cabrera, E.; Hernández-Reyes, Y.; Hernández-Fernaud, J.R.; Gillespie, D.A.; Salido, E.; Hernández-Porto, M.; Freire, R. The Nucleocapsid protein triggers the main humoral immune response in COVID-19 patients. Biochem. Biophys. Res. Commun. 2021, 543, 45–49. [Google Scholar] [CrossRef]
- Mangalmurti, N.; Hunter, C.A. Cytokine Storms: Understanding COVID-19. Immunity 2020, 53, 19–25. [Google Scholar] [CrossRef]
- Hojyo, S.; Uchida, M.; Tanaka, K.; Hasebe, R.; Tanaka, Y.; Murakami, M.; Hirano, T. How COVID-19 induces cytokine storm with high mortality. Inflamm. Regen. 2020, 40, 37. [Google Scholar] [CrossRef]
- Ackermann, M.; Verleden, S.E.; Kuehnel, M.; Haverich, A.; Welte, T.; Laenger, F.; Vanstapel, A.; Werlein, C.; Stark, H.; Tzankov, A.; et al. Pulmonary Vascular Endothelialitis, Thrombosis, and Angiogenesis in COVID-19. N. Engl. J. Med. 2020, 383, 120–128. [Google Scholar] [CrossRef]
- Mehta, P.; McAuley, D.F.; Brown, M.; Sanchez, E.; Tattersall, R.S.; Manson, J.J. COVID-19: Consider cytokine storm syndromes and immunosuppression. Lancet 2020, 395, 1033–1034. [Google Scholar] [CrossRef]
- Gao, Y.; Li, T.; Han, M.; Li, X.; Wu, D.; Xu, Y.; Zhu, Y.; Liu, Y.; Wang, X.; Wang, L. Diagnostic utility of clinical laboratory data determinations for patients with the severe COVID-19. J. Med. Virol. 2020, 92, 791–796. [Google Scholar] [CrossRef]
- Manjili, R.H.; Zarei, M.; Habibi, M.; Manjili, M.H. COVID-19 as an Acute Inflammatory Disease. J. Immunol. 2020, 205, 12–19. [Google Scholar] [CrossRef] [PubMed]
- Jia, F.; Wang, G.; Xu, J.; Long, J.; Deng, F.; Jiang, W. Role of tumor necrosis factor-α in the mortality of hospitalized patients with severe and critical COVID-19 pneumonia. Aging 2021, 13, 23895–23912. [Google Scholar] [CrossRef] [PubMed]
- AlSamman, M.; Caggiula, A.; Ganguli, S.; Misak, M.; Pourmand, A. Non-respiratory presentations of COVID-19, a clinical review. Am. J. Emerg. Med. 2020, 38, 2444–2454. [Google Scholar] [CrossRef] [PubMed]
- Smatti, M.K.; Cyprian, F.S.; Nasrallah, G.K.; Al Thani, A.A.; Almishal, R.O.; Yassine, H.M. Viruses and Autoimmunity: A Review on the Potential Interaction and Molecular Mechanisms. Viruses 2019, 11, 762. [Google Scholar] [CrossRef] [PubMed]
- Halpert, G.; Shoenfeld, Y. SARS-CoV-2, the autoimmune virus. Autoimmun. Rev. 2020, 19, 102695. [Google Scholar] [CrossRef]
- Zvaifler, N.J.; Firestein, G.S. Pannus and pannocytes. Alternative models of joint destruction in rheumatoid arthritis. Arthritis Rheum. 1994, 37, 783–789. [Google Scholar] [CrossRef]
- Chauhan, K.; Jandu, J.S.; Goyal, A.; Al-Dhahir, M.A. Rheumatoid Arthritis. In StatPearls; StatPearls Publishing LLC.: Treasure Island, FL, USA, 2022. [Google Scholar]
- Huber, L.C.; Distler, O.; Tarner, I.; Gay, R.E.; Gay, S.; Pap, T. Synovial fibroblasts: Key players in rheumatoid arthritis. Rheumatology 2006, 45, 669–675. [Google Scholar] [CrossRef]
- Pal, A.; Roongta, R.; Mondal, S.; Sinha, D.; Sinhamahapatra, P.; Ghosh, A.; Chattopadhyay, A. Does post-COVID reactive arthritis exist? Experience of a tertiary care centre with a review of the literature. Reumatol. Clin. 2022, in press. [Google Scholar] [CrossRef]
- Perrot, L.; Hemon, M.; Busnel, J.M.; Muis-Pistor, O.; Picard, C.; Zandotti, C.; Pham, T.; Roudier, J.; Desplat-Jego, S.; Balandraud, N. First flare of ACPA-positive rheumatoid arthritis after SARS-CoV-2 infection. Lancet. Rheumatol. 2021, 3, e6–e8. [Google Scholar] [CrossRef]
- Zeidler, H. Post-Corona-Virus-Disease-19 arthritis. Manifestation under the clinical picture of a reactive arthritis. Z. Fur Rheumatol. 2021, 80, 555–558. [Google Scholar] [CrossRef]
- Derksen, V.; van der Woude, D. Response to: ‘Correspondence on ‘Onset of rheumatoid arthritis after COVID-19: Coincidence or connected?’’ by Roongta et al. Ann. Rheum. Dis. 2021. [Google Scholar] [CrossRef] [PubMed]
- Roongta, R.; Chattopadhyay, A.; Ghosh, A. Correspondence on ‘Onset of rheumatoid arthritis after COVID-19: Coincidence or connected?’. Ann. Rheum. Dis. 2021, 80, 1096–1098. [Google Scholar] [CrossRef] [PubMed]
- Derksen, V.F.A.M.; Kissel, T.; Lamers-Karnebeek, F.B.G.; van der Bijl, A.E.; Venhuizen, A.C.; Huizinga, T.W.J.; Toes, R.E.M.; Roukens, A.H.E.; van der Woude, D. Onset of rheumatoid arthritis after COVID-19: Coincidence or connected? Ann. Rheum. Dis. 2021, 80, 1096–1098. [Google Scholar] [CrossRef] [PubMed]
- Grainger, R.; Kim, A.H.J.; Conway, R.; Yazdany, J.; Robinson, P.C. COVID-19 in people with rheumatic diseases: Risks, outcomes, treatment considerations. Nat. Rev. Rheumatol. 2022, 18, 191–204. [Google Scholar] [CrossRef]
- Stoermer, K.A.; Morrison, T.E. Complement and viral pathogenesis. Virology 2011, 411, 362–373. [Google Scholar] [CrossRef]
- Okroj, M.; Heinegård, D.; Holmdahl, R.; Blom, A.M. Rheumatoid arthritis and the complement system. Ann. Med. 2007, 39, 517–530. [Google Scholar] [CrossRef]
- Pouw, R.B.; Ricklin, D. Tipping the balance: Intricate roles of the complement system in disease and therapy. Semin. Immunopathol. 2021, 43, 757–771. [Google Scholar] [CrossRef]
- Dunkelberger, J.R.; Song, W.-C. Complement and its role in innate and adaptive immune responses. Cell Res. 2010, 20, 34–50. [Google Scholar] [CrossRef]
- Merle, N.S.; Church, S.E.; Fremeaux-Bacchi, V.; Roumenina, L.T. Complement System Part I—Molecular Mechanisms of Activation and Regulation. Front. Immunol. 2015, 6, 262. [Google Scholar] [CrossRef]
- Ricklin, D.; Hajishengallis, G.; Yang, K.; Lambris, J.D. Complement: A key system for immune surveillance and homeostasis. Nat. Immunol. 2010, 11, 785–797. [Google Scholar] [CrossRef] [Green Version]
- Ma, L.; Sahu, S.K.; Cano, M.; Kuppuswamy, V.; Bajwa, J.; McPhatter, J.; Pine, A.; Meizlish, M.L.; Goshua, G.; Chang, C.H.; et al. Increased complement activation is a distinctive feature of severe SARS-CoV-2 infection. Sci. Immunol. 2021, 6, eabh2259. [Google Scholar] [CrossRef]
- De Nooijer, A.H.; Grondman, I.; Janssen, N.A.F.; Netea, M.G.; Willems, L.; van de Veerdonk, F.L.; Giamarellos-Bourboulis, E.J.; Toonen, E.J.M.; Joosten, L.A.B. Complement Activation in the Disease Course of Coronavirus Disease 2019 and Its Effects on Clinical Outcomes. J. Infect. Dis. 2021, 223, 214–224. [Google Scholar] [CrossRef] [PubMed]
- Huber, S.; Massri, M.; Grasse, M.; Fleischer, V.; Kellnerová, S.; Harpf, V.; Knabl, L.; Knabl, L., Sr.; Heiner, T.; Kummann, M.; et al. Systemic Inflammation and Complement Activation Parameters Predict Clinical Outcome of Severe SARS-CoV-2 Infections. Viruses 2021, 13, 2376. [Google Scholar] [CrossRef] [PubMed]
- Holter, J.C.; Pischke, S.E.; de Boer, E.; Lind, A.; Jenum, S.; Holten, A.R.; Tonby, K.; Barratt-Due, A.; Sokolova, M.; Schjalm, C.; et al. Systemic complement activation is associated with respiratory failure in COVID-19 hospitalized patients. Proc. Natl. Acad. Sci. USA 2020, 117, 25018–25025. [Google Scholar] [CrossRef] [PubMed]
- Gao, T.; Hu, M.; Zhang, X.; Li, H.; Zhu, L.; Liu, H.; Dong, Q.; Zhang, Z.; Wang, Z.; Hu, Y.; et al. Highly pathogenic coronavirus N protein aggravates lung injury by MASP-2-mediated complement over-activation. medRxiv 2020. [Google Scholar] [CrossRef]
- Yu, J.; Yuan, X.; Chen, H.; Chaturvedi, S.; Braunstein, E.M.; Brodsky, R.A. Direct activation of the alternative complement pathway by SARS-CoV-2 spike proteins is blocked by factor D inhibition. Blood 2020, 136, 2080–2089. [Google Scholar] [CrossRef]
- Jensen, E.C. Quantitative Analysis of Histological Staining and Fluorescence Using ImageJ. Anat. Rec. 2013, 296, 378–381. [Google Scholar] [CrossRef]
- Lafyatis, R.; Remmers, E.F.; Roberts, A.B.; Yocum, D.E.; Sporn, M.B.; Wilder, R.L. Anchorage-independent growth of synoviocytes from arthritic and normal joints. Stimulation by exogenous platelet-derived growth factor and inhibition by transforming growth factor-beta and retinoids. J. Clin. Investig. 1989, 83, 1267–1276. [Google Scholar] [CrossRef]
- Nygaard, G.; Firestein, G.S. Restoring synovial homeostasis in rheumatoid arthritis by targeting fibroblast-like synoviocytes. Nat. Rev. Rheumatol. 2020, 16, 316–333. [Google Scholar] [CrossRef]
- Korb, A.; Pavenstädt, H.; Pap, T. Cell death in rheumatoid arthritis. Apoptosis Int. J. Program. Cell Death 2009, 14, 447–454. [Google Scholar] [CrossRef]
- Yi, X.; Liu, M.; Luo, Q.; Zhuo, H.; Cao, H.; Wang, J.; Han, Y. Toxic effects of dimethyl sulfoxide on red blood cells, platelets, and vascular endothelial cells in vitro. FEBS Open Bio 2017, 7, 485–494. [Google Scholar] [CrossRef] [PubMed]
- Häupl, T.; Yahyawi, M.; Lübke, C.; Ringe, J.; Rohrlach, T.; Burmester, G.R.; Sittinger, M.; Kaps, C. Gene expression profiling of rheumatoid arthritis synovial cells treated with antirheumatic drugs. J. Biomol. Screen. 2007, 12, 328–340. [Google Scholar] [CrossRef] [PubMed]
- You, S.; Yoo, S.-A.; Choi, S.; Kim, J.-Y.; Park, S.-J.; Ji, J.D.; Kim, T.-H.; Kim, K.-J.; Cho, C.-S.; Hwang, D.; et al. Identification of key regulators for the migration and invasion of rheumatoid synoviocytes through a systems approach. Proc. Natl. Acad. Sci. USA 2014, 111, 550–555. [Google Scholar] [CrossRef] [PubMed]
- Pessler, F.; Ogdie, A.; Diaz-Torne, C.; Dai, L.; Yu, X.; Einhorn, E.; Gay, S.; Schumacher, H.R. Subintimal Ki-67 as a synovial tissue biomarker for inflammatory arthropathies. Ann. Rheum. Dis. 2008, 67, 162–167. [Google Scholar] [CrossRef]
- You, S.; Koh, J.H.; Leng, L.; Kim, W.U.; Bucala, R. The Tumor-Like Phenotype of Rheumatoid Synovium: Molecular Profiling and Prospects for Precision Medicine. Arthritis Rheumatol. 2018, 70, 637–652. [Google Scholar] [CrossRef]
- Sartain, S.E.; Turner, N.A.; Moake, J.L. Brain microvascular endothelial cells exhibit lower activation of the alternative complement pathway than glomerular microvascular endothelial cells. J. Biol. Chem. 2018, 293, 7195–7208. [Google Scholar] [CrossRef]
- Ahearn, J.M.; Fearon, D.T. Structure and Function of the Complement Receptors, CR1 (CD35) and CR2 (CD21). In Advances in Immunology; Dixon, F.J., Ed.; Academic Press: Cambridge, MA, USA, 1989; Volume 46, pp. 183–219. [Google Scholar]
- Wymann, S.; Dai, Y.; Nair, A.G.; Cao, H.; Powers, G.A.; Schnell, A.; Martin-Roussety, G.; Leong, D.; Simmonds, J.; Lieu, K.G.; et al. A novel soluble complement receptor 1 fragment with enhanced therapeutic potential. J. Biol. Chem. 2021, 296, 100200. [Google Scholar] [CrossRef]
- Athanasou, N.; Quinn, J. Immunocytochemical analysis of human synovial lining cells: Phenotypic relation to other marrow derived cells. Ann. Rheum. Dis. 1991, 50, 311–315. [Google Scholar] [CrossRef]
- Collard, C.D.; Bukusoglu, C.; Agah, A.; Colgan, S.P.; Reenstra, W.R.; Morgan, B.P.; Stahl, G.L. Hypoxia-induced expression of complement receptor type 1 (CR1, CD35) in human vascular endothelial cells. Am. J. Physiol. 1999, 276, C450–C458. [Google Scholar] [CrossRef]
- Ni Choileain, S.; Astier, A.L. CD46 plasticity and its inflammatory bias in multiple sclerosis. Arch. Immunol. Ther. Exp. 2011, 59, 49–59. [Google Scholar] [CrossRef] [Green Version]
- Cardone, J.; Le Friec, G.; Kemper, C. CD46 in innate and adaptive immunity: An update. Clin. Exp. Immunol. 2011, 164, 301–311. [Google Scholar] [CrossRef] [PubMed]
- Dho, S.H.; Lim, J.C.; Kim, L.K. Beyond the Role of CD55 as a Complement Component. Immune Netw. 2018, 18, e11. [Google Scholar] [CrossRef] [PubMed]
- Karpus, O.; Kiener, H.; Niederreiter, B.; Yilmaz-Elis, A.; van der Kaa, J.; Ramaglia, V.; Arens, R.; Smolen, J.; Botto, M.; Tak, P.; et al. CD55 deposited on synovial collagen fibers protects from immune complex-mediated arthritis. Arthritis Res. Ther. 2015, 17, 6. [Google Scholar] [CrossRef] [PubMed]
- Hamann, J.; Vogel, B.; van Schijndel, G.M.; van Lier, R.A. The seven-span transmembrane receptor CD97 has a cellular ligand (CD55, DAF). J. Exp. Med. 1996, 184, 1185–1189. [Google Scholar] [CrossRef]
- Hamann, J.; Wishaupt, J.O.; van Lier, R.A.; Smeets, T.J.; Breedveld, F.C.; Tak, P.P. Expression of the activation antigen CD97 and its ligand CD55 in rheumatoid synovial tissue. Arthritis Rheum. 1999, 42, 650–658. [Google Scholar] [CrossRef]
- Hoek, R.M.; de Launay, D.; Kop, E.N.; Yilmaz-Elis, A.S.; Lin, F.; Reedquist, K.A.; Verbeek, J.S.; Medof, M.E.; Tak, P.P.; Hamann, J. Deletion of either CD55 or CD97 ameliorates arthritis in mouse models. Arthritis Rheum. 2010, 62, 1036–1042. [Google Scholar] [CrossRef]
- Andoh, A.; Fujiyama, Y.; Sumiyoshi, K.; Sakumoto, H.; Okabe, H.; Bamba, T. Tumour necrosis factor-alpha up-regulates decay-accelerating factor gene expression in human intestinal epithelial cells. Immunology 1997, 90, 358–363. [Google Scholar] [CrossRef]
- Li, W.; Tada, T.; Miwa, T.; Okada, N.; Ito, J.; Okada, H.; Tateyama, H.; Eimoto, T. mRNA expression of complement components and regulators in rat arterial smooth muscle cells. Microbiol. Immunol. 1999, 43, 585–593. [Google Scholar] [CrossRef]
- Mason, J.C.; Yarwood, H.; Sugars, K.; Morgan, B.P.; Davies, K.A.; Haskard, D.O. Induction of decay-accelerating factor by cytokines or the membrane-attack complex protects vascular endothelial cells against complement deposition. Blood 1999, 94, 1673–1682. [Google Scholar] [CrossRef]
- Konttinen, Y.T.; Ceponis, A.; Meri, S.; Vuorikoski, A.; Kortekangas, P.; Sorsa, T.; Sukura, A.; Santavirta, S. Complement in acute and chronic arthritides: Assessment of C3c, C9, and protectin (CD59) in synovial membrane. Ann. Rheum. Dis. 1996, 55, 888–894. [Google Scholar] [CrossRef] [Green Version]
- Das, N.; Anand, D.; Biswas, B.; Kumari, D.; Gandhi, M. The membrane complement regulatory protein CD59 and its association with rheumatoid arthritis and systemic lupus erythematosus. Curr. Med. Res. Pract. 2019, 9, 182–188. [Google Scholar] [CrossRef]
- Chu, W.-M. Tumor necrosis factor. Cancer Lett. 2013, 328, 222–225. [Google Scholar] [CrossRef] [PubMed]
- Cao, M.; Ong, M.T.Y.; Yung, P.S.H.; Tuan, R.S.; Jiang, Y. Role of synovial lymphatic function in osteoarthritis. Osteoarthr. Cartil. 2022, 30, 1186–1197. [Google Scholar] [CrossRef] [PubMed]
- Chu, C.Q.; Field, M.; Feldmann, M.; Maini, R.N. Localization of tumor necrosis factor alpha in synovial tissues and at the cartilage-pannus junction in patients with rheumatoid arthritis. Arthritis Rheum. 1991, 34, 1125–1132. [Google Scholar] [CrossRef]
- Tohidnezhad, M.; Bayer, A.; Rasuo, B.; Hock, J.V.P.; Kweider, N.; Fragoulis, A.; Sönmez, T.T.; Jahr, H.; Pufe, T.; Lippross, S. Platelet-Released Growth Factors Modulate the Secretion of Cytokines in Synoviocytes under Inflammatory Joint Disease. Mediat. Inflamm. 2017, 2017, 1046438. [Google Scholar] [CrossRef]
- Mrosewski, I.; Jork, N.; Gorte, K.; Conrad, C.; Wiegand, E.; Kohl, B.; Ertel, W.; John, T.; Oberholzer, A.; Kaps, C.; et al. Regulation of osteoarthritis-associated key mediators by TNFα and IL-10: Effects of IL-10 overexpression in human synovial fibroblasts and a synovial cell line. Cell Tissue Res. 2014, 357, 207–223. [Google Scholar] [CrossRef]
- Tanaka, T.; Narazaki, M.; Kishimoto, T. IL-6 in inflammation, immunity, and disease. Cold Spring Harb. Perspect. Biol. 2014, 6, a016295. [Google Scholar] [CrossRef]
- Yoshida, Y.; Tanaka, T. Interleukin 6 and rheumatoid arthritis. BioMed Res. Int. 2014, 2014, 698313. [Google Scholar] [CrossRef]
- Hirano, T.; Matsuda, T.; Turner, M.; Miyasaka, N.; Buchan, G.; Tang, B.; Sato, K.; Shimizu, M.; Maini, R.; Feldmann, M.; et al. Excessive production of interleukin 6/B cell stimulatory factor-2 in rheumatoid arthritis. Eur. J. Immunol. 1988, 18, 1797–1801. [Google Scholar] [CrossRef]
- Baumann, H.; Kushner, I. Production of interleukin-6 by synovial fibroblasts in rheumatoid arthritis. Am. J. Pathol. 1998, 152, 641–644. [Google Scholar]
- Kunisch, E.; Gandesiri, M.; Fuhrmann, R.; Roth, A.; Winter, R.; Kinne, R.W. Predominant activation of MAP kinases and pro-destructive/pro-inflammatory features by TNF alpha in early-passage synovial fibroblasts via TNF receptor-1: Failure of p38 inhibition to suppress matrix metalloproteinase-1 in rheumatoid arthritis. Ann. Rheum. Dis. 2007, 66, 1043–1051. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ogata, A.; Kato, Y.; Higa, S.; Yoshizaki, K. IL-6 inhibitor for the treatment of rheumatoid arthritis: A comprehensive review. Mod. Rheumatol. 2019, 29, 258–267. [Google Scholar] [CrossRef] [PubMed]
- Alivernini, S.; Cingolani, A.; Gessi, M.; Paglionico, A.; Pasciuto, G.; Tolusso, B.; Fantoni, M.; Gremese, E. Comparative analysis of synovial inflammation after SARS-CoV-2 infection. Ann. Rheum. Dis. 2021, 80, e91. [Google Scholar] [CrossRef] [PubMed]


| Primer | Company | Sequence | Assay ID | Amplicon Length (bp) |
|---|---|---|---|---|
| GAPDH | ABI | * | Hs99999905_m1 | 93 |
| CD46 | ABI | * | Hs00387246_m1 | 94 |
| CD55 | ABI | * | Hs00167090_m1 | 62 |
| CD59 | ABI | * | Hs00174141_m1 | 70 |
| C3aR1 | ABI | * | Hs00269693_s1 | 82 |
| C5aR1 | ABI | * | Hs00704891_s1 | 68 |
| TNFα | ABI | * | Hs00174128_m1 | 80 |
| IL-6 | ABI | * | Hs00174131_m1 | 95 |
| Specificity and Species | Company | Catalogue Number | Stock Concentration | Used Dilution |
|---|---|---|---|---|
| Mouse anti-human C5aR1 | GeneTex, Eching, Germany | GTX74845 | 1 mg/mL | 1:50 |
| Goat anti-human CD55 | R&D systems, Minneapolis, MN, USA | AF2009 | 200 µg/mL | 1:50 |
| Rabbit anti-human IL-6 | Novus Biologicals, Centennial, CO, USA | NB600-1131 | + | 1:50 |
| Goat anti-human TNFα | Peprotech, Princeton, NJ, USA | 500-P31AG | 0.5 mg/mL | 1:50 |
| Mouse anti-human Ki67 | Chemicon International Inc., Temecula, CA, USA | MAB4190 | 1 mg/mL | 1:50 |
| Donkey anti-mouse- Alexa Fluor 488 | Invitrogen, Waltham, MA, USA | A21202 | 2 mg/mL | 1:200 |
| Donkey anti-goat- Alexa Fluor 488 | Life Technologies, Carlsbad, CA, USA | A11055 | 2 mg/mL | 1:200 |
| Donkey anti-rabbit- Alexa Fluor 555 | Life Technologies, Carlsbad, CA, USA | A31572 | 2 mg/mL | 1:200 |
| Donkey anti-goat- cyanine (Cy)3 | Dianova, Hamburg, Germany | 705-165-147 | 1.5 mg/mL | 1:200 |
| Donkey anti-mouse-Cy3 | Dianova, Hamburg, Germany | 715-166-150 | 1.5 mg/mL | 1:200 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Franke, V.; Meyer, S.; Schulze-Tanzil, G.G.; Braun, T.; Kokozidou, M.; Fischlein, T.; Silawal, S. Complement Regulation in Immortalized Fibroblast-like Synoviocytes and Primary Human Endothelial Cells in Response to SARS-CoV-2 Nucleocapsid Protein and Pro-Inflammatory Cytokine TNFα. Life 2022, 12, 1527. https://doi.org/10.3390/life12101527
Franke V, Meyer S, Schulze-Tanzil GG, Braun T, Kokozidou M, Fischlein T, Silawal S. Complement Regulation in Immortalized Fibroblast-like Synoviocytes and Primary Human Endothelial Cells in Response to SARS-CoV-2 Nucleocapsid Protein and Pro-Inflammatory Cytokine TNFα. Life. 2022; 12(10):1527. https://doi.org/10.3390/life12101527
Chicago/Turabian StyleFranke, Vincent, Sophie Meyer, Gundula Gesine Schulze-Tanzil, Tobias Braun, Maria Kokozidou, Theodor Fischlein, and Sandeep Silawal. 2022. "Complement Regulation in Immortalized Fibroblast-like Synoviocytes and Primary Human Endothelial Cells in Response to SARS-CoV-2 Nucleocapsid Protein and Pro-Inflammatory Cytokine TNFα" Life 12, no. 10: 1527. https://doi.org/10.3390/life12101527





