Integracides: Tetracyclic Triterpenoids from Fusarium sp.—Their 5-Lipoxygenase Inhibitory Potential and Structure–Activity Relation Using In Vitro and Molecular Docking Studies
Abstract
1. Introduction
2. Results
2.1. Purification of Metabolites
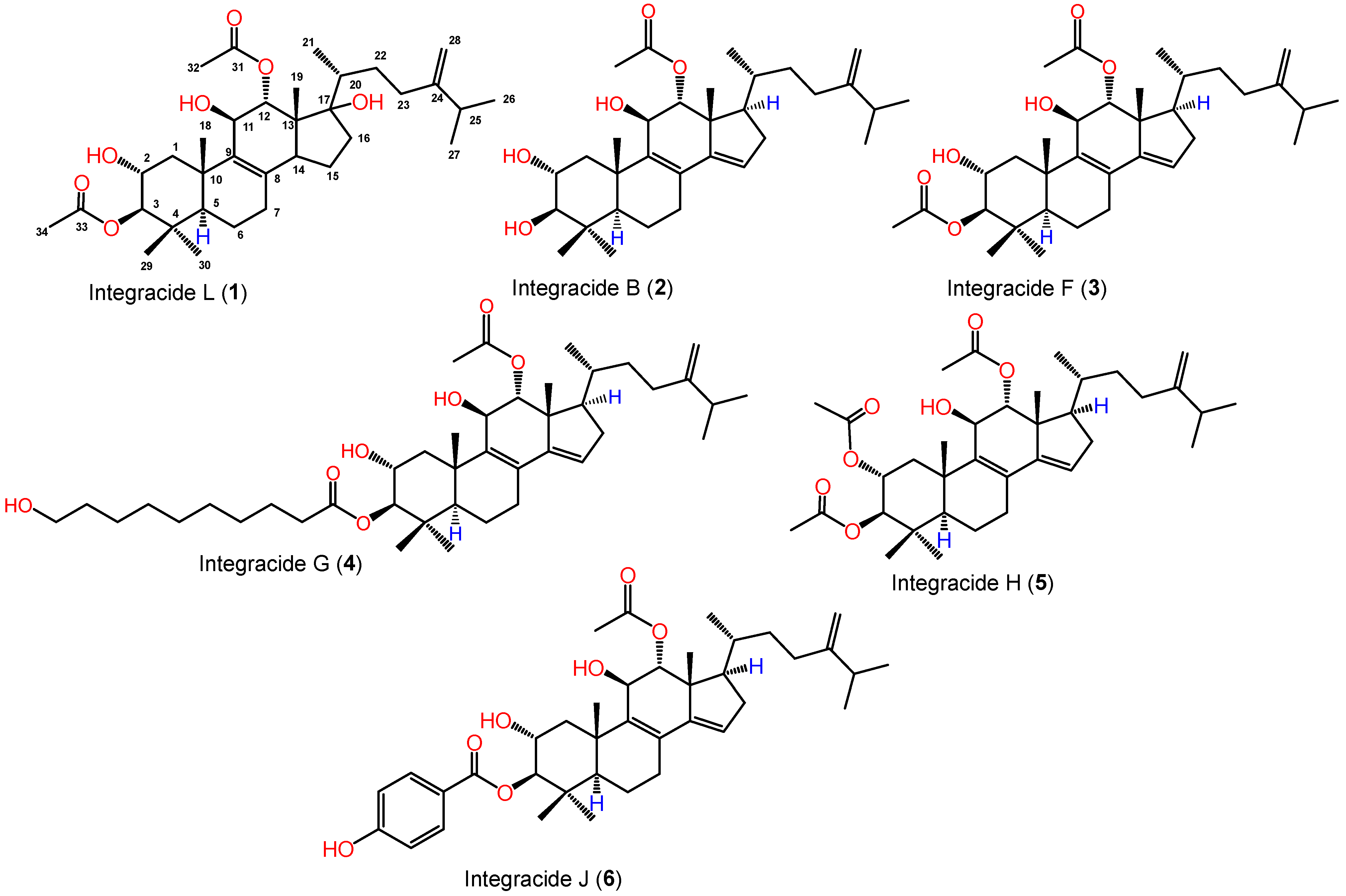
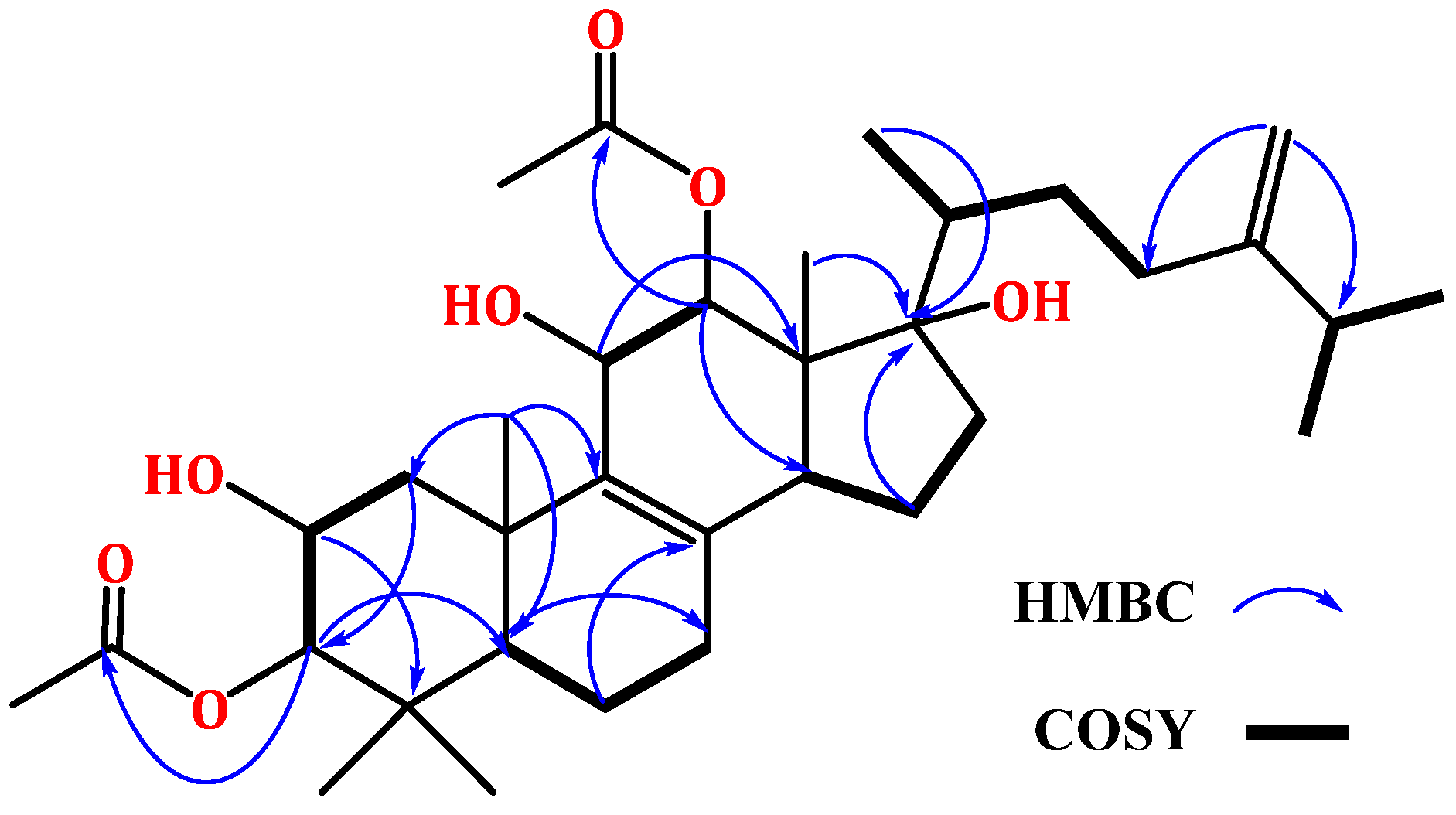
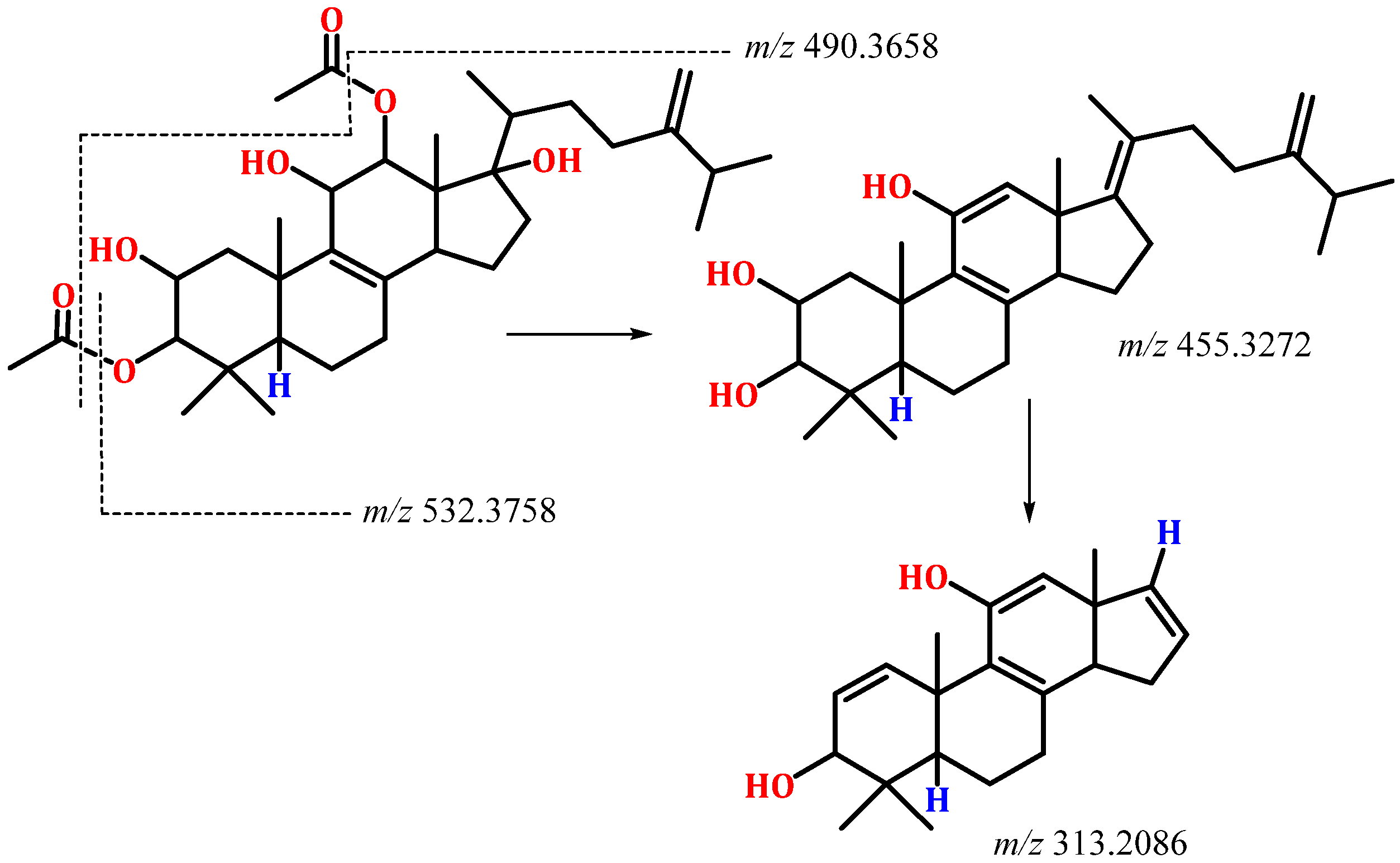
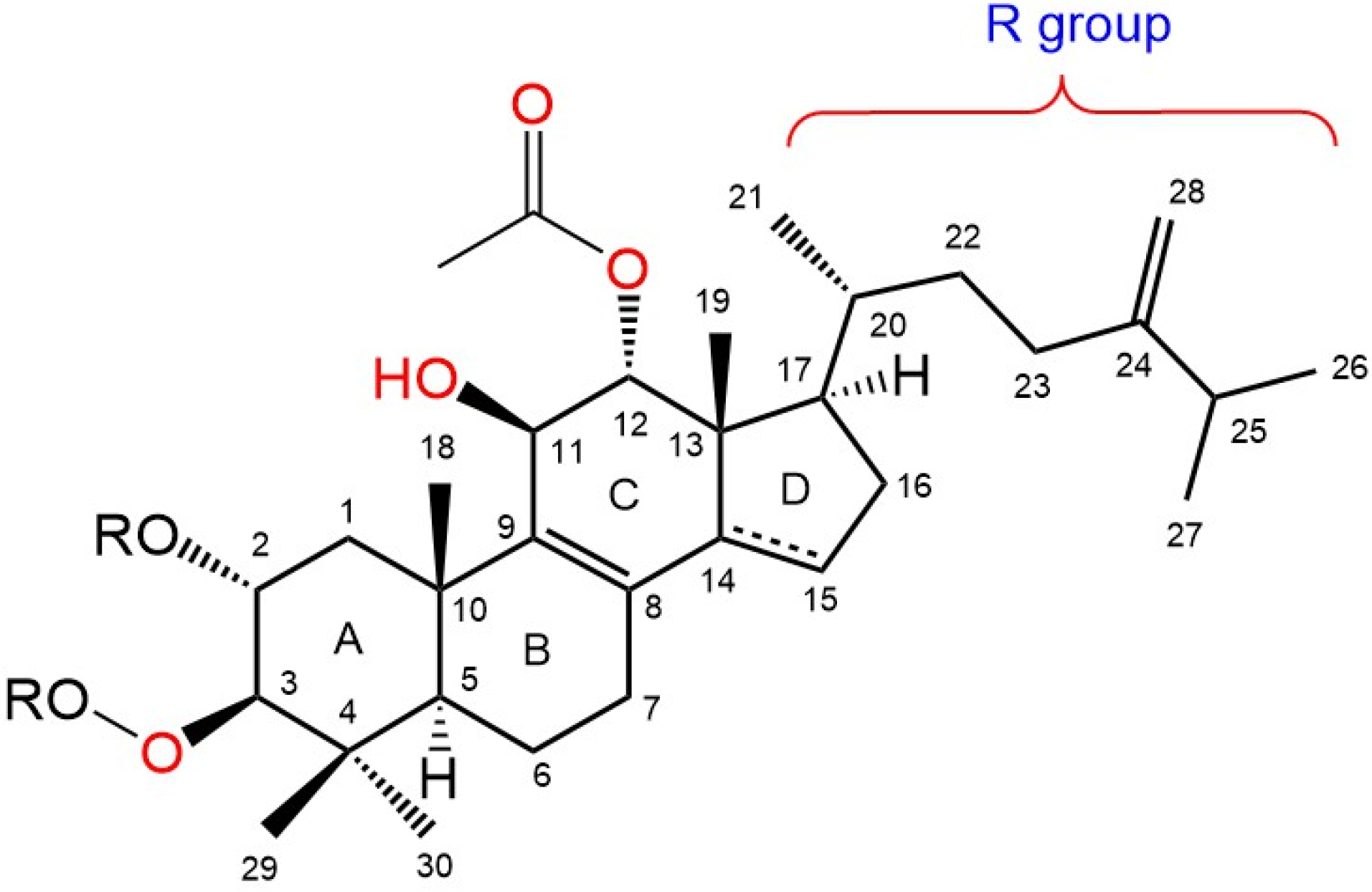
2.2. Structural Elucidation of Integracide L
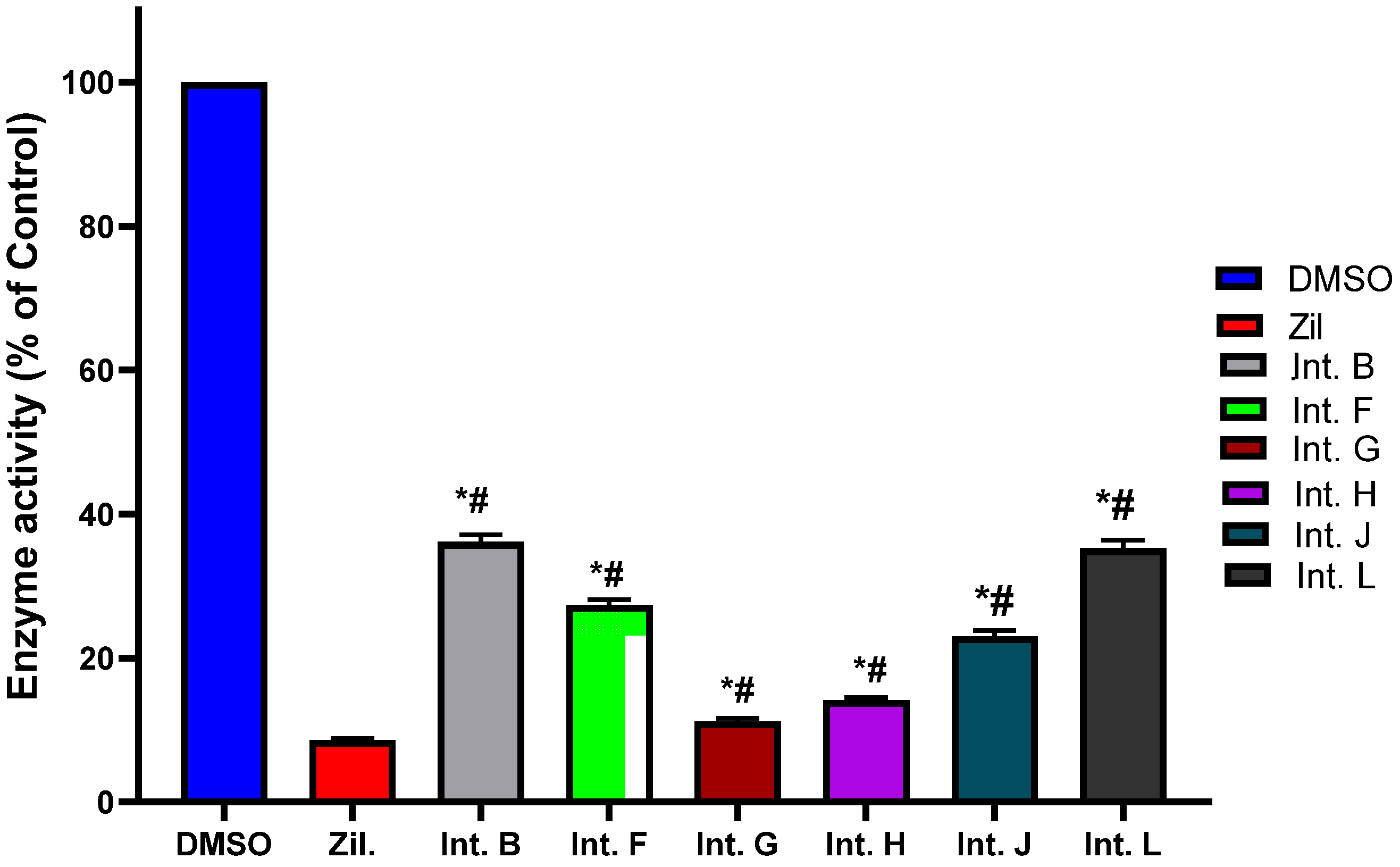
2.3. 5-LOX Inhibitory Activity
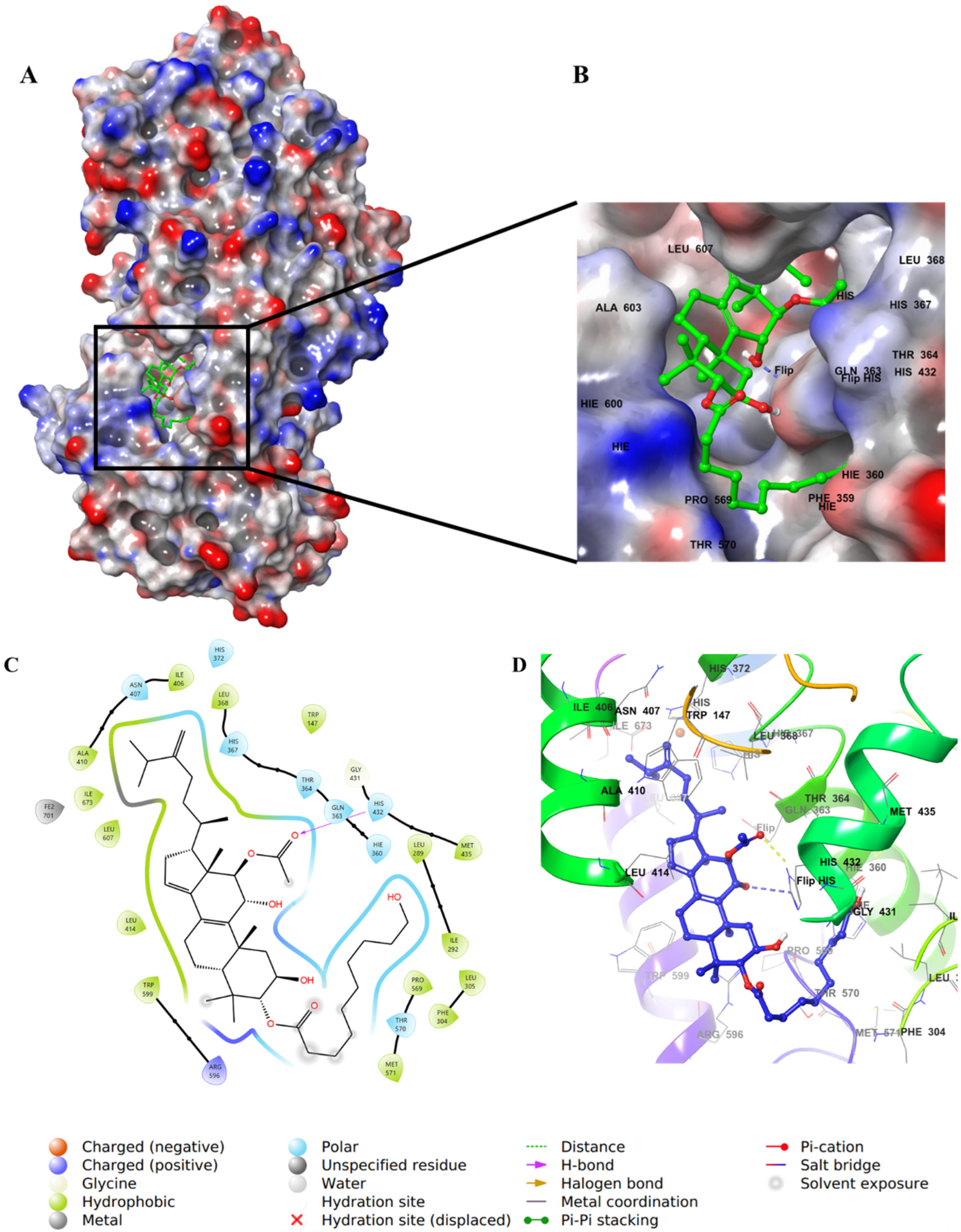
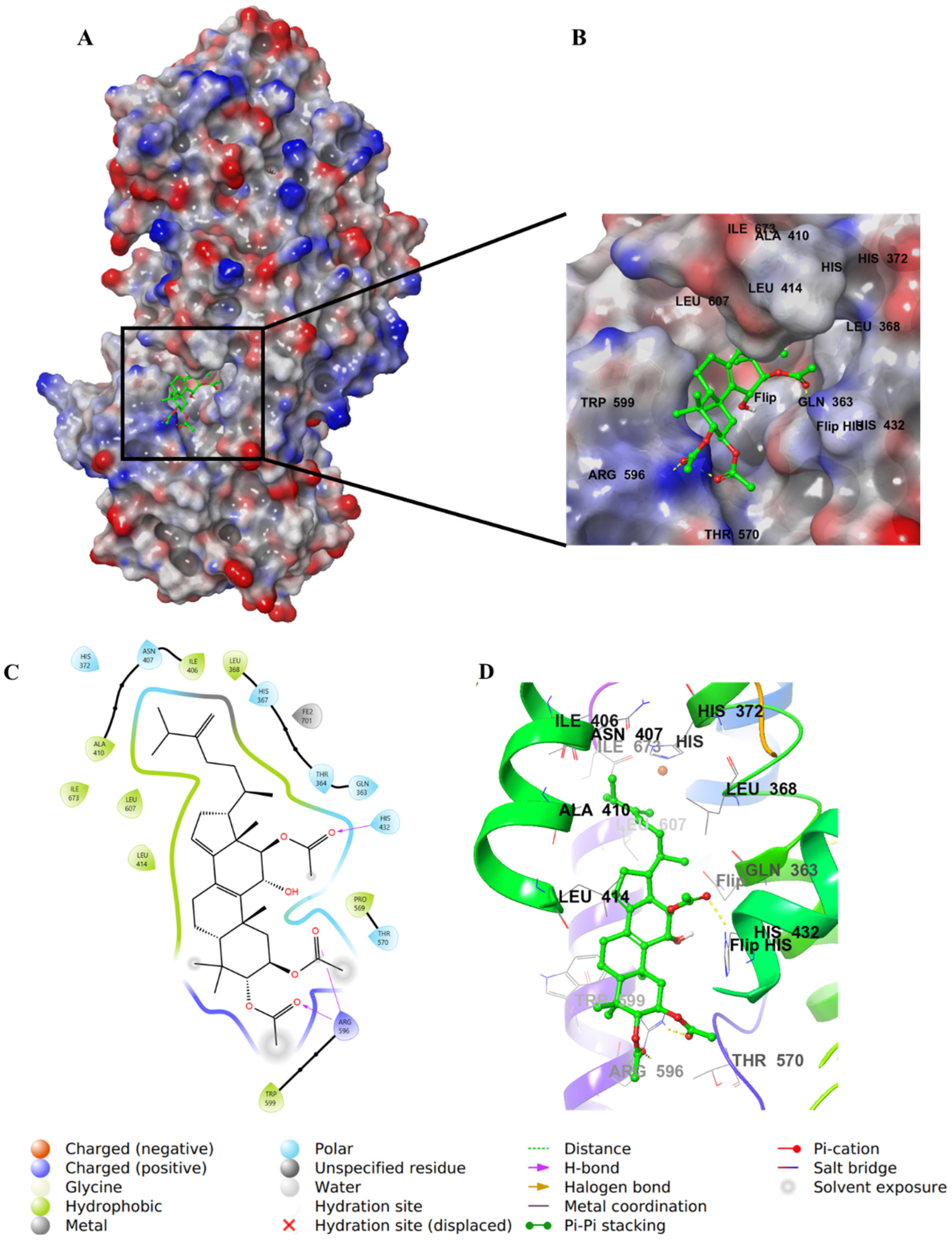
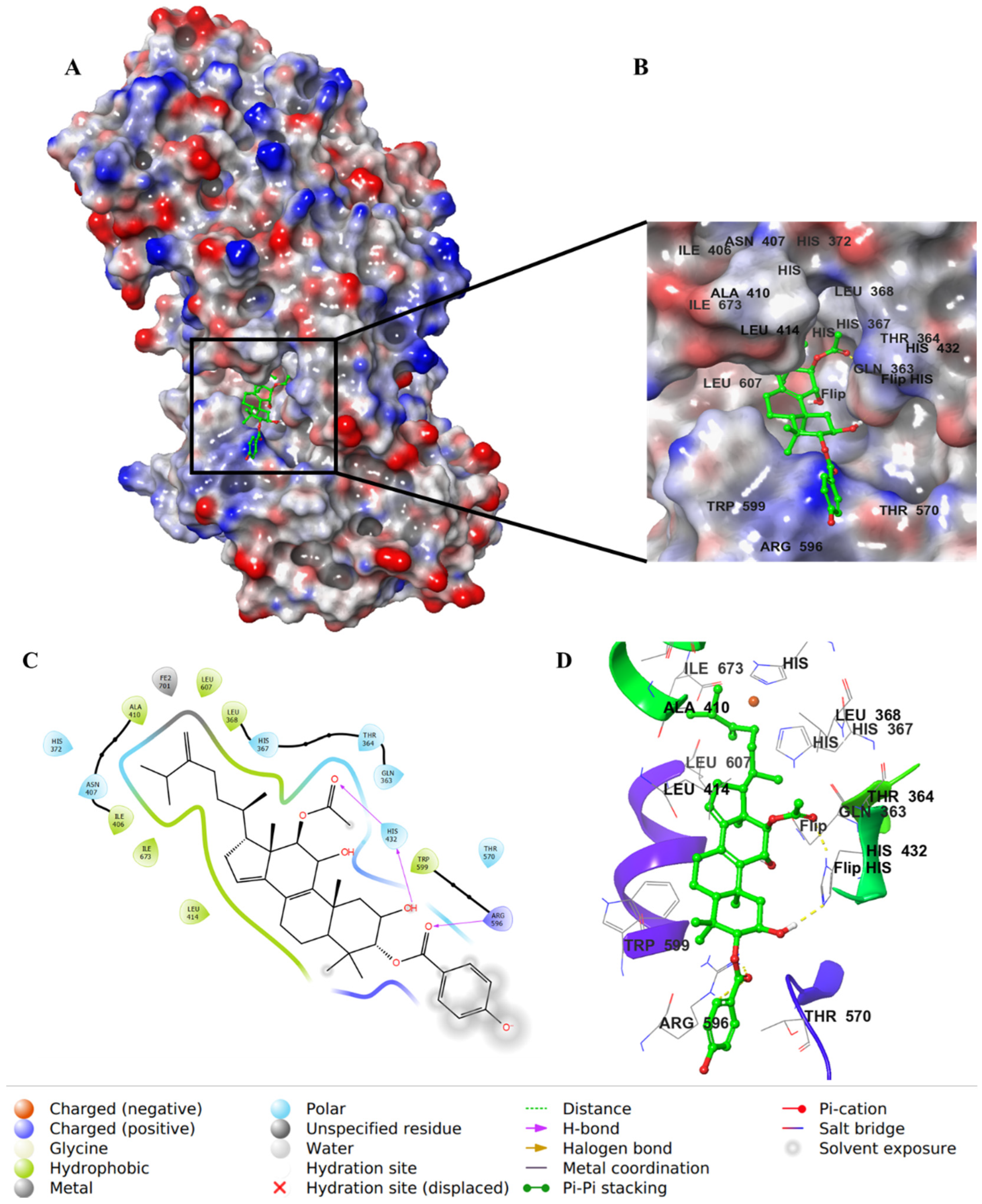
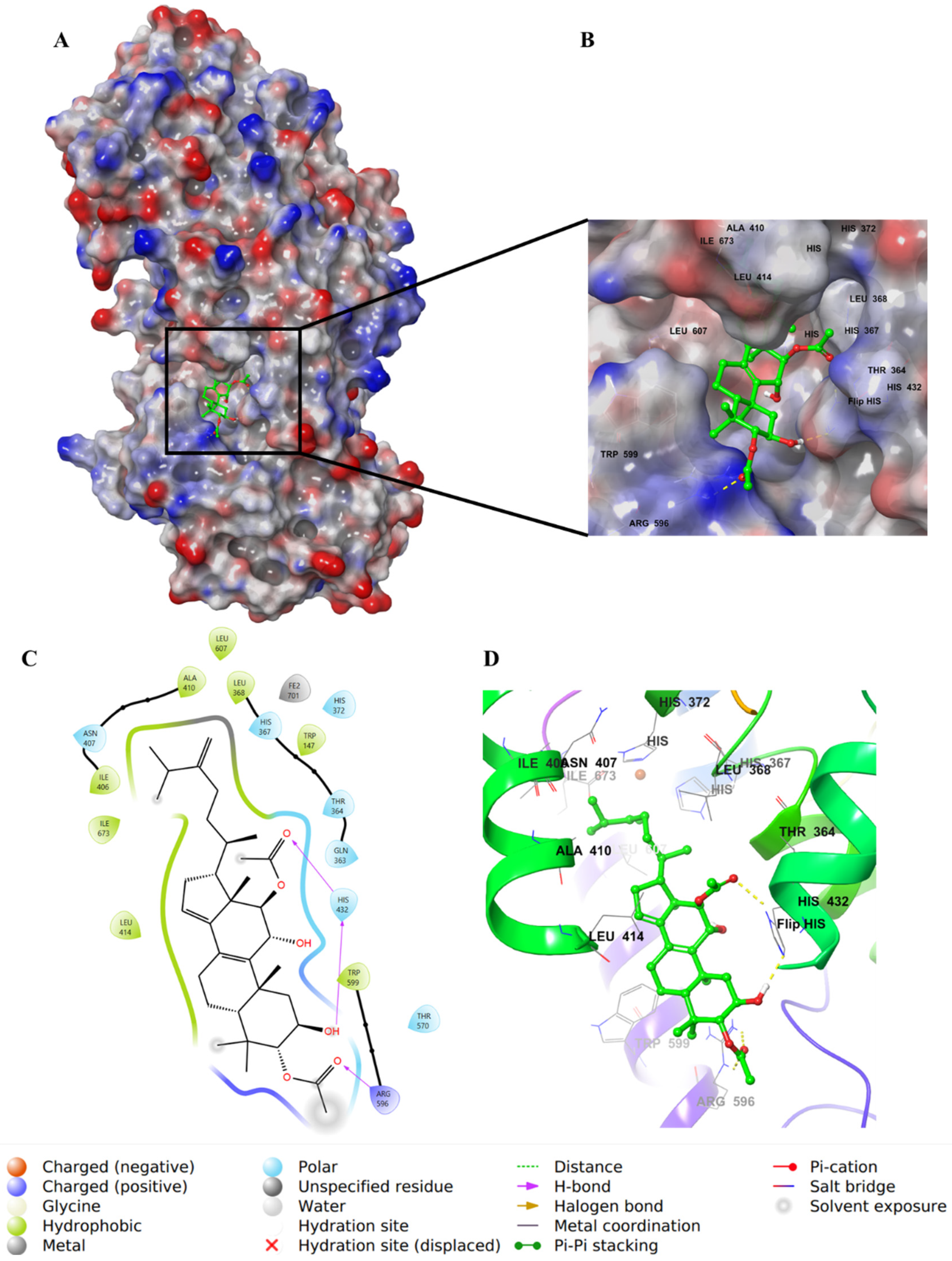
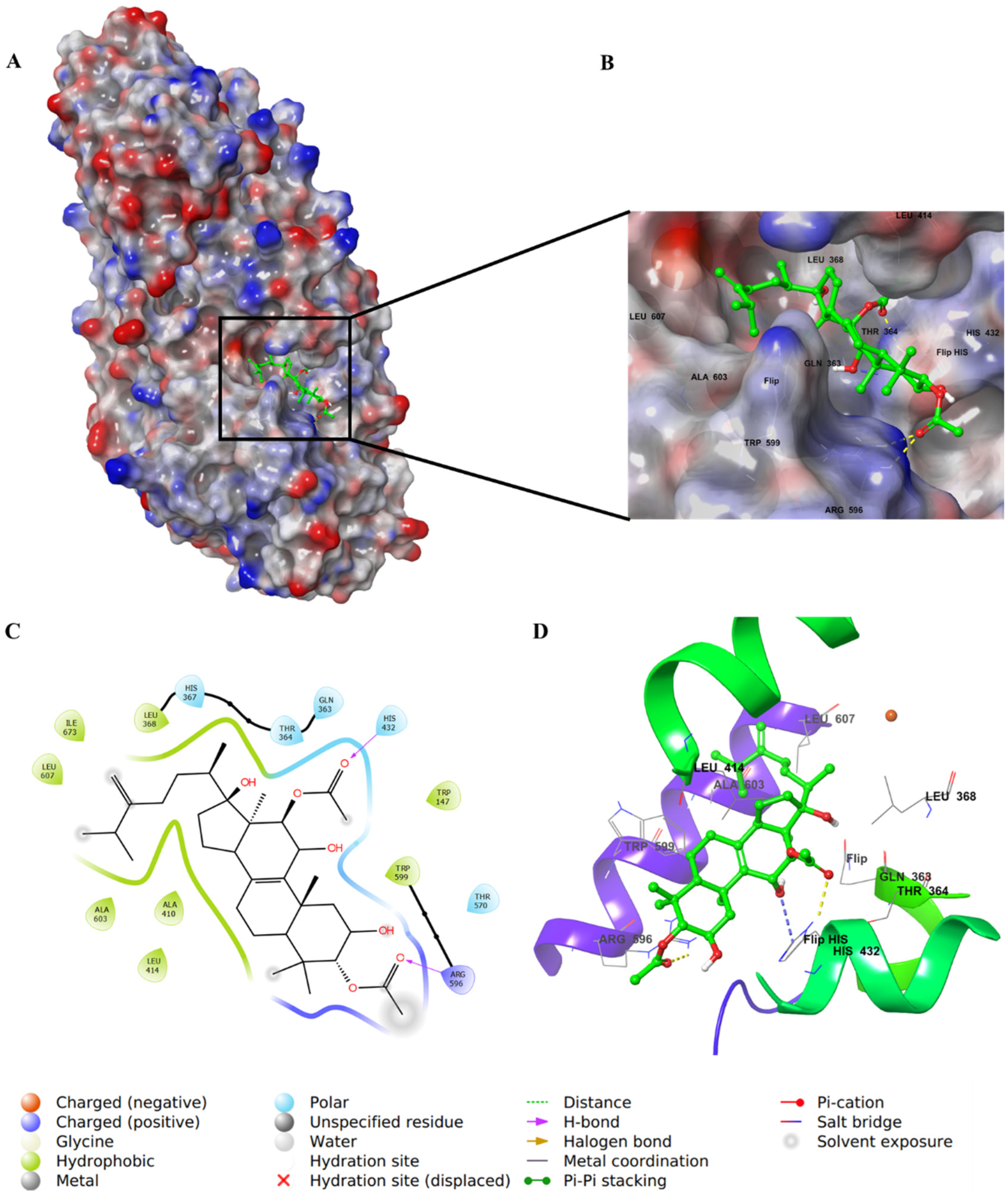
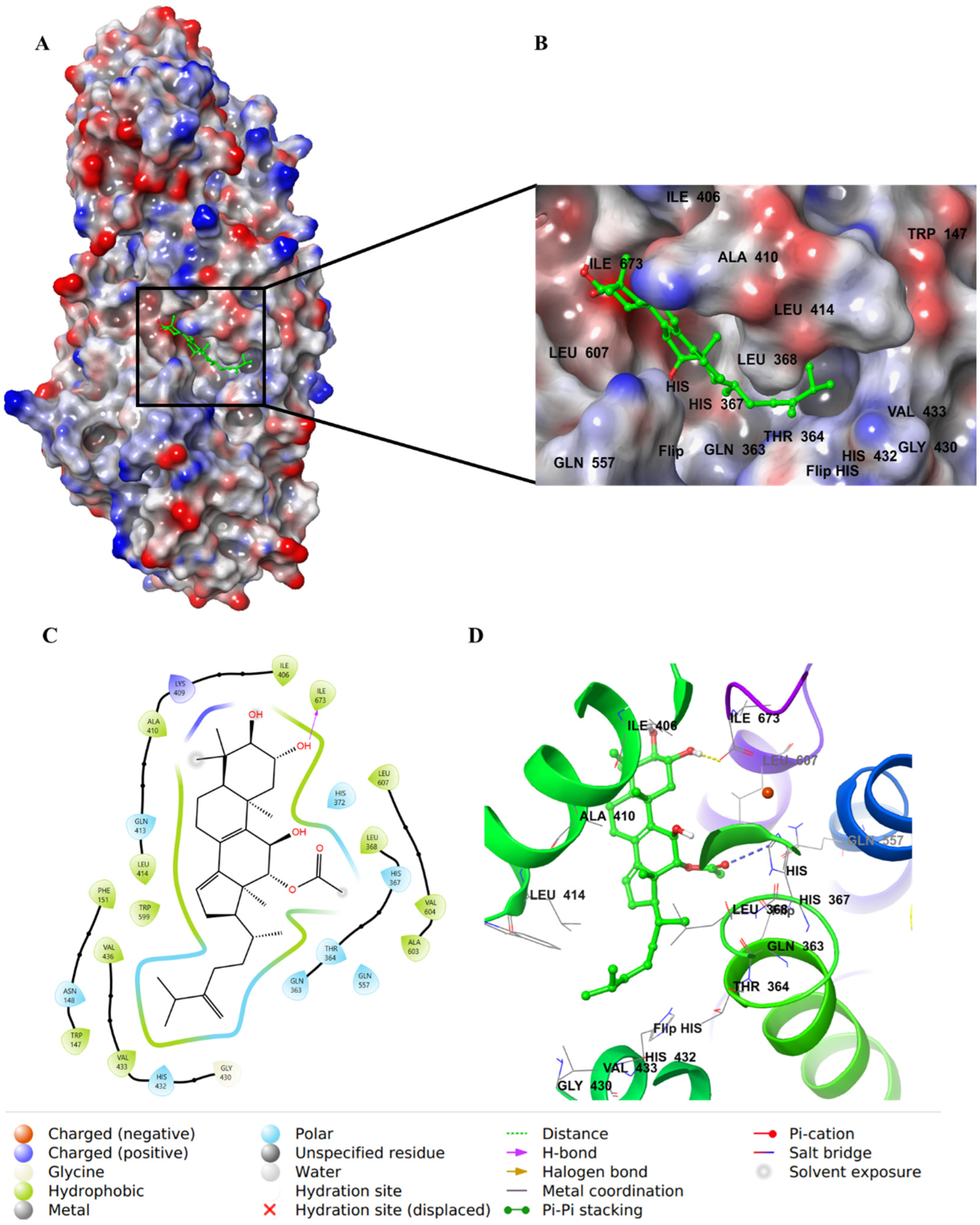
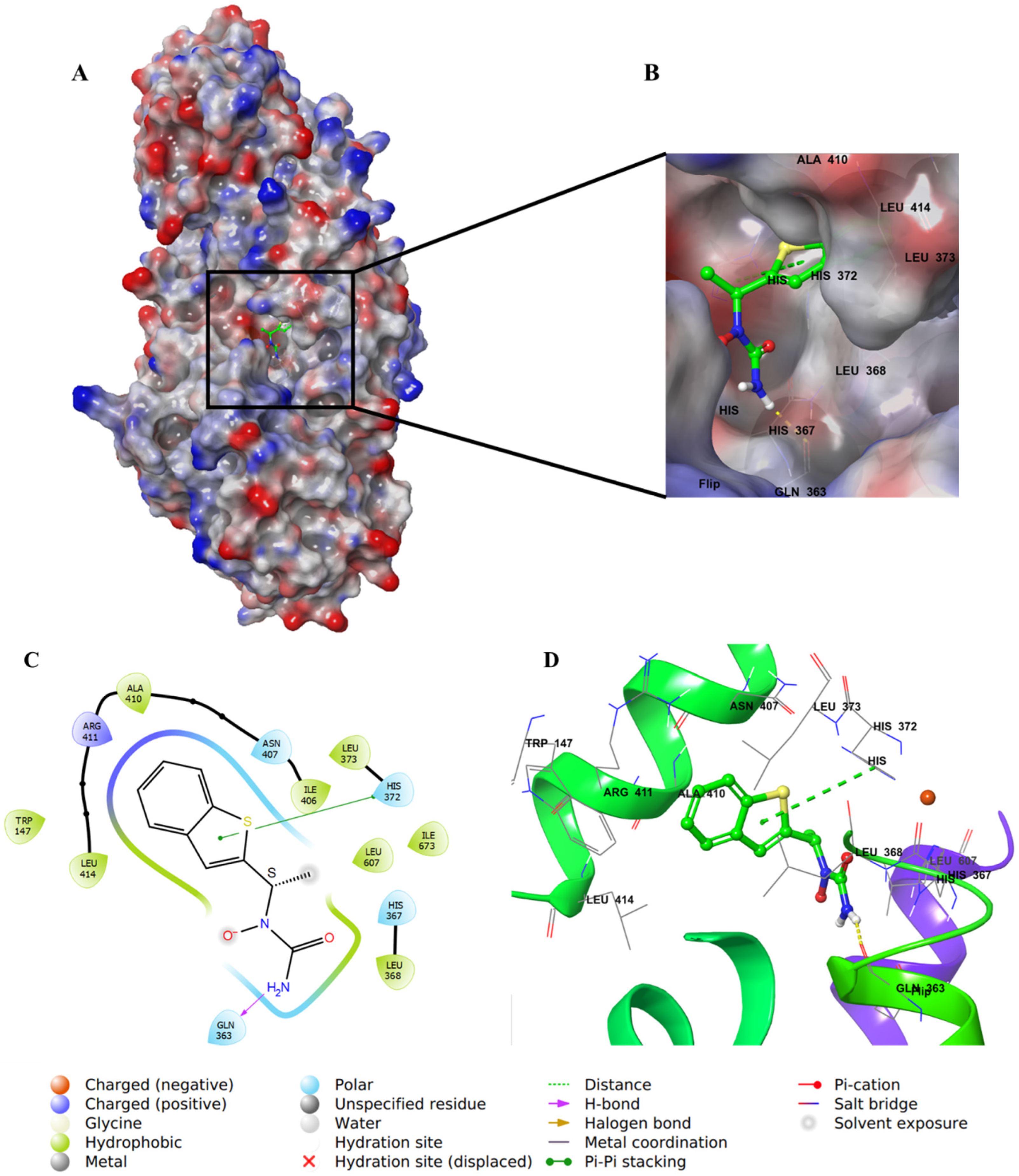
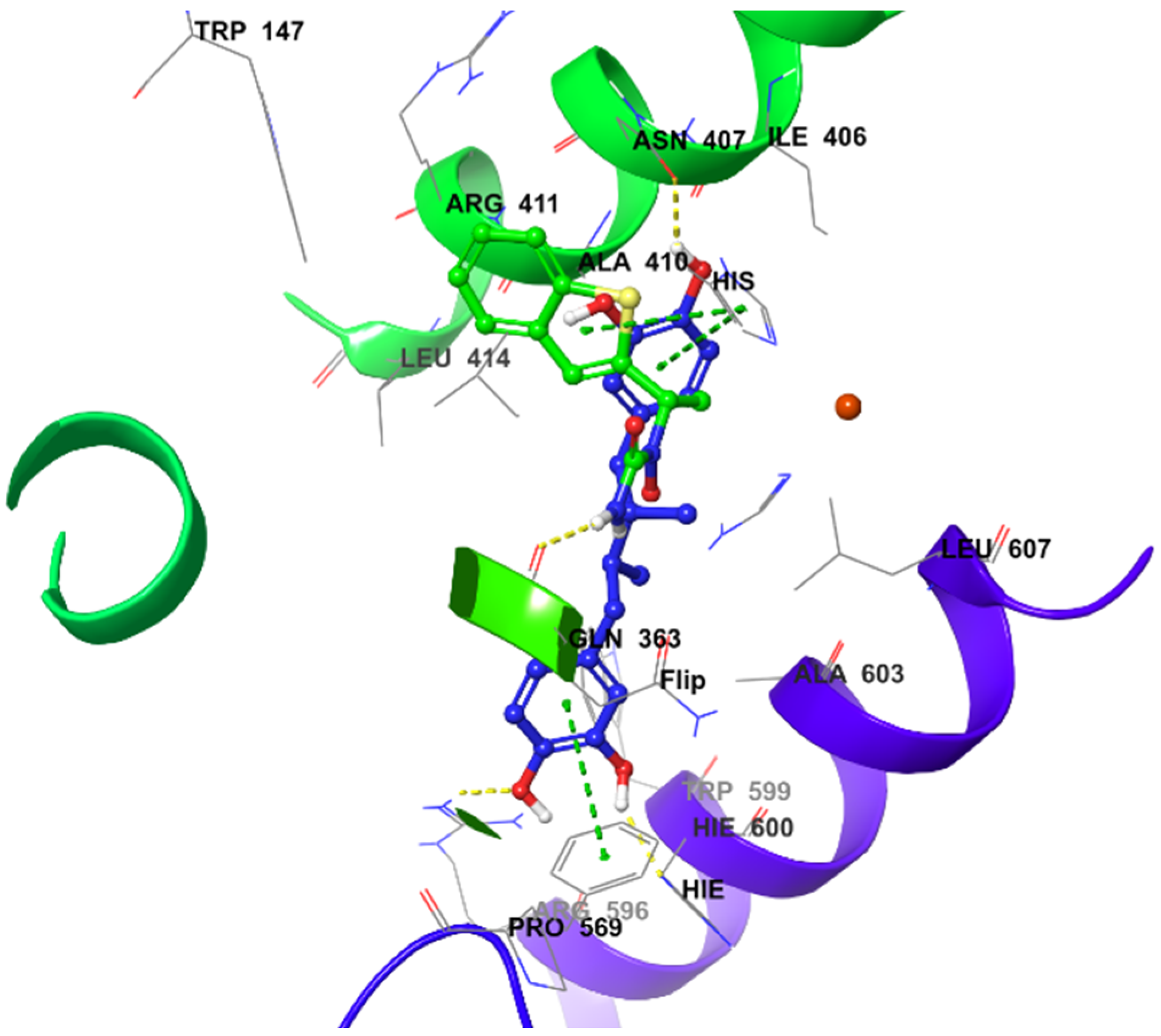
2.4. Molecular Docking Studies
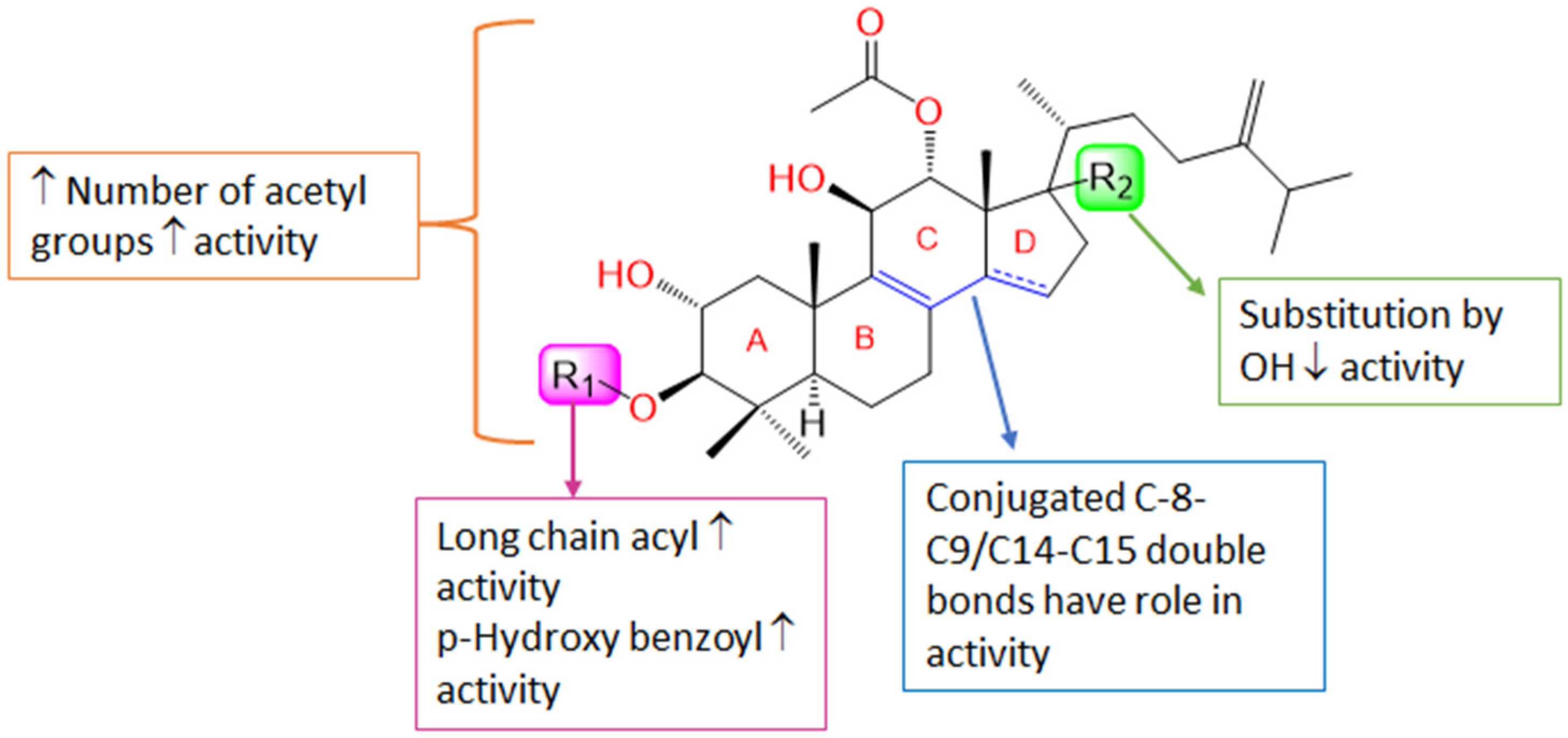
3. Discussion
4. Material and Methods
4.1. General Experimental Procedures
4.2. Cultivation of Fungal Material
4.3. Extraction and Isolation
NMR Spectral Data of Integracide L (1)
4.4. 5-Lipoxygenase Inhibitory Assay
4.5. Statistical Analysis
4.6. Molecular Docking Study
4.6.1. Ligand and Protein Preparation
4.6.2. Grid Generation and Molecular Docking
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- White, M. Mediators of Inflammation and the Inflammatory Process. J. Allergy Clin. Immunol. 1999, 103, S378–S381. [Google Scholar] [CrossRef] [PubMed]
- Sinha, S.; Doble, M.; Manju, S.L. 5-Lipoxygenase as a drug target: A review on trends in inhibitors structural design, SAR and mechanism based approach. Bioorg. Med. Chem. 2019, 27, 3745–3759. [Google Scholar] [CrossRef] [PubMed]
- Al-Attas, A.; El-Shaer, N.S.; Mohamed, G.A.; Ibrahim, S.; Esmat, A. New Anti-Inflammatory Sesquiterpenes from the Rhizomes of Costus speciosus. J. Ethnopharmacol. 2015, 176, 365–374. [Google Scholar] [CrossRef] [PubMed]
- Lewis, R.A.; Austen, K.F.; Soberman, R.J. Leukotrienes and Other Products of the 5-Lipoxygenase Pathway: Biochemistry and Relation to Pathobiology in Human Diseases. N. Engl. J. Med. 1990, 323, 645–655. [Google Scholar]
- Henderson, W.R. The Role of Leukotrienes in Inflammation. Ann. Intern. Med. 1994, 121, 684–697. [Google Scholar] [CrossRef] [PubMed]
- Su, C.; Oliw, E.H. Manganese Lipoxygenase: Purification and Characterization. J. Biol. Chem. 1998, 273, 13072–13079. [Google Scholar] [CrossRef] [PubMed]
- Steele, V.E.; Holmes, C.A.; Hawk, E.T.; Kopelovich, L.; Lubet, R.A.; Crowell, J.A.; Sigman, C.C.; Kelloff, G.J. Lipoxygenase Inhibitors as Potential Cancer Chemopreventives. Cancer Epidemiol. Biomark. Prev. 1999, 8, 467–483. [Google Scholar]
- Charlier, C.; Michaux, C. Dual Inhibition of Cyclooxygenase-2 (COX-2) and 5-Lipoxygenase (5-LOX) as a New Strategy to Provide Safer Non-Steroidal Anti-Inflammatory Drugs. Eur. J. Med. Chem. 2003, 38, 645–659. [Google Scholar] [CrossRef]
- Misra, S.; Ghatak, S.; Patil, N.; Dandawate, P.; Ambike, V.; Adsule, S.; Unni, D.; Swamy, K.V.; Padhye, S. Novel Dual Cyclooxygenase and Lipoxygenase Inhibitors Targeting hyaluronan–CD44v6 Pathway and Inducing Cytotoxicity in Colon Cancer Cells. Bioorg. Med. Chem. 2013, 21, 2551–2559. [Google Scholar] [CrossRef][Green Version]
- Tomy, M.J.; Sharanya, C.S.; Dileep, K.V.; Prasanth, S.; Sabu, A.; Sadasivan, C.; Haridas, M. Derivatives Form Better Lipoxygenase Inhibitors than Piperine: In Vitro and in Silico Study. Chem. Biol. Drug Des. 2015, 85, 715–721. [Google Scholar] [CrossRef]
- Shankar, A.; Sharma, K.K. Fungal secondary metabolites in food and pharmaceuticals in the era of multi-omics. Appl. Microbiol. Biotechnol. 2022, 106, 3465–3488. [Google Scholar] [CrossRef] [PubMed]
- Manganyi, M.C.; Ateba, C.N. Untapped Potentials of Endophytic Fungi: A Review of Novel Bioactive Compounds with Biological Applications. Microorganisms 2020, 8, 1934. [Google Scholar] [CrossRef]
- Slusarczyk, J.; Adamska, E.; Czerwik-Marcinkowska, J. Fungi and Algae as Sources of Medicinal and Other Biologically Active Compounds: A Review. Nutrients 2021, 13, 3178. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, G.A.; Ibrahim, S.R.M. Untapped Potential of Marine-Associated Cladosporium Species: An Overview on Secondary Metabolites, Biotechnological Relevance, and Biological Activities. Mar. Drugs 2021, 19, 645. [Google Scholar] [CrossRef] [PubMed]
- Adebayo, E.A.; Azeez, M.A.; Alao, M.B.; Oke, A.M.; Aina, D.A. Fungi as veritable tool in current advances in nanobiotechnology. Heliyon 2021, 7, e08480. [Google Scholar] [CrossRef]
- Mohamed, A.G.T. Stachybotrys chartarum: A Novel Biological Agent for the Extracellular Synthesis of Silver Nanoparticles and their Antimicrobial Activity. Indones. J. Biotechnol. 2013, 18, 75–82. [Google Scholar] [CrossRef]
- Macías-Rubalcava, M.L.; Sánchez-Fernández, R.E. Secondary metabolites of endophytic Xylaria species with potential applications in medicine and agriculture. World J. Microbiol. Biotechnol. 2017, 33, 15. [Google Scholar] [CrossRef]
- Yu, H.; Zhang, L.; Li, L.; Zheng, C.; Guo, L.; Li, W.; Sun, P.; Qin, L. Recent developments and future prospects of antimicrobial metabolites produced by endophytes. Microbiol. Res. 2010, 165, 437–449. [Google Scholar] [CrossRef]
- Kaul, S.; Gupta, S.; Ahmed, M.; Dhar, M.K. Endophytic fungi from medicinal plants: A treasure hunt for bioactive metabolites. Phytochem. Rev. 2012, 11, 487–505. [Google Scholar] [CrossRef]
- Ibrahim, S.R.; Mohamed, G.A.; Ross, S.A. Integracides F and G: New Tetracyclic Triterpenoids from the Endophytic Fungus Fusarium Sp. Phytochem. Lett. 2016, 15, 125–130. [Google Scholar] [CrossRef]
- Adeleke, B.S.; Babalola, O.O. The plant endosphere-hidden treasures: A review of fungal endophytes. Biotechnol. Genet. Eng. Rev. 2021, 37, 154–177. [Google Scholar] [CrossRef] [PubMed]
- De Souza, J.J.; Vieira, I.J.C.; Rodrigues-Filho, E.; Braz-Filho, R. Terpenoids from Endophytic Fungi. Molecules 2011, 16, 10604–10618. [Google Scholar] [CrossRef] [PubMed]
- Noor, A.O.; Almasri, D.M.; Bagalagel, A.A.; Abdallah, H.M.; Mohamed, S.G.A.; Mohamed, G.A.; Ibrahim, S.R.M. Naturally occurring isocoumarins derivatives from endophytic fungi: Sources, isolation, structural characterization, biosynthesis, and biological activities. Molecules 2020, 25, 395. [Google Scholar] [CrossRef]
- Zhang, H.W.; Song, Y.C.; Tan, R.X. Biology and Chemistry of Endophytes. Nat. Prod. Rep. 2006, 23, 753–771. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Zhao, J.; Xu, L.; Huang, Y.; Ma, Z.; Wang, J.; Jiang, W. Antimicrobial Compounds Produced by Plant Endophytic Fungi. Fungicides: Chemistry, Environmental Impact and Health Effects; De Costa, P., Bezerra, P., Eds.; Nova Science Publishers: New York, NY, USA, 2009; pp. 91–119. [Google Scholar]
- Parthasarathy, R.; Chandrika, M.; Rao, H.Y.; Kamalraj, S.; Jayabaskaran, C.; Pugazhendhi, A. Molecular Profiling of Marine Endophytic Fungi from Green Algae: Assessment of Antibacterial and Anticancer Activities. Process Biochem. 2020, 96, 11–20. [Google Scholar] [CrossRef]
- Preethi, K.; Manon Mani, V.; Lavanya, N. Endophytic fungi: A potential source of bioactive compounds for commercial and therapeutic applications. In Endophytes; Patil, R.H., Maheshwari, V.L., Eds.; Springer: Singapore, 2021; pp. 247–272. [Google Scholar]
- Rai, N.; Kumari Keshri, P.; Verma, A.; Kamble, S.C.; Mishra, P.; Barik, S.; Kumar Singh, S.; Gautam, V. Plant Associated Fungal Endophytes as a Source of Natural Bioactive Compounds. Mycology 2021, 12, 139–159. [Google Scholar] [CrossRef]
- Summerell, B.A.; Salleh, B.; Leslie, J.F. A Utilitarian Approach to Identifying Fusarium Species. Plant Dis. 2003, 87, 117–128. [Google Scholar] [CrossRef]
- Ibrahim, S.R.; Abdallah, H.M.; Mohamed, G.A.; Ross, S.A. Integracides H-J: New Tetracyclic Triterpenoids from the Endophytic Fungus Fusarium Sp. Fitoterapia 2016, 112, 161–167. [Google Scholar] [CrossRef]
- Singh, S.B.; Ondeyka, J.G.; Schleif, W.A.; Felock, P.; Hazuda, D.J. Chemistry and Structure—Activity Relationship of HIV-1 Integrase Inhibitor Integracide B and Related Natural Products. J. Nat. Prod. 2003, 66, 1338–1344. [Google Scholar] [CrossRef]
- Singh, S.B.; Zink, D.L.; Dombrowski, A.W.; Polishook, J.D.; Ondeyka, J.G.; Hirshfield, J.; Felock, P.; Hazuda, D.J. Integracides: Tetracyclic Triterpenoid Inhibitors of HIV-1 Integrase Produced by Fusarium Sp. Bioorg. Med. Chem. 2003, 11, 1577–1582. [Google Scholar] [CrossRef]
- Singh, S.B. A New Mild PTSA-Catalyzed Method for Sulfate Ester Hydrolysis and Acid-Catalyzed Rearrangement of 12-Acetyl-Diene-11-Ol Tetracyclic Triterpenoids Involving an Angular Methyl Migration. Tetrahedron Lett. 2000, 41, 6973–6976. [Google Scholar] [CrossRef]
- Brill, G.M.; Kati, W.M.; Montgomery, D.; Karwowski, J.P.; Humphrey, P.E.; Jackson, M.; Clement, J.J.; Kadam, S.; Chen, R.H.; McAlpine, J.B. Novel Triterpene Sulfates from Fusarium compactum using a Rhinovirus 3C Protease Inhibitor Screen. J. Antibiot. 1996, 49, 541–546. [Google Scholar] [CrossRef] [PubMed]
- Tabata, N.; Tomoda, H.; Yamaguchi, Y.; Masuma, R.; BAMBERGER, M.J.; Omura, S. Inhibition of Cholesteryl Ester Transfer Protein by Fungal Metabolites, L681, 512. J. Antibiot. 1999, 52, 1042–1045. [Google Scholar] [CrossRef][Green Version]
- Olsson, M.H.; Søndergaard, C.R.; Rostkowski, M.; Jensen, J.H. PROPKA3: Consistent Treatment of Internal and Surface Residues in Empirical P K a Predictions. J. Chem. Theory Comput. 2011, 7, 525–537. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, N.C.; Gerstmeier, J.; Schexnaydre, E.E.; Börner, F.; Garscha, U.; Neau, D.B.; Werz, O.; Newcomer, M.E. Structural and Mechanistic Insights into 5-Lipoxygenase Inhibition by Natural Products. Nat. Chem. Biol. 2020, 16, 783–790. [Google Scholar] [CrossRef] [PubMed]
- Madhavi Sastry, G.; Adzhigirey, M.; Day, T.; Annabhimoju, R.; Sherman, W. Protein and Ligand Preparation: Parameters, Protocols, and Influence on Virtual Screening Enrichments. J. Comput. Aided Mol. Des. 2013, 27, 221–234. [Google Scholar] [CrossRef]
- Friesner, R.A.; Banks, J.L.; Murphy, R.B.; Halgren, T.A.; Klicic, J.J.; Mainz, D.T.; Repasky, M.P.; Knoll, E.H.; Shelley, M.; Perry, J.K. Glide: A New Approach for Rapid, Accurate Docking and Scoring. 1. Method and Assessment of Docking Accuracy. J. Med. Chem. 2004, 47, 1739–1749. [Google Scholar] [CrossRef]
- Schrödinger Release 2021-4: Glide; Schrödinger, LLC: New York, NY, USA, 2021.
- Friesner, R.A.; Murphy, R.B.; Repasky, M.P.; Frye, L.L.; Greenwood, J.R.; Halgren, T.A.; Sanschagrin, P.C.; Mainz, D.T. Extra Precision Glide: Docking and Scoring Incorporating a Model of Hydrophobic Enclosure for Protein—Ligand Complexes. J. Med. Chem. 2006, 49, 6177–6196. [Google Scholar] [CrossRef]
- Neau, D.B.; Bender, G.; Boeglin, W.E.; Bartlett, S.G.; Brash, A.R.; Newcomer, M.E. Crystal Structure of a Lipoxygenase in Complex with Substrate. J. Biol. Chem. 2014, 289, 31905–31913. [Google Scholar] [CrossRef]
- Ge, J.; Liu, Z.; Zhong, Z.; Wang, L.; Zhuo, X.; Li, J.; Jiang, X.; Ye, X.; Xie, T.; Bai, R. Natural Terpenoids with Anti-Inflammatory Activities: Potential Leads for Anti-Inflammatory Drug Discovery. Bioorg. Chem. 2022, 124, 105817. [Google Scholar] [CrossRef]
- Xu, J.; Yi, M.; Ding, L.; He, S. A Review of Anti-Inflammatory Compounds from Marine Fungi, 2000–2018. Mar. Drugs 2019, 17, 636. [Google Scholar] [CrossRef] [PubMed]
- Antony, T.; Chakraborty, K. Anti-inflammatory polyether triterpenoids from the marine macroalga Gracilaria salicornia: Newly described natural leads attenuate pro-inflammatory 5-lipoxygenase and cyclooxygenase-2. Algal Res. 2020, 47, 101791. [Google Scholar] [CrossRef]
- Koeberle, A.; Henkel, A.; Verhoff, M.; Tausch, L.; König, S.; Fischer, D.; Kather, N.; Seitz, S.; Paul, M.; Jauch, J.; et al. Triterpene acids from frankincense and semi-synthetic derivatives that inhibit 5-lipoxygenase and cathepsin G. Molecules 2018, 23, 506. [Google Scholar] [CrossRef] [PubMed]
- Braca, A.; Dal Piaz, F.; Marzocco, S.; Vassallo, A.; De Tommasi, N. Triterpene derivatives as inhibitors of protein involved in the inflammatory process: Molecules interfering with phospholipase A2, cycloxygenase, and lipoxygenase. Curr. Drug Targets 2011, 12, 302–321. [Google Scholar] [CrossRef]
- Mbarik, M.; Poirier, S.J.; Doiron, J.; Selka, A.; Barnett, D.A.; Cormier, M.; Touaibia, M.; Surette, M.E. Phenolic Acid Phenethylesters and their Corresponding Ketones: Inhibition of 5-lipoxygenase and Stability in Human Blood and HepaRG Cells. Pharmacol. Res. Perspect. 2019, 7, e00524. [Google Scholar] [CrossRef]
- Ibrahim, S.R.M.; Mohamed, G.A.; Khayat, M.T.A.; Zayed, M.F.; El-Kholy, A.A.S. Anti-Inflammatory Terpenoids from Cyperus rotundus Rhizomes. Pak. J. Pharm. Sci. 2018, 31 (Suppl. 4), 1449–1456. [Google Scholar]
- Carter, G.W.; Young, P.R.; Albert, D.H.; Bouska, J.; Dyer, R.; Bell, R.L.; Summers, J.B.; Brooks, D.W. 5-Lipoxygenase Inhibitory Activity of Zileuton. J. Pharmacol. Exp. Ther. 1991, 256, 929–937. [Google Scholar]
- Mohamed, G.A. Tagenols A and B: New Lipoxygenase Inhibitor Flavonols from Tagetes minuta. Phytochem. Lett. 2016, 16, 141–145. [Google Scholar] [CrossRef]
- Noreen, Y.; Ringbom, T.; Perera, P.; Danielson, H.; Bohlin, L. Development of a Radiochemical Cyclooxygenase-1 and-2 in Vitro Assay for Identification of Natural Products as Inhibitors of Prostaglandin Biosynthesis. J. Nat. Prod. 1998, 61, 2–7. [Google Scholar] [CrossRef]
- Schrödinger Release 2021-4: LigPrep; Schrödinger, LLC: New York, NY, USA, 2021.

| No. | δH [mult., J (Hz)] | δC (mult.) | HMBC |
|---|---|---|---|
| 1 | 2.65 m 1.11 m | 42.6 CH2 | 2, 3, 5 |
| 2 | 3.73 dd (10.3, 4.2) | 66.8 CH | 1, 3, 5 |
| 3 | 3.62 d (10.3) | 88.0 CH | 2, 4, 29, 30, 33 |
| 4 | - | 39.2 C | - |
| 5 | 1.03 dd (12.8, 3.4) | 51.1 CH | 3, 4, 7, 10, 18 |
| 6 | 1.66 m 1.52 m | 18.0 CH2 | 5, 7, 8 |
| 7 | 2.21 m 1.46 m | 26.8 CH2 | 5, 8, 9 |
| 8 | - | 127.4 C | - |
| 9 | - | 143.5 C | - |
| 10 | - | 38.5 C | - |
| 11 | 4.25 brs | 67.9 CH | 8, 9, 12 |
| 12 | 5.00 d (3.4) | 78.3 CH | 9, 11, 13, 14, 19, 31 |
| 13 | - | 47.6 C | - |
| 14 | 2.06 t (6.2) | 39.8 CH | - |
| 15 | 1.68 m 1.45 m | 31.3 CH2 | 8, 13, 14, 17 |
| 16 | 2.28 m 2.18 m | 33.5 CH2 | 14, 15, 17 |
| 17 | - | 86.0 C | 13, 16, 22 |
| 18 | 1.20 s | 22.1 CH3 | 1, 5, 9, 10 |
| 19 | 0.61 s | 13.2 CH3 | 12, 13, 14, 17 |
| 20 | 1.67 m | 33.0 CH | 17, 21, 23 |
| 21 | 1.22 d (6.3) | 21.8 CH3 | 17, 20, 22 |
| 22 | 2.08 m 1.87 m | 31.7 CH2 | 21, 24 |
| 23 | 2.41 m 1.81 m | 35.0 CH2 | 24, 25, 28 |
| 24 | - | 155.4 C | - |
| 25 | 2.19 m | 32.3 CH | 23, 24, 26, 27 |
| 26 | 0.98 d (6.8) | 21.6 CH3 | 24, 25, 27 |
| 27 | 0.96 d (6.8) | 21.5 CH3 | 24, 25, 26 |
| 28 | 4.70 brs 4.66 brs | 106.9 CH2 | 23, 24, 25 |
| 29 | 0.73 s | 17.9 CH3 | 3, 4, 5, 30 |
| 30 | 0.91 s | 28.5 CH3 | 3, 4, 5, 29 |
| 31 | - | 169.5 C | - |
| 32 | 1.98 s | 20.6 CH3 | 31 |
| 33 | - | 170.1 C | - |
| 34 | 2.05 s | 21.0 CH3 | 33 |
| 2-OH | 5.30 brs | - | 1, 2, 3 |
| 11-OH | 5.21 d (6.8) | - | 9, 11 |
| 17-OH | 5.39 s | - | 13, 17 |
| Compound | XP Gscore | Glide Gscore | Glide Emodel |
|---|---|---|---|
| NDGA | −7.856 | −7.856 | −51.600 |
| Integracide G (4) | −6.708 | −6.708 | −66.318 |
| Integracide F (3) | −6.453 | −6.453 | −47.304 |
| Integracide J (6) | −6.169 | −6.169 | −47.646 |
| Integracide H (5) | −5.221 | −5.221 | −46.847 |
| Integracide L (1) | −5.109 | −5.109 | −34.117 |
| Integracide B (2) | −5.051 | −5.051 | −35.498 |
| Zileuton | −4.766 | −4.766 | −31.899 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khayat, M.T.; Mohammad, K.A.; Mohamed, G.A.; Safo, M.K.; Ibrahim, S.R.M. Integracides: Tetracyclic Triterpenoids from Fusarium sp.—Their 5-Lipoxygenase Inhibitory Potential and Structure–Activity Relation Using In Vitro and Molecular Docking Studies. Life 2022, 12, 2095. https://doi.org/10.3390/life12122095
Khayat MT, Mohammad KA, Mohamed GA, Safo MK, Ibrahim SRM. Integracides: Tetracyclic Triterpenoids from Fusarium sp.—Their 5-Lipoxygenase Inhibitory Potential and Structure–Activity Relation Using In Vitro and Molecular Docking Studies. Life. 2022; 12(12):2095. https://doi.org/10.3390/life12122095
Chicago/Turabian StyleKhayat, Maan T., Khadijah A. Mohammad, Gamal A. Mohamed, Martin K. Safo, and Sabrin R. M. Ibrahim. 2022. "Integracides: Tetracyclic Triterpenoids from Fusarium sp.—Their 5-Lipoxygenase Inhibitory Potential and Structure–Activity Relation Using In Vitro and Molecular Docking Studies" Life 12, no. 12: 2095. https://doi.org/10.3390/life12122095
APA StyleKhayat, M. T., Mohammad, K. A., Mohamed, G. A., Safo, M. K., & Ibrahim, S. R. M. (2022). Integracides: Tetracyclic Triterpenoids from Fusarium sp.—Their 5-Lipoxygenase Inhibitory Potential and Structure–Activity Relation Using In Vitro and Molecular Docking Studies. Life, 12(12), 2095. https://doi.org/10.3390/life12122095










