1. Introduction
The intestine is an outstanding model with which to study cellular coordination and cooperation and how they are regulated to achieve functional tissue. The intestine’s main functions are to digest and absorb nutrients and water during propulsion of food along the gastrointestinal tract. In order to maintain its vital functions, a high degree of coordination between the different cell types and cellular components is required. The epithelium limits interactions between luminal residents, such as pathogens or the microbiota, and the host, including immune cells, and cooperates so that epithelial-cell-type composition does not change, and regeneration occurs. The enteric nervous system cooperates with smooth muscle cells (SMCs) to control motility, and intestinal vasculature is crucial for nutrient absorption and recruitment of immune cells.
Recently, in searching for the source of cells which provide the signals required for epithelial functional support, we identified mesenchymal cells expressing the transcription factor FOXL1 as an important source of Wnt proteins without which intestinal stem and progenitor cells cannot proliferate and support regeneration [
1,
2]. Using a FOXL1-Cre transgenic line in conjunction with genetic lineage labeling using the
Rosa26-mTmG allele [
3], in which Cre-marked cells produce a membrane targeted version of green fluorescent protein (GFP), we unexpectedly found that FOXL1-expressing cells belong to a novel type of mesenchymal cells called telocytes (TCs) that form a continuous comprehensive three-dimensional network of contact with the entire intestinal epithelium, from the crypt base to the tip of the villi. In support of our work, recent studies from different research groups used diverse markers to label TCs and demonstrated their importance as a critical stem cell niche component that supports intestinal crypts [
4,
5,
6].
TCs are characterized by cytoplasmic processes that are separated from the epithelium by sub-micrometer distances. Intestinal subepithelial TCs express the surface membrane platelet-derived growth factor receptor α (PDGFRα), while the transcription factor FOXL1 is expressed in their nuclei. TCs, primarily described using electron microscopy (EM), were observed by Popescu and Faussone-Pellegrini to be peculiar cells in the intestinal muscle layers which are structurally similar but distinct from the interstitial Cajal cells (ICCs) [
7,
8,
9,
10,
11,
12,
13,
14,
15]. PDGFRa+ cells, located in the muscle layers together with SMCs and ICCs, were previously reported to form an electrical syncytium that coordinates motility [
16].
Here, we used our transgenic mouse line FOXL1-Cre: Rosa-mTmG, in which FOXL1 Cre drives the expression of a membrane-bound GFP and performed whole mounting as well as section staining for c-kit to label ICCs, SMA for SMCs and PDGFRa or CD34 for TCs, to characterize FOXL1 lineage cells in the muscle layers and understand their relations and interactions to the above-mentioned cell types. We found that, in the muscle layers, FOXL1 lineage GFP+ cells had two main cellular characteristics: (i) elongated multinucleated SMA+ muscle cells, intermingled in parallel or perpendicular to muscle fibers. (ii) A TC-like structure displaying a flat cell body with multiple cell processes, expressing PDGFRa and CD34. These findings may suggest a mutual origin between SMCs and TCs linked by FOXL1 lineages.
2. Materials and Methods
2.1. FOXL1Cre: Rosa-mTmG Mice
Mice were obtained by crossing a FOXL1-Cre transgenic line in conjunction with genetic lineage labeling using the
Rosa26-mTmG allele [
3] as described previously [
1,
2].
2.2. Immunofluorescence Staining
Mouse intestines were rinsed in PBS and fixed with 4% paraformaldehyde overnight at 4 °C, washed in PBS, and immersed in 30% sucrose overnight at 4 °C, then embedded in OCT in order to create cryoblocks. Blocks were serially sectioned at 10 µm.
The following antibodies were used: goat anti-PDGFRa (R&D cat:AB1066; 1:200), chicken anti-GFP (Novos Bio cat no: NB100–1614; 1:500), goat anti-GFP (Abcam cat no: AB6673; 1:200), rat anti c-KIT (CD117; Invitrogen Thermo Fisher cat no: 14-1172-82; 1:500).
2.3. Clearing Mouse Intestine Using X-ClarityTM
Mice were sacrificed and their duodenums dissected and fixed at 4%PFA for 24 h while shaking at 4 °C. Following washes with PBS for additional 24 h at 4 °C, tissue was incubated in hydrogel solution (according to manufacturer’s protocol) for 24 h at 4 °C. Following a further 3 h incubation at 37 °C, the hydrogel-embedded tissue was placed in an X-CLARITY ETC chamber (LOGOS Biosystems) for electrophoretic tissue clearing for 7 h. The cleared duodenum was immunostained with goat anti-PDGFRa (1:100), chicken anti-GFP (1:100), rat anti-c-KIT (1:200), goat anti-GFP (1:500), or rabbit anti-SMA (Abcam 1:600). Primary and secondary antibodies were incubated for 48 h at 4 °C. The stained intestine was placed en bloc on an image slide using a 0.5 mm depth adhesive silicone isolator, mounted in X-CLARITY mounting solution and imaged using confocal scanning. Z-stack projections were compiled.
3. Results
Here, we used a transgenic reporter mouse in which FOXL1 Cre drives the expression of a membrane-tag GFP in order to understand the identity of FOXL1 lineage cells and their relations to ICCs, SMCs and PDGFRa+ CD34+ TCs.
3.1. The Distribution of FOXL1 Lineage GFP+ Cells in the Tunica Muscularis
The intestinal tunica muscularis contains two layers of smooth muscles. The inner layer consists of circular muscles, whereas the outer muscle fibers are oriented longitudinally. In order to give a broad view on the distribution of FOXL1 GFP+ lineage cells throughout the tunica muscularis, we used our transgenic mouse line FOXL1 Cre: Rosa-mTmG, cleared a whole duodenum (
Figure 1a,b) and stained for GFP (green) to follow FOXL1+ lineage cells, using a c-kit to label ICCs (red) and PDGFRa (gray) to label the previously described TCs. Interestingly, most of the FOXL1 lineage GFP+ cells at the muscle layers were negative for PDGFRa expression. FOXL1 lineage cells showed a striated elongated structure and were distributed in parallel or perpendicular to longitudinal muscle fibers negative for c-kit or PDGFRa expression. Staining for SMA to label SMCs (
Figure 1c red) confirmed that FOXl1 lineage cells distributed in the muscle layers belong to a fraction of multinucleated muscle cells (in
Figure 1d asterisks point to nuclei).
3.2. FOXL1 Lineage GFP+ Cells in Relation to Interstital Cajal Cells (ICCs)
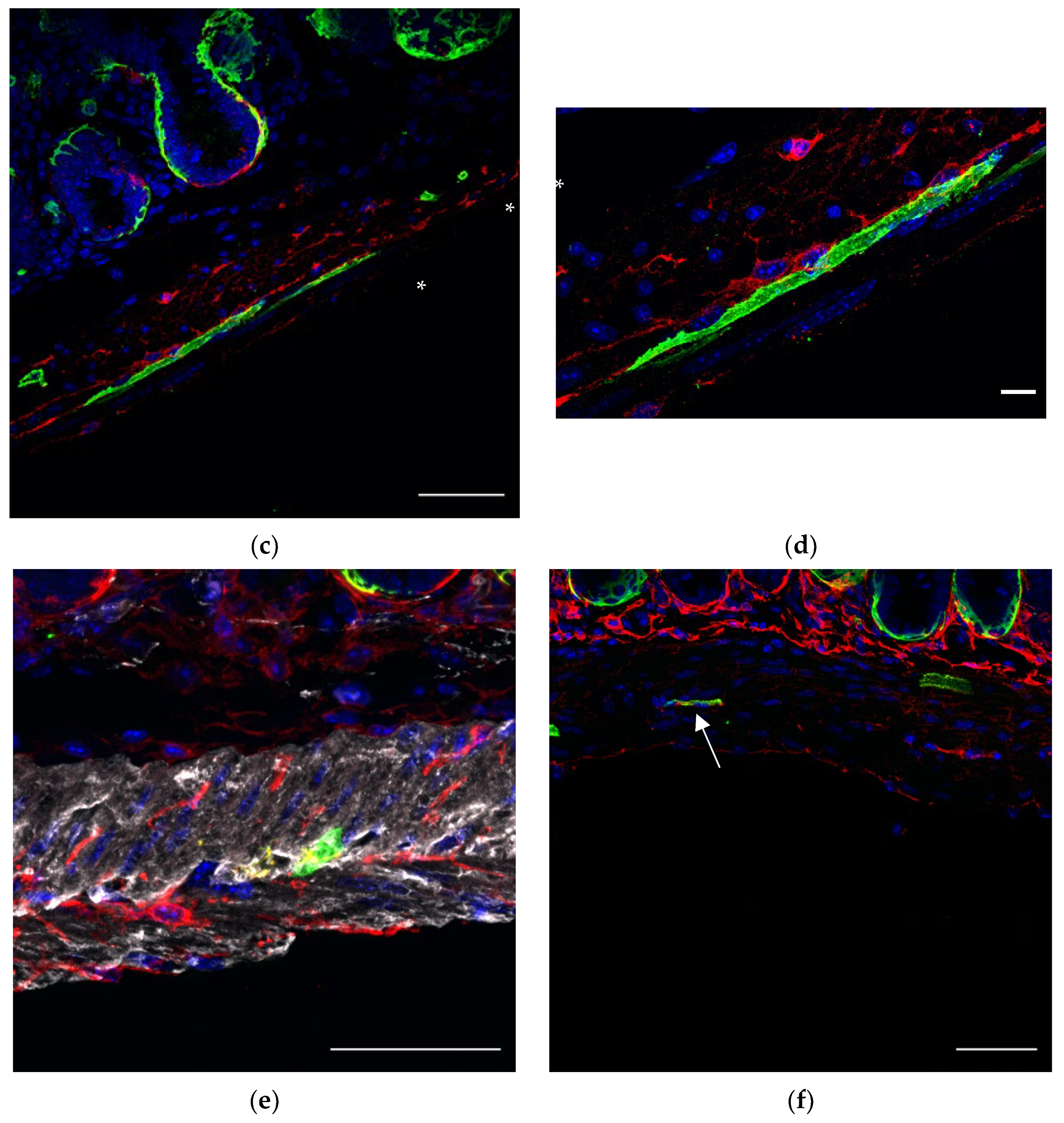
ICCs form networks throughout the intestinal wall, close to nerve endings and SMCs to regulate motility. In order to appreciate the relations between GFP+ FOXL1 lineage cells to ICCs or SMCs, we used cryosections of our transgenic line FOXL1 Cre: Rosa-mTmG and co-stained for GFP, c-kit and smooth muscle actin (SMA) to label FOXL1 lineage cells, ICCs and SMCs, respectively. In addition to GFP+ SMA+ muscle cells, we noticed that a small fraction of the GFP+ cells showed typical TC-like characteristics (
Figure 2b arrows), cells with small cell body and multicellular processes. Close interactions between GFP+ cells ICCs and SMCs were observed (
Figure 2c–e). The overlapping staining in yellow (
Figure 2e asterisks) represents sites of cell–cell contact suggesting potential communication between GFP+ cells ICCs and SMCs.
3.3. FOXL1 Lineage GFP+ in Relation to PDGFRa+ or CD34+ TCs
In order to confirm whether FOXL1 lineage GFP+ cells which exhibit typical TC-like characteristics indeed belong to TCs and to characterize the relations between GFP+ cells and TCs, we co-stained sections of our transgenic lines for PDGFRa or CD34 (
Figure 3). Whereas most of the subepithelial network of TCs consist of double positive GFP-, PDGFRa-expressing cells (
Figure 3c arrows), most of the GFP+ cells along the muscle layer showed SMC characteristics negative for PDGFRa (
Figure 3c asterisk). Notably, close association between GFP+ and PDGFRa+ cells was observed, suggesting potential communication between the cells. As we mentioned earlier, a small fraction of GFP+ cells along the muscle layers showed the typical TC-like structure of cells with multicellular processes; in order to test whether this fraction of cells belongs to TCs, we stained for PDGFRa or CD34. Indeed, GFP+ cells were positive for PDGFRa (
Figure 3e red) and CD34 (
Figure 3f red), representing a small fraction of the overall PDGFRa+ or CD34+ cells.
4. Discussion
TCs have been described primarily by EM, defined by their structure as mesenchymal cells with a small cell body and long processes, called telopodes. These telopodes may reach a length of 100 microns and consist of dilated segments containing mitochondria, rER and caveolae [
11,
12,
13,
14,
15]. The attempts to try and find specific markers which label TCs by performing immunofluorescent staining using markers such as SMA, CD34 or PDGFRa proved challenging as TCs are heterogeneous in their gene expression profile according to the organ, their tissue location, condition, etc. [
2,
3,
7,
8,
9,
10,
11,
12,
13,
14].
In the gut, TCs were considered as CD34-positive c-kit-negative, PDGFRa-positive c-kit-negative or double PDGFRa CD34-positive c-kit-negative, although PDGFRa and CD34 also label endothelial cells, emphasizing the complexity in trying to find one specific marker to label TCs [
9].
We recently identified a subepithelial network of FOXL1+ cells along the entire intestinal crypt–villus axis [
1,
2]. Using a transgenic reporter mouse line in which FOXL1 Cre drives the expression of a membrane-tag GFP, which allows us to visualize the structure of FOXL1+ cells, and based on EM and immune-staining for PDGFRa, we found that the subepithelial FOXL1+ cells are mesenchymal TCs. Most of the subepithelial network of TCs express both PDGFRa and FOXL1, although a portion of the cells were PDGFRa+ only, distinct from the FOXL1+ lineage cells and a portion of the FOXL1+ lineage cells were PDGFRa-negative.
In addition to FOXL1+ TCs’ subepithelial distribution, we also observed GFP+ cells along the muscle layers. We characterized FOXL1 lineage GFP+ cells along the muscle layers of the duodenum in order to understand their identity and relation to ICCs and the previously described PDGFRa+ or CD34+ TCs.
We found that GFP-labeled FOXL1 lineage cells located along the muscle layers have two main cellular features: (i) a smooth striated elongated fiber-like structure SMA-expressing muscle cell and (ii) a typical TC structure of a small cell body with multiple cytoplasmic processes expressing PDGFRa or CD34. Cells were oriented in parallel and perpendicular to muscle fibers, ICCs or PDGFRa+ cells and showed close association with each cell type. Notably, along the muscle layers the GFP+ FOXL1+ lineage cells represented a small fraction from the SMA+ SMCs or the PDGFRa+/CD34+ TCs.
Interestingly, similar to our findings using a FOXL1 cre driven reporter mouse line, PDGFRa+ lineage tracing experiments revealed similar cell progeny in the intestine with the highest level of recombination of PDGFRa+ becoming GFP in SMCs [
17]. A possible explanation for these cells is that FOXL1+ PDGFRa+ cells share a mutual cell origin of a progenitor population with differentiation capabilities to SMCs. However, at the moment we cannot exclude possible miss-expression of the transgene.
A detailed lineage-tracing of FOXL1+ cells and their descendants over time together with transcriptional analysis at a single cell level in combination with spatial tissue characterization would clarify the identity of FOXL1 progeny. The FOXL1 progeny of SMCs and TCs may suggest mutual origin for the two cell types.
The close association between GFP+ SMCs and PDGFRa+ cells and ICCs suggest potential interactions between the cells and a possible role in regulating intestinal motility.
Author Contributions
Conceptualization M.S. and M.S.-C.; methodology, M.S.; writing original draft preparation, M.S. and M.S.-C.; writing-review and editing, M.S. and M.S.-C.; visualization, M.S.; supervision, M.S.-C.; funding acquisition, M.S.-C. All authors have read and agreed to the published version of the manuscript.
Funding
This work is supported by the Israel Science Foundation grant #1997/19.
Institutional Review Board Statement
All animal procedures were approved by the animal care and use committee of the Hebrew University (Protocol MD 19-15621-2). Every effort was made to minimize the numbers of animals and suffering.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We thank Shoshkes lab members for helpful suggestions and assistance.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Aoki, R.; Carmel, M.S.; Gao, N.; Shin, S.; May, C.L.; Golson, M.L.; Zahm, A.M.; Ray, M.; Wiser, C.L.; Wright, C.V.; et al. Foxl1-Expressing Mesenchymal Cells Constitute the Intestinal Stem Cell Niche. Cell. Mol. Gastroenterol. Hepatol. 2015, 2, 175–188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shoshkes-Carmel, M.; Wang, Y.J.; Wangensteen, K.J.; Tóth, B.; Kondo, A.; Massassa, E.E.; Itzkovitz, S.; Kaestner, K.H. Subepithelial telocytes are an important source of Wnts that supports intestinal crypts. Nature 2018, 557, 242–246. [Google Scholar] [CrossRef] [PubMed]
- Halpern, K.B.; Massalha, H.; Zwick, R.K.; Moor, A.E.; Castillo-Azofeifa, D.; Rozenberg, M.; Farack, L.; Egozi, A.; Miller, D.R.; Averbukh, I.; et al. Lgr5+ telocytes are a signaling source at the intestinal villus tip. Nat Commun. 2020, 11, 1936. [Google Scholar] [CrossRef] [Green Version]
- Muzumdar, M.D.; Tasic, B.; Miyamichi, K.; Li, L.; Luo, L. A global double-fluorescent Cre reporter mouse. Genesis 2007, 45, 593–605. [Google Scholar] [CrossRef] [PubMed]
- Stzepourginski, I.; Nigro, G.; Jacob, J.-M.; Dulauroy, S.; Sansonetti, P.J.; Eberl, G.; Peduto, L. CD34+ mesenchymal cells are a major component of the intestinal stem cells niche at homeostasis and after injury. Proc. Natl. Acad. Sci. USA 2017, 114, E506–E513. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Greicius, G.; Kabiri, Z.; Sigmundsson, K.; Liang, C.; Bunte, R.; Singh, M.; Virshup, D.M. PDGFRα+ pericryptal stromal cells are the critical source of Wnts and RSPO3 for murine intestinal stem cells in vivo. Proc. Natl. Acad. Sci. USA 2018, 115, E3173–E3181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Degirmenci, B.; Valenta, T.; Dimitrieva, S.; Hausmann, G.; Basler, K. GLI1-expressing mesenchymal cells form the essential Wnt-secreting niche for colon stem cells. Nature 2018, 558, 449–453. [Google Scholar] [CrossRef] [PubMed]
- El Maadawi, Z.M. A Tale of Two Cells: Telocyte and stem cell unique relationship. In Telocytes: Connecting Cells; Wang, X., Cretoiu, D., Eds.; Springer: Singapore, 2016; pp. 359–376. [Google Scholar]
- Vannucchi, M.-G.; Faussone-Pellegrini, M.-S. The telocyte subtypes. In Telocytes: Connecting Cells; Wang, X., Cretoiu, D., Eds.; Springer: Singapore, 2016; pp. 115–126. [Google Scholar]
- Vannucchi, M.-G.; Traini, C.; Manetti, M.; Ibba-Manneschi, L.; Faussone-Pellegrini, M.-S. Telocytes express PDGFRα in the human gastrointestinal tract. J. Cell Mol. Med. 2013, 17, 1099–1108. [Google Scholar] [CrossRef] [PubMed]
- Popescu, L.M.; Faussone-Pellegrini, M.S. Telocytes—A case of serendipity: The winding way from interstitial cells of Cajal (ICC), via interstitial Cajal-like Cells (ICLC) to telocytes. J. Cell Mol. Med. 2010, 14, 729–740. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rusu, M.C.; Jianu, A.M.; Mirancea, N.; Didilescu, A.C.; Manoiu, V.S.; Paduraru, D. Tracheal telocytes. J. Cell Mol. Med. 2012, 16, 401–405. [Google Scholar] [CrossRef] [PubMed]
- Rusu, M.C.; Mirancea, N.; Manoiu, V.S.; Valcu, M.; Nicolescu, M.I.; Paduraru, D. Skin telocytes. Ann. Anat. 2012, 194, 359–367. [Google Scholar] [CrossRef] [PubMed]
- Rusu, M.C.; Nicolescu, M.I.; Jianu, A.M.; Lighezan, R.; Manoiu, V.S.; Paduraru, D. Esophageal telocytes and hybrid mor-phologies. Cell Biol. Int. 2012, 36, 1079–1088. [Google Scholar] [CrossRef] [PubMed]
- Popescu, L.M.; Gherghiceanu, M.; Suciu, L.C.; Manole, C.G.; Hinescu, M.E. Telocytes and putative stem cells in the lungs: Electron microscopy, electron tomography and laser scanning microscopy. Cell Tissue Res. 2011, 345, 391–403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanders, K.M.; Koh, S.D.; Ro, S.; Ward, S.M. Regulation of gastrointestinal motility–insights from smooth muscle biology. Nat. Rev. Gastroenterol. Hepatol. 2012, 9, 633–645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Rourke, M.; Cullen, C.L.; Auderset, L.; Pitman, K.A.; Achatz, D.; Gasperini, R.; Young, K.M. Evaluating Tissue-Specific Recombination in a Pdgfrα-CreERT2 Transgenic Mouse Line. PLoS ONE 2016, 11, 0162858. [Google Scholar] [CrossRef] [PubMed]
| Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).