Robust Profiling of Cytochrome P450s (P450ome) in Notable Aspergillus spp.
Abstract
:1. Introduction
2. Materials and Methods
2.1. Selection of Aspergillus Species, Sequence Retrieval, and Validation
2.2. Identification of CYP450 Protein Gene Families and Clans in Eight Aspergillus Species
2.3. Evolutionary Relatedness of Cyp Protein Genes Amongst Eight Aspergillus Species
2.4. Prediction of Subcellular Localization of Cyp Protein Genes
2.5. Gene Clusters Associated with Secondary Metabolite Synthesis
3. Results
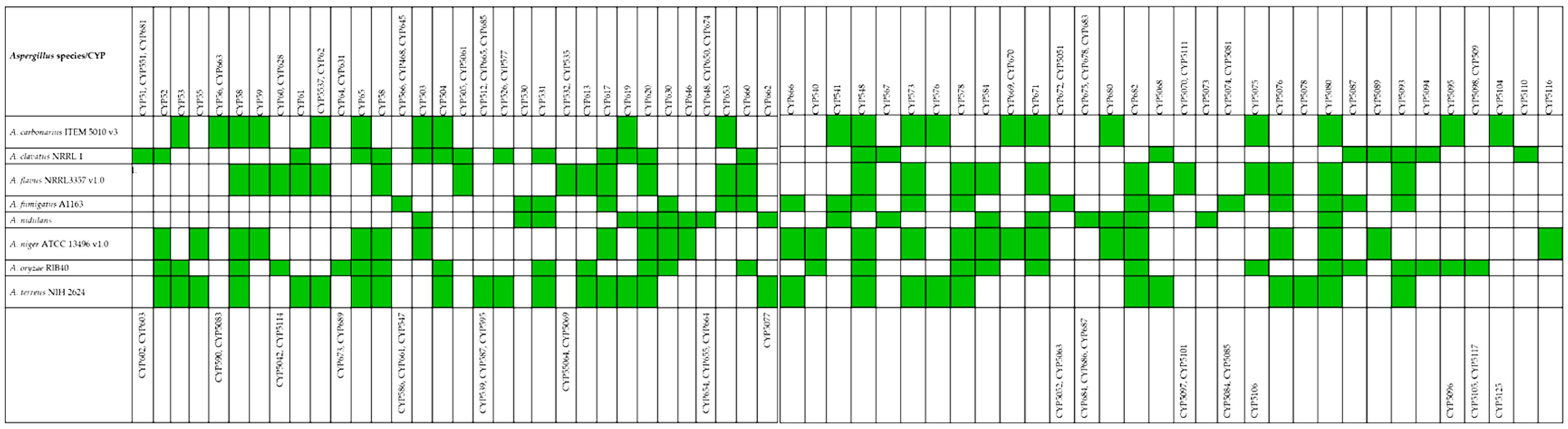
3.1. Diverseness and Widespread of Cyp450 Families and Clans in Eighth Aspergillus Species
3.2. Phylogenetic Distribution of Eight Aspergillus spp. in Cytochrome P450 Families and Clans
3.3. Prediction of Subcellular Localization Analysis of CYP450 Protein Genes of Eight Aspergillus Species
3.4. CYP450s Implicated with Secondary Metabolism-Related Gene Clusters in Aspergillus Species
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Samson, R.A.; Visagie, C.M.; Houbraken, J.; Hong, S.B.; Hubka, V.; Klaassen, C.H.; Perrone, G.; Seifert, K.A.; Susca, A.; Frisvad, J.C.; et al. Phylogeny, identification and nomenclature of the genus Aspergillus. Stud. Mycol. 2014, 78, 141–173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, L.; Li, X.; Dong, L.; Wang, B.; Pan, L. Profiling of chromatin accessibility identifies transcription factor binding sites across the genome of Aspergillus species. BMC Biol. 2021, 19, 189. [Google Scholar] [CrossRef] [PubMed]
- de Vries, R.P.; Riley, R.; Wiebenga, A.; Aguilar-Osorio, G.; Anderluh, G.; Asadollahi, M.; Askin, M.; Amillis, S.; Uchima, C.A.; Grigoriev, I.V. Comparative genomics reveals high biological diversity and specific adaptations in the industrially and medically important fungal genus Aspergillus. Genome Biol. 2017, 18, 28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zarafi, A.B.; Dauda, W.P. Exploring the importance of fungi in agricultural biotechnology. Int. J. Agric. Sci. Vet. Med. 2019, 7, 1–12. [Google Scholar]
- Elkadi, O.A.; Hassan, R.; Elanany, M.; Byrne, H.J.; Ramadan, M.A. Identification of Aspergillus species in human blood plasma by infrared spectroscopy and machine learning. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2021, 248, 119259. [Google Scholar] [CrossRef] [PubMed]
- Latgé, J.-P.; Chamilos, G. Aspergillus fumigatus and Aspergillosis in 2019. Clin. Microbiol. Rev. 2019, 33, e00140-18. [Google Scholar] [CrossRef]
- Taniwaki, M.H.; Pitt, J.I.; Magan, N. Aspergillus species and mycotoxins: Occurrence and importance in major food commodities. Curr. Opin. Food Sci. 2018, 23, 38–43. [Google Scholar] [CrossRef] [Green Version]
- Ji, C.; Fan, Y.; Zhao, L. Review on biological degradation of mycotoxins. Anim. Nutr. 2016, 2, 127–133. [Google Scholar] [CrossRef]
- Díaz Nieto, C.H.; Granero, A.M.; Zon, M.A.; Fernández, H. Sterigmatocystin: A mycotoxin to be seriously considered. Food Chem. Toxicol. 2018, 118, 460–470. [Google Scholar] [CrossRef]
- Gomi, K. ASPERGILLUS|Aspergillus oryzae. In Tortorello, Encyclopedia of Food Microbiology, 2nd ed.; Batt, C.A., Ed.; Elsevier Press: Amsterdam, The Netherlands, 2014; pp. 92–96. [Google Scholar] [CrossRef]
- Braia, M.; Cabezudo, I.; Barrera, V.L.; Anselmi, P.; Meini, M.R.; Romanini, D. An optimization approach to the bioconversion of flour mill waste to α-amylase enzyme by Aspergillus oryzae. Process Biochem. 2021, 111, 102–108. [Google Scholar] [CrossRef]
- Tamayo-Ordóñez, M.C.; Contreras-Esquivel, J.C.; Ayil-Gutiérrez, B.A.; De la Cruz-Arguijo, E.A.; Tamayo-Ordóñez, F.A.; Ríos-González, L.J.; Tamayo-Ordóñez, Y.J. Interspecific evolutionary relationships of alpha-glucuronidase in the genus Aspergillus. Fungal Biol. 2021, 125, 560–575. [Google Scholar] [CrossRef] [PubMed]
- Sadeghi, H.M.M.; Rabbani, M.; Fazeli, S. Optimization of alkaline phosphatase gene expression in E. coli. Res. Pharm. Sci. 2008, 3, 35–39. [Google Scholar]
- Suresh, N.; Das, A. Purification and characterization of phosphatases from Aspergillus fumigatus for applications in industrial uses. Rasayan J. Chem. 2014, 7, 118–128. [Google Scholar]
- Huarte-Bonnet, C.; Kumar, S.; Saparrat, M.C.; Girotti, J.R.; Santana, M.; Hallsworth, J.E.; Pedrini, N. Insights into hydrocarbon assimilation by eurotialean and hypocrealean fungi: Roles for CYP52 and CYP53 clans of cytochrome P450 genes. Appl. Biochem. Biotechnol. 2014, 184, 1047–1060. [Google Scholar] [CrossRef]
- Črešnar, B.; Petrič, Š. Cytochrome P450 enzymes in the fungal kingdom. Biochim. Biophys. Acta-Proteins Proteom. 2011, 1814, 29–35. [Google Scholar] [CrossRef]
- Guengerich, F.P.; Munro, A. Unusual Cytochrome P450 Enzymes and Reactions. J. Biol. Chem. 2013, 288, 17065–17073. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Guo, J.; Cheng, F.; Li, S. Cytochrome P450 enzymes in fungal natural product biosynthesis. Nat. Prod. Rep. 2021, 38, 1072–1099. [Google Scholar] [CrossRef]
- Zhao, W.-Y.; Yi, J.; Chang, Y.-B.; Sun, C.-P.; Ma, X.-C. Recent studies on terpenoids in Aspergillus fungi: Chemical diversity, biosynthesis, and bioactivity. Phytochemistry 2021, 193, 113011. [Google Scholar] [CrossRef]
- Syed, K.; Yadav, J.S. P450 monooxygenases (P450ome) of the model white rot fungus Phanerochaetechrysosporium. Crit. Rev. Microbiol. 2012, 38, 339–363. [Google Scholar] [CrossRef] [Green Version]
- Newsome, A.W.; Nelson, D.; Corran, A.; Kelly, S.L.; Kelly, D.E. The cytochrome P 450 complement (CYP ome) of Mycosphaerellagraminicola. Biotechnol. Appl. Biochem. 2013, 60, 52–64. [Google Scholar] [CrossRef]
- Lah, L.; Haridas, S.; Bohlmann, J.; Breuil, C. The cytochromes P450 of Grosmanniaclavigera: Genome organization, phylogeny, and expression in response to pine host chemicals. Fungal Genet. Biol. 2013, 50, 72–81. [Google Scholar] [CrossRef] [PubMed]
- Chadha, S.; Mehetre, S.T.; Bansal, R.; Kuo, A.; Aerts, A.; Grigoriev, I.V.; Druzhinina, I.S.; Mukherjee, P.K. Genome-wide analysis of cytochrome p450s of Trichoderma spp.: Annotation and evolutionary relationships. Fungal Biol. Biotechnol. 2018, 5, 12. [Google Scholar] [CrossRef] [PubMed]
- Palmer-Brown, W.; Miranda-CasoLuengo, R.; Wolfe, K.H.; Byrne, K.P.; Murphy, C.D. The CYPome of the model xenobiotic-biotransforming fungus Cunninghamella elegans. Sci. Rep. 2019, 9, 9240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dauda, W.P.; Glen, E.; Abraham, P.; Adetunji, C.O.; Morumda, D.; Abraham, S.E.; Wabba, G.P.; Ogwuche, I.O.; Azameti, M.K. Comparative Phylogenomic Analysis of Cytochrome P450 Monooxygenases From Fusarium Species. Search. Sq. 2021. Preprint from Research Square, 02 December 2021. [Google Scholar] [CrossRef]
- Dauda, W.P.; Abraham, P.; Fasogbon, I.V.; Adetunji, C.O.; Banwo, O.O.; Kashina, B.D.; Alegbejo, M.D. Cassava mosaic virus in Africa: Functional analysis of virus coat proteins based on evolutionary processes and protein structure. Gene Rep. 2021, 24, 101239. [Google Scholar] [CrossRef]
- Hansen, C.C.; Nelson, D.R.; Møller, B.L.; Werck-Reichhart, D. Plant cytochrome P450 plasticity and evolution. Mol. Plant 2021, 14, 1244–1265. [Google Scholar] [CrossRef]
- Syed, K.; Shale, K.; Pagadala, N.S.; Tuszynski, J. Systematic identification and evolutionary analysis of catalytically versatile cytochrome p450 monooxygenase families enriched in model basidiomycete fungi. PLoS ONE 2021, 22, e86683. [Google Scholar] [CrossRef]
- Sello, M.M.; Jafta, N.; Nelson, D.R.; Chen, W.; Yu, J.-H.; Parvez, M.; Kgosiemang, I.K.R.; Monyaki, R.; Raselemane, S.C.; Qhanya, L.B.; et al. Diversity and evolution of cytochrome P450 monooxygenases in Oomycetes. Sci. Rep. 2015, 5, 11572. [Google Scholar] [CrossRef] [Green Version]
- Feyereisen, R. Arthropod CYPomes illustrate the tempo and mode in P450 evolution. Biochim. Biophys. Acta-Proteins Proteom. 2011, 1814, 19–28. [Google Scholar] [CrossRef]
- Rampersad, S.N. Pathogenomics and management of Fusarium diseases in plants. Pathogens 2020, 9, 340. [Google Scholar] [CrossRef]
- Werck-Reichhart, D.; Feyereisen, R. Cytochromes P450: A success story. Genome Biol. 2000, 1, reviews3003.1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nelson, D.R.; Koymans, L.; Kamataki, T.; Stegeman, J.J.; Feyereisen, R.; Waxman, D.J.; Waterman, M.R.; Gotoh, O.; Coon, M.J.; Estabrook, R.W.; et al. P450 superfamily: Update on new sequences, gene mapping, accession numbers and nomenclature. Pharmacogenetics 1996, 1, 1–42. [Google Scholar] [CrossRef] [PubMed]
- Bayram, Ö.; Braus, G.H. Coordination of secondary metabolism and development in fungi: The velvet family of regulatory proteins. FEMS Microbiol. Rev. 2012, 36, 1–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoffmeister, D.; Keller, N.P. Natural products of filamentous fungi: Enzymes, genes, and their regulation. Nat. Prod. Rep. 2017, 24, 393–416. [Google Scholar] [CrossRef]
- Wiemann, P.; Keller, N.P. Strategies for mining fungal natural products. J. Ind. Microbiol. Biotechnol. 2014, 41, 301–313. [Google Scholar] [CrossRef]
- Keller, N.P.; Turner, G.; Bennett, J.W. Fungal secondary metabolism—From biochemistry to genomics. Nat. Rev. Microbiol. 2005, 3, 937–947. [Google Scholar] [CrossRef]
- Strieker, M.; Tanović, A.; Marahiel, M.A. Nonribosomal peptide synthetases: Structures and dynamics. Curr. Opin. Struct. Biol. 2010, 20, 234–240. [Google Scholar] [CrossRef]
- Crawford, J.M.; Townsend, C.A. New insights into the formation of fungal aromatic polyketides. Nat. Rev. Microbiol. 2010, 8, 879–889. [Google Scholar] [CrossRef] [Green Version]
- Brakhage, A.A.; Schroeckh, V. Fungal secondary metabolites–strategies to activate silent gene clusters. Fungal Genet. Biol. 2011, 48, 15–22. [Google Scholar] [CrossRef]
- Shi-Kunne, X.; de Jové, R.P.; Depotter, J.R.L.; Ebert, M.K.; Seidl, M.F.; Thomma, B.P.H.J. In silico prediction and characterisation of secondary metabolite clusters in the plant pathogenic fungus Verticillium dahliae. FEMS Microbiol. Lett. 2019, 366, fnz081. [Google Scholar] [CrossRef] [Green Version]
- Cox, R.J. Polyketides, proteins and genes in fungi: Programmed nano-machines begin to reveal their secrets. Org. Biomol. Chem. 2007, 5, 2010–2026. [Google Scholar] [CrossRef] [PubMed]
- Drott, M.T.; Bastos, R.W.; Rokas, A.; Ries, L.N.A.; Gabaldón, T.; Goldman, G.H.; Keller, N.P.; Greco, C. Diversity of Secondary Metabolism in Aspergillus nidulans Clinical Isolates. mSphere 2020, 5, e00156-20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brakhage, A.A. Regulation of fungal secondary metabolism. Nat. Rev. Microbiol. 2012, 11, 21–32. [Google Scholar] [CrossRef] [PubMed]
- Yim, G.; Huimi Wang, H.; Davies Frs, J. Antibiotics as signalling molecules. Philos. Trans. R. Soc. B Biol. Sci. 2007, 362, 1195–1200. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Aspergillus Species | Genome Size (Mb) | Number of Predicted Genes | Total Proteins Sequences | Total Cyp Proteins | Protein with Complete Sequences | Family Type | Clan Type | Families with No FCPD Matches |
|---|---|---|---|---|---|---|---|---|
| A. carbonarius ITEM 5010 v3 | 36.29 | 11,624 | 222 | 114 | 35 | 29 | 22 | 1 |
| A. clavatus NRRL 1 | 27.86 | 9121 | 145 | 91 | 35 | 28 | 20 | - |
| A. flavus NRRL3357 v1.0 | 37.75 | 13,715 | 231 | 139 | 45 | 36 | 28 | - |
| A. fumigatus A1163 | 29.21 | 9916 | 130 | 75 | 32 | 31 | 27 | 2 |
| A. nidulans A1146 | 30.48 | 10,680 | 184 | 103 | 30 | 29 | 20 | - |
| A. niger ATCC 13496 v1.0 | 35.69 | 12,197 | 232 | 133 | 34 | 26 | 19 | - |
| A. oryzae RIB40 | 37.88 | 12,030 | 249 | 140 | 48 | 37 | 22 | - |
| A. terreus NIH 2624 | 29.33 | 10,406 | 183 | 101 | 47 | 35 | 24 | - |
| Aspergillus spp. | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Clan | CYP Proteins in Clan | Families | A. Carbonarius | A. Clavatus | A. Flavus | A. Fumigatus | A. Nidulans | A. Niger | A. Oryzae | A. Terreus | Total |
| CYP51 | 2 | Cyp51 | - | 2 | - | - | - | - | - | - | 2 |
| CYP52 | 15 | Cyp52 | - | 1 | - | - | - | 1 | 1 | 3 | 6 |
| Cyp539 | - | - | - | - | - | - | - | 1 | 1 | ||
| Cyp584 | - | - | 1 | - | 1 | 1 | 1 | - | 4 | ||
| Cyp655 | - | - | - | - | 1 | - | - | - | 1 | ||
| Cyp5087 | - | 1 | - | 1 | - | - | 1 | - | 3 | ||
| CYP53 | 4 | Cyp53 | 1 | - | - | - | - | - | 1 | 2 | 4 |
| CYP54 | 6 | Cyp602 | - | 1 | - | - | - | - | - | - | 1 |
| Cyp503 | 1 | 1 | - | - | 1 | 1 | - | - | 4 | ||
| Cyp5085 | - | - | - | 1 | - | - | - | - | 1 | ||
| CYP55 | 2 | Cyp55 | - | - | - | - | - | 1 | - | 1 | 2 |
| CYP56 | 3 | Cyp56 | 1 | - | - | - | - | - | - | - | 1 |
| Cyp661 | - | - | - | 1 | - | - | - | - | 1 | ||
| Cyp5099 | - | - | - | - | - | - | 1 | - | 1 | ||
| CYP58 | 29 | Cyp58 | 1 | - | 1 | - | - | 2 | 3 | 3 | 10 |
| Cyp551 | - | 1 | - | - | - | - | - | - | 1 | ||
| Cyp680 | 1 | - | - | - | 1 | 1 | - | - | 3 | ||
| Cyp681 | - | 1 | - | - | - | - | - | - | 1 | ||
| Cyp682 | - | - | 1 | 1 | 2 | 1 | 1 | 1 | 7 | ||
| Cyp5094 | - | 1 | - | - | - | - | 1 | - | 2 | ||
| Cyp5095 | 1 | - | - | - | - | - | 1 | - | 2 | ||
| Cyp5096 | 1 | - | - | - | - | - | 1 | - | 2 | ||
| Cyp5105 | - | - | - | - | - | - | 1 | - | 1 | ||
| CYP59 | 8 | Cyp59 | 1 | - | 2 | - | - | 1 | - | - | 4 |
| Cyp586 | - | - | - | 1 | - | - | - | - | 1 | ||
| Cyp587 | - | - | - | - | - | - | - | 1 | 1 | ||
| Cyp662 | - | - | - | - | 1 | - | - | 1 | 2 | ||
| CYP61 | 6 | Cyp61 | - | 1 | 2 | - | - | - | - | 3 | 6 |
| CYP62 | 4 | Cyp62 | 1 | - | 1 | - | - | - | - | 1 | 3 |
| Cyp684 | - | - | - | - | 1 | - | - | - | 1 | ||
| CYP65 | 18 | Cyp60 | - | - | 1 | - | - | - | 1 | - | 2 |
| Cyp65 | 4 | 2 | - | - | - | 2 | 2 | 2 | 12 | ||
| Cyp567 | - | 1 | - | - | 1 | - | - | - | 2 | ||
| Cyp5117 | - | - | - | - | - | - | 2 | - | 2 | ||
| CYP68 | 12 | Cyp68 | - | 1 | 1 | - | - | 2 | 2 | 1 | 7 |
| Cyp595 | - | - | - | - | - | - | - | 1 | 1 | ||
| Cyp650 | - | - | - | - | 1 | - | - | - | 1 | ||
| Cyp5061 | - | 1 | 1 | - | - | - | - | - | 2 | ||
| Cyp5073 | - | - | - | - | 1 | - | - | - | 1 | ||
| CYP504 | 5 | Cyp504 | 2 | 1 | - | - | - | - | 1 | 1 | 5 |
| CYP505 | 6 | Cyp505 | - | 2 | 1 | - | - | - | - | - | 3 |
| Cyp541 | 1 | - | - | 1 | 1 | - | - | - | 3 | ||
| CYP507 | 1 | Cyp535 | - | - | 1 | - | - | - | - | - | 1 |
| CYP512 | 1 | Cyp512 | - | - | - | - | - | - | - | 1 | 1 |
| CYP526 | 2 | Cyp526 | - | 1 | - | - | - | - | - | 1 | 2 |
| CYP530 | 17 | Cyp530 | - | - | - | 1 | 1 | - | - | - | 2 |
| Cyp619 | 1 | 2 | - | - | 1 | - | - | 1 | 5 | ||
| Cyp663 | 1 | - | - | - | - | - | - | - | 1 | ||
| Cyp665 | - | - | - | - | - | - | - | 1 | 1 | ||
| Cyp5068 | - | 1 | - | 1 | - | - | - | 1 | 3 | ||
| Cyp5069 | - | - | 1 | - | - | - | - | - | 1 | ||
| Cyp5093 | - | 1 | 1 | - | - | - | - | 1 | 3 | ||
| Cyp5098 | - | - | - | - | - | - | 1 | - | 1 | ||
| CYP531 | 23 | Cyp531 | - | 2 | - | 1 | 1 | - | 2 | 2 | 8 |
| Cyp532 | - | - | 1 | - | - | - | - | - | 1 | ||
| Cyp631 | - | - | - | - | - | - | 1 | - | 1 | ||
| Cyp674 | - | - | - | - | 1 | - | - | - | 1 | ||
| Cyp675 | - | - | - | - | 1 | - | - | - | 1 | ||
| Cyp5077 | - | - | - | - | 1 | - | - | 1 | 2 | ||
| Cyp5078 | - | - | - | - | - | - | - | 1 | 1 | ||
| Cyp5080 | 2 | - | 1 | 1 | 1 | 1 | 1 | - | 7 | ||
| Cyp5104 | 1 | - | - | - | - | - | - | - | 1 | ||
| CYP533 | 9 | Cyp64 | - | - | - | - | - | - | 1 | - | 1 |
| Cyp620 | - | 1 | 2 | - | 1 | 2 | 1 | 1 | 8 | ||
| CYP537 | 6 | Cyp537 | 2 | - | 1 | - | - | - | - | 1 | 4 |
| Cyp577 | - | 1 | - | - | - | - | - | 1 | 2 | ||
| CYP540 | 2 | Cyp540 | - | - | - | - | - | 1 | 1 | - | 2 |
| CYP547 | 11 | Cyp547 | - | - | - | 1 | - | - | - | - | 1 |
| Cyp617 | - | 2 | 1 | 2 | - | 2 | - | 2 | 9 | ||
| Cyp5070 | - | - | 1 | - | - | - | - | - | 1 | ||
| CYP548 | 12 | Cyp548 | 1 | 2 | 2 | 1 | - | 1 | 2 | 1 | 10 |
| Cyp5114 | - | - | 1 | - | - | - | 1 | - | 2 | ||
| CYP550 | 6 | Cyp660 | - | 1 | 2 | 1 | - | - | 2 | - | 6 |
| CYP566 | 1 | Cyp566 | - | - | - | 1 | - | - | - | - | 1 |
| CYP572 | 5 | Cyp573 | 1 | - | 1 | 1 | - | 1 | - | 1 | 5 |
| CYP574 | 16 | Cyp628 | - | - | 1 | - | - | - | 1 | - | 2 |
| Cyp669 | 1 | - | - | - | - | 1 | - | - | 2 | ||
| Cyp670 | 1 | - | - | - | - | 1 | - | - | 2 | ||
| Cyp671 | 1 | - | 1 | - | 1 | 2 | 1 | - | 6 | ||
| Cyp5076 | - | - | 1 | 1 | - | 1 | - | 1 | 4 | ||
| CYP576 | 2 | Cyp576 | 1 | - | - | - | - | - | - | 1 | 2 |
| CYP578 | 8 | Cyp578 | - | - | 2 | 1 | - | 2 | 1 | 2 | 8 |
| CYP589 | 5 | Cyp5075 | 1 | - | 2 | - | - | - | 2 | - | 5 |
| CYP590 | 1 | Cyp590 | 1 | - | - | - | - | - | - | - | 1 |
| CYP603 | 1 | Cyp603 | - | 1 | - | - | - | - | - | - | 1 |
| CYP613 | 6 | Cyp613 | - | - | 1 | - | - | - | 2 | 1 | 4 |
| Cyp685 | - | - | - | - | - | - | - | 2 | 2 | ||
| CYP630 | 4 | Cyp630 | - | - | - | 1 | 1 | 1 | 1 | - | 4 |
| CYP645 | 1 | Cyp645 | - | - | - | 1 | - | - | - | - | 1 |
| CYP646 | 3 | Cyp646 | - | - | - | - | 1 | 2 | - | - | 3 |
| CYP648 | 1 | Cyp648 | - | - | - | - | 1 | - | - | - | 1 |
| CYP653 | 4 | Cyp653 | 1 | - | 1 | 1 | - | - | - | - | 3 |
| Cyp654 | - | - | - | - | 1 | - | - | - | 1 | ||
| CYP659 | 2 | Cyp5090 | - | - | - | 1 | - | - | - | - | 1 |
| Cyp5111 | - | - | 1 | - | - | - | - | - | 1 | ||
| CYP664 | 1 | Cyp664 | - | - | - | - | 1 | - | - | - | 1 |
| CYP666 | 3 | Cyp666 | - | - | - | - | - | 1 | 1 | 1 | 3 |
| CYP672 | 1 | Cyp672 | - | - | - | 1 | - | - | - | - | 1 |
| CYP673 | 1 | Cyp673 | - | - | - | - | - | - | 1 | - | 1 |
| CYP677 | 1 | Cyp5064 | - | - | 1 | - | - | - | - | - | 1 |
| CYP678 | 1 | Cyp678 | - | - | - | - | 1 | - | - | - | 1 |
| CYP683 | 1 | Cyp683 | - | - | - | - | 1 | - | - | - | 1 |
| CYP687 | 1 | Cyp687 | - | - | - | - | 1 | - | - | - | 1 |
| CYP698 | 1 | Cyp698 | - | - | - | - | - | - | 1 | - | 1 |
| CYP5042 | 4 | Cyp5042 | - | - | 3 | - | - | - | 1 | - | 4 |
| CYP5052 | 1 | Cyp5052 | - | - | - | 1 | - | - | - | - | 1 |
| CYP5063 | 1 | Cyp5063 | - | - | - | 1 | - | - | - | - | 1 |
| CYP5081 | 1 | Cyp5081 | - | - | - | 1 | - | - | - | - | 1 |
| CYP5083 | 1 | Cyp5083 | 1 | - | - | - | - | - | - | - | 1 |
| CYP5084 | 1 | Cyp5084 | - | - | - | 1 | - | - | - | - | 1 |
| CYP5089 | 2 | Cyp5089 | - | 1 | - | - | - | 1 | - | - | 2 |
| CYP5101 | 1 | Cyp5101 | - | - | 1 | - | - | - | - | - | 1 |
| CYP5071 | 3 | Cyp5106 | 1 | - | 1 | - | - | - | 1 | - | 3 |
| CYP5110 | 1 | Cyp5110 | - | 1 | - | - | - | - | - | - | 1 |
| CYP5116 | 1 | Cyp5116 | - | - | - | - | - | 1 | - | - | 1 |
| Absent | Cyp468 | - | - | - | 1 | - | - | - | - | 1 | |
| Absent | Cyp5051 | - | - | - | 1 | - | - | - | - | 1 | |
| Absent | Cyp5125 | 1 | - | - | - | - | - | - | - | 1 | |
| Total CYP families in each Aspergillus sp. | 35 | 35 | 45 | 32 | 30 | 34 | 48 | 47 | 306 | ||
| Phylogenetic Clade | Sequence Entry | CYP Families | CYP Clans | Putative Functions |
|---|---|---|---|---|
| I | 46 | Cyp65, Cyp60, Cyp5117, Cyp567, Cyp672, Cyp5083, Cyp566, Cyp548, Cyp5114, Cyp671, Cyp628, Cyp670, Cyp669, Cyp5076, Cyp578 | CYP65, CYP672, CYP5083, CYP566, CYP548, CYP574, CYP578 | Primary metabolism, Secondary metabolism |
| II | 5 | Cyp5084, Cyp680, Cyp673, Cyp535, Cyp567 | CYP5084, CYP673, CYP58, CYP65, CYP507 | Secondary/xenobiotic metabolism |
| III | 7 | Cyp630, Cyp53 | CYP630, CYP53 | Xenobiotic/primary metabolism |
| IV | 27 | Cyp573, Cyp5080, Cyp5104, Cyp675, Cyp674, Cyp531, Cyp5077, Cyp631, Cyp5078, Cyp532 | CYP572, CYP531 | Xenobiotic metabolism |
| V | 53 | Cyp578, Cyp577, Cyp537, Cyp683, Cyp5089, Cyp53, Cyp62, Cyp684, Cyp678, Cyp5064, Cyp680, Cyp5095, Cyp5096, Cyp682, Cyp551, Cyp5094, Cyp58, Cyp681, Cyp5105, Cyp5097 | CYP578, CYP58, CYP537, CYP683, CYP53, CYP62, CYP678, CYP677, CYP5097, CYP5089 | Secondary/xenobiotic Metabolism |
| VI | 3 | Cyp5081, Cyp576 | CYP5081, CYP576, | |
| VII | 8 | Cyp59, Cyp587, Cyp586, Cyp662 | CYP59 | Xenobiotic metabolism |
| VIII | 17 | Cyp617, Cyp547, Cyp5070, Cyp526, Cyp540, Cyp468 | CYP547, CYP540, CYP526, | Primary/xenobiotic metabolism |
| IX | 30 | Cyp5075, Cyp5106, Cyp590, Cyp666, Cyp56, Cyp5099, Cyp661, Cyp655, Cyp5087, Cyp52, Cyp5051, Cyp539, Cyp548, Cyp584 | CYP5075, CYP56, CYP5071, CYP666, CYP52, CYP590, Cyp548 | Secondary metabolism |
| X | 7 | Cyp613, Cyp686, Cyp685 | CYP613 | Xenobiotic metabolism |
| XI | 38 | Cyp5098, Cyp5052, Cyp5068, Cyp663, Cyp5063, Cyp5097, Cyp5069, Cyp5093, Cyp665, Cyp530, Cyp664, Cyp619, Cyp5042, Cyp620, Cyp64, Cyp504 | CYP530, CYP533, CYP504, CYP5052, CYP5097, CYP5063, CYP5042, CYP664 | Xenobiotic metabolism |
| XII | 10 | Cyp61, Cyp5116, Cyp55, Cyp51 | CYP61, CYP55, CYP51, CYP5116 | Primary metabolism |
| XIII | 45 | Cyp660, Cyp5090, Cyp5101, Cyp5111, Cyp5100, Cyp5106, Cyp646, Cyp5110, Cyp653, Cyp503, Cyp698, Cyp602, Cyp5093, Cyp654, Cyp5125, Cyp503, Cyp512, Cyp5085, Cyp648, Cyp603, Cyp595, Cyp68, Cyp5061, Cyp5074, Cyp5073, Cyp650 | CYP550, CYP659, CYP5101, CYP5070, CYP54, CYP639, CYP648, CYP646, CYP68, CYP5110, CYP603, CYP512, CYP530, CYP653, CYP698, CYP608, | Secondary/xenobiotic Metabolism |
| XIV | 7 | Cyp505, Cyp541 | CYP505, CYP5125 | Primary metabolism |
| XV | 3 | Cyp682, Cyp645, Cyp687 | CYP645, CYP687, CYP58 | Secondary/xenobiotic metabolism |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dauda, W.P.; Abraham, P.; Glen, E.; Adetunji, C.O.; Ghazanfar, S.; Ali, S.; Al-Zahrani, M.; Azameti, M.K.; Alao, S.E.L.; Zarafi, A.B.; et al. Robust Profiling of Cytochrome P450s (P450ome) in Notable Aspergillus spp. Life 2022, 12, 451. https://doi.org/10.3390/life12030451
Dauda WP, Abraham P, Glen E, Adetunji CO, Ghazanfar S, Ali S, Al-Zahrani M, Azameti MK, Alao SEL, Zarafi AB, et al. Robust Profiling of Cytochrome P450s (P450ome) in Notable Aspergillus spp. Life. 2022; 12(3):451. https://doi.org/10.3390/life12030451
Chicago/Turabian StyleDauda, Wadzani Palnam, Peter Abraham, Elkanah Glen, Charles Oluwaseun Adetunji, Shakira Ghazanfar, Shafaqat Ali, Majid Al-Zahrani, Mawuli Kwamla Azameti, Sheik Emmanuel Laykay Alao, Afiniki Bawa Zarafi, and et al. 2022. "Robust Profiling of Cytochrome P450s (P450ome) in Notable Aspergillus spp." Life 12, no. 3: 451. https://doi.org/10.3390/life12030451
APA StyleDauda, W. P., Abraham, P., Glen, E., Adetunji, C. O., Ghazanfar, S., Ali, S., Al-Zahrani, M., Azameti, M. K., Alao, S. E. L., Zarafi, A. B., Abraham, M. P., & Musa, H. (2022). Robust Profiling of Cytochrome P450s (P450ome) in Notable Aspergillus spp. Life, 12(3), 451. https://doi.org/10.3390/life12030451








