The Highest Density of Phosphorylated Histone H1 Appeared in Prophase and Prometaphase in Parallel with Reduced H3K9me3, and HDAC1 Depletion Increased H1.2/H1.3 and H1.4 Serine 38 Phosphorylation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cultivation of Mouse Embryonic Stem Cells and Tumor Cells
2.2. Immunodetection and Confocal Microscopy of Histone Markers
2.3. Proteomic Analysis of Histone H1 Phosphorylation Status
2.3.1. Histone Preparation for Liquid Chromatography-Tandem Mass Spectrometry LC–MS/MS Analysis
2.3.2. LC–MS/MS Analysis, Database Search, and Data Evaluation
3. Results
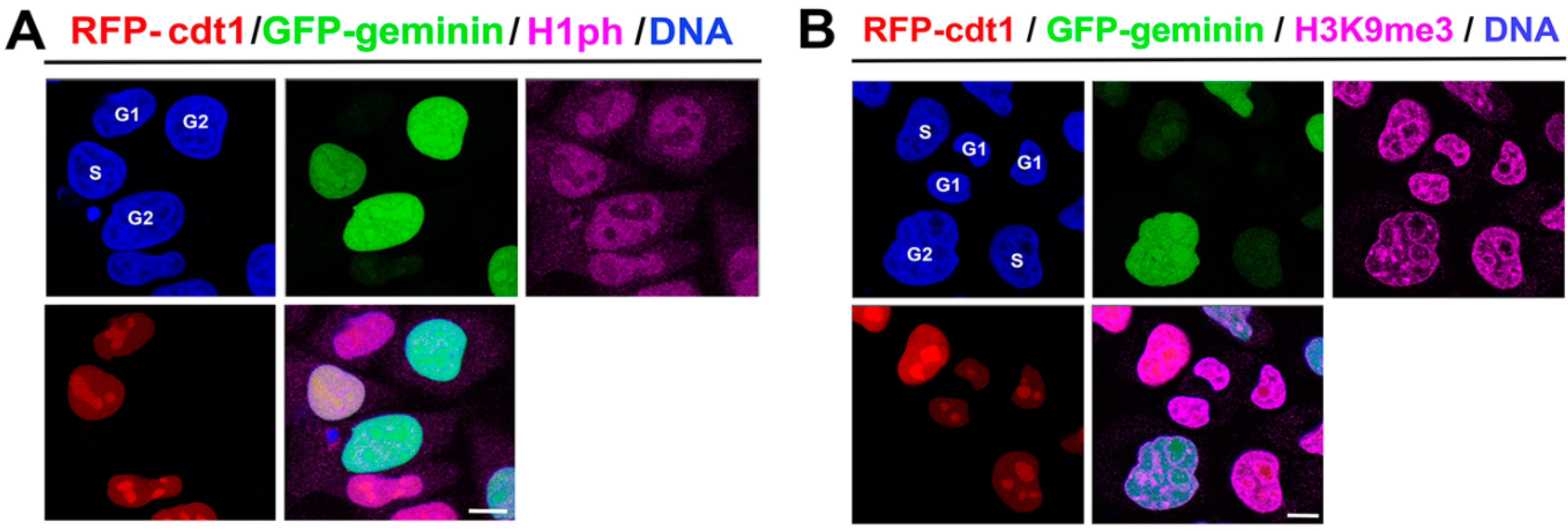
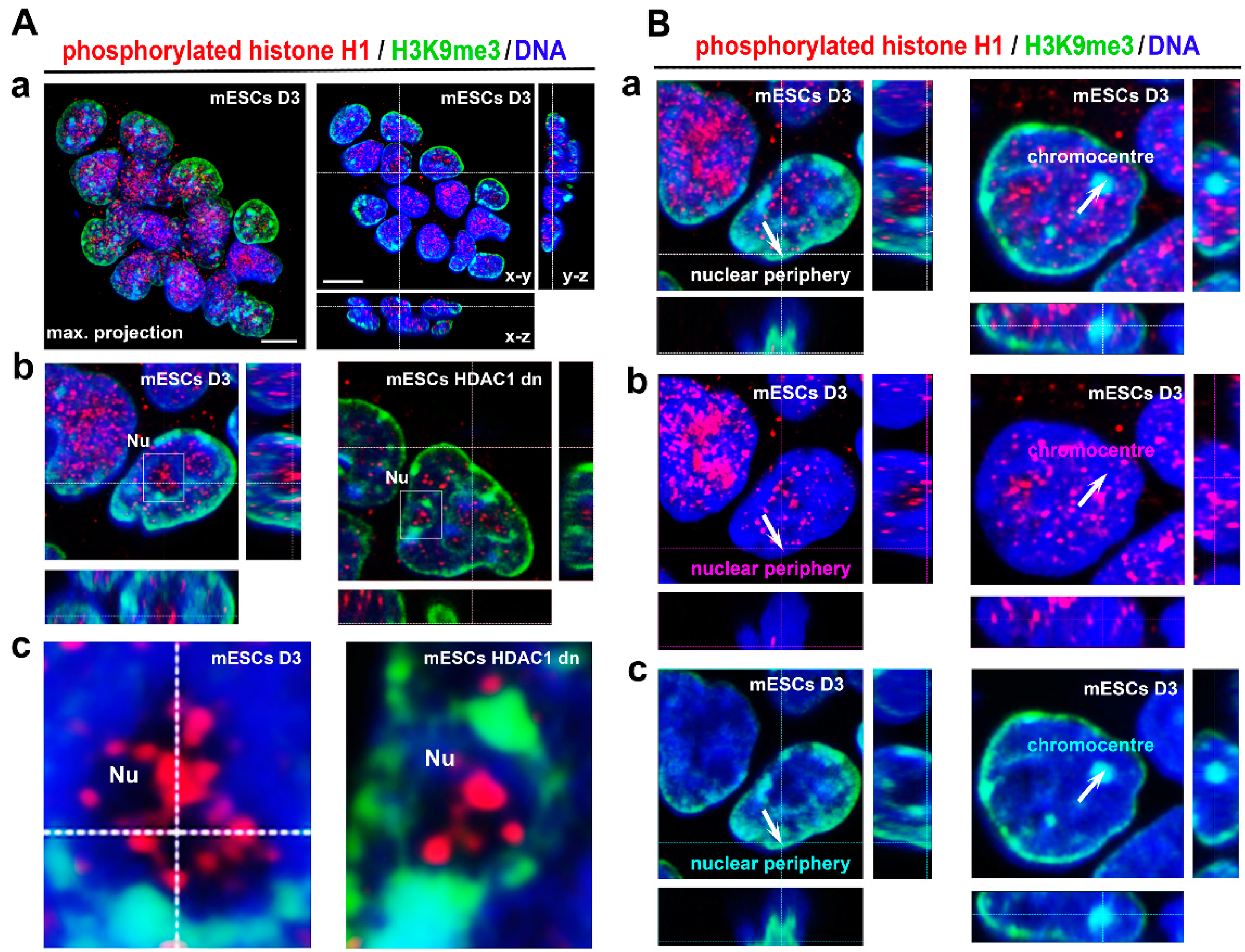
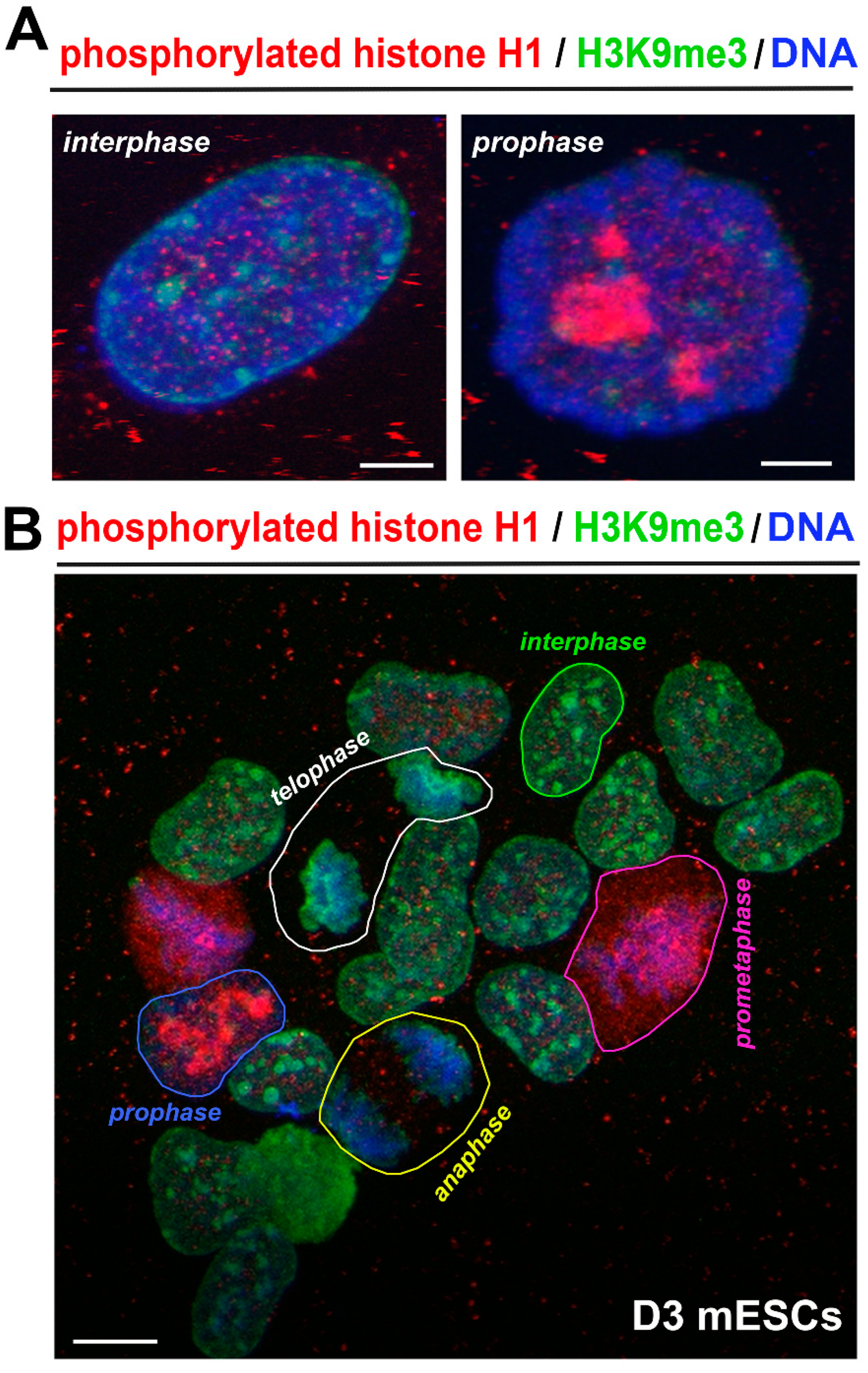
3.1. Localization of Phosphorylated Histone H1 and H3K9me3 Is Cell Cycle Specific
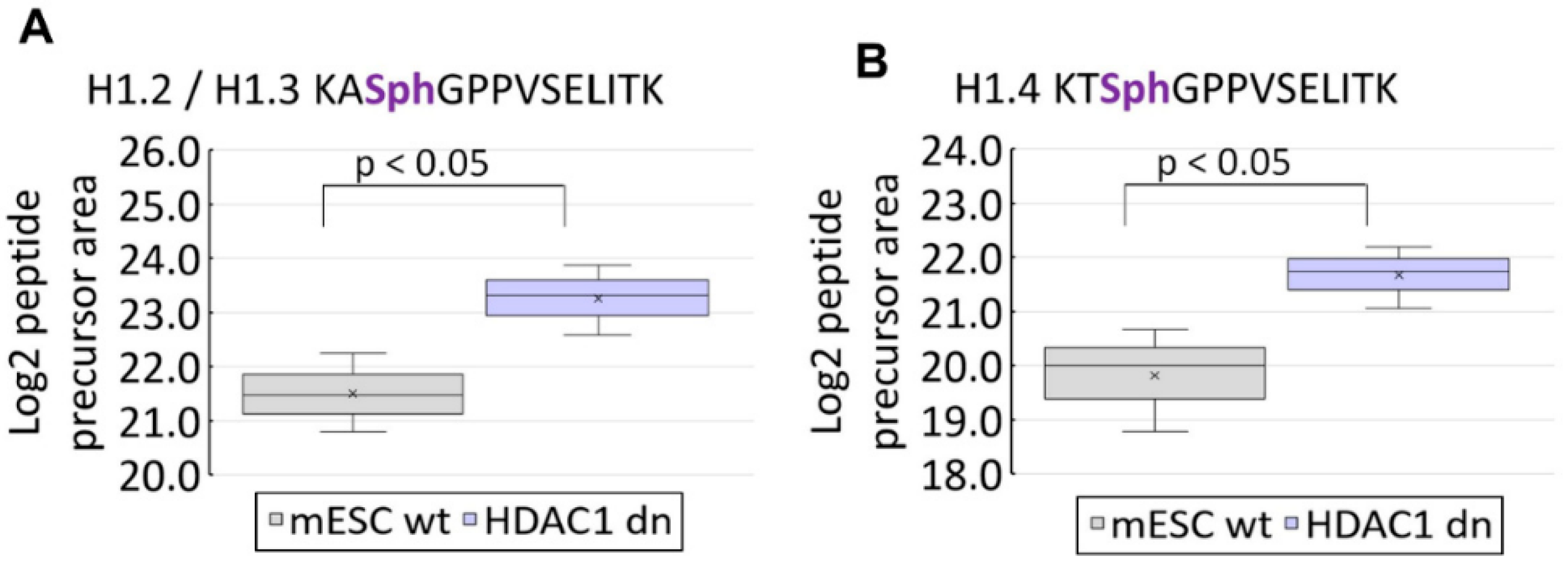
3.2. Post-Translational Modifications of Histone H1 Studied by Mass Spectrometry
4. Discussion
5. Conclusions and Future Directions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Thoma, F.; Koller, T.; Klug, A. Involvement of histone H1 in the organization of the nucleosome and of the salt-dependent superstructures of chromatin. J. Cell Biol. 1979, 83, 403–427. [Google Scholar] [CrossRef] [PubMed]
- Ramakrishnan, V.; Finch, J.T.; Graziano, V.; Lee, P.L.; Sweet, R.M. Crystal structure of globular domain of histone H5 and its implications for nucleosome binding. Nature 1993, 362, 219–223. [Google Scholar] [CrossRef] [PubMed]
- Garcia, B.A.; Busby, S.A.; Barber, C.M.; Shabanowitz, J.; Allis, C.D.; Hunt, D.F. Characterization of phosphorylation sites on histone H1 isoforms by tandem mass spectrometry. J. Proteome Res. 2004, 3, 1219–1227. [Google Scholar] [CrossRef] [PubMed]
- Happel, N.; Doenecke, D. Histone H1 and its isoforms: Contribution to chromatin structure and function. Gene 2009, 431, 1–12. [Google Scholar] [CrossRef]
- Millan-Arino, L.; Islam, A.B.; Izquierdo-Bouldstridge, A.; Mayor, R.; Terme, J.M.; Luque, N.; Sancho, M.; Lopez-Bigas, N.; Jordan, A. Mapping of six somatic linker histone H1 variants in human breast cancer cells uncovers specific features of H1.2. Nucleic Acids Res. 2014, 42, 4474–4493. [Google Scholar] [CrossRef]
- Li, J.Y.; Patterson, M.; Mikkola, H.K.; Lowry, W.E.; Kurdistani, S.K. Dynamic distribution of linker histone H1.5 in cellular differentiation. PLoS Genet. 2012, 8, e1002879. [Google Scholar] [CrossRef]
- Izzo, A.; Schneider, R. The role of linker histone H1 modifications in the regulation of gene expression and chromatin dynamics. Biochim. Biophys. Acta 2016, 1859, 486–495. [Google Scholar] [CrossRef]
- Gurley, L.R.; Valdez, J.G.; Buchanan, J.S. Characterization of the mitotic specific phosphorylation site of histone H1. Absence of a consensus sequence for the p34cdc2/cyclin B kinase. J. Biol. Chem. 1995, 270, 27653–27660. [Google Scholar] [CrossRef] [Green Version]
- Talasz, H.; Helliger, W.; Puschendorf, B.; Lindner, H. In vivo phosphorylation of histone H1 variants during the cell cycle. Biochemistry 1996, 35, 1761–1767. [Google Scholar] [CrossRef]
- Kratzmeier, M.; Albig, W.; Hanecke, K.; Doenecke, D. Rapid dephosphorylation of H1 histones after apoptosis induction. J. Biol. Chem. 2000, 275, 30478–30486. [Google Scholar] [CrossRef] [Green Version]
- Kim, K.; Jeong, K.W.; Kim, H.; Choi, J.; Lu, W.; Stallcup, M.R.; An, W. Functional interplay between p53 acetylation and H1.2 phosphorylation in p53-regulated transcription. Oncogene 2012, 31, 4290–4301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chubb, J.E.; Rea, S. Core and linker histone modifications involved in the DNA damage response. Subcell. Biochem. 2010, 50, 17–42. [Google Scholar] [CrossRef] [PubMed]
- Ohe, Y.; Hayashi, H.; Iwai, K. Human spleen histone H1. Isolation and amino acid sequence of a main variant, H1b. J. Biochem. 1986, 100, 359–368. [Google Scholar] [CrossRef] [PubMed]
- Kuzmichev, A.; Jenuwein, T.; Tempst, P.; Reinberg, D. Different EZH2-containing complexes target methylation of histone H1 or nucleosomal histone H3. Mol. Cell 2004, 14, 183–193. [Google Scholar] [CrossRef]
- Terme, J.M.; Millan-Arino, L.; Mayor, R.; Luque, N.; Izquierdo-Bouldstridge, A.; Bustillos, A.; Sampaio, C.; Canes, J.; Font, I.; Sima, N.; et al. Dynamics and dispensability of variant-specific histone H1 Lys-26/Ser-27 and Thr-165 post-translational modifications. FEBS Lett. 2014, 588, 2353–2362. [Google Scholar] [CrossRef] [Green Version]
- Kamieniarz, K.; Izzo, A.; Dundr, M.; Tropberger, P.; Ozretic, L.; Kirfel, J.; Scheer, E.; Tropel, P.; Wisniewski, J.R.; Tora, L.; et al. A dual role of linker histone H1.4 Lys 34 acetylation in transcriptional activation. Genes Dev. 2012, 26, 797–802. [Google Scholar] [CrossRef] [Green Version]
- Doetschman, T.C.; Eistetter, H.; Katz, M.; Schmidt, W.; Kemler, R. The in vitro development of blastocyst-derived embryonic stem cell lines: Formation of visceral yolk sac, blood islands and myocardium. J. Embryol. Exp. Morphol. 1985, 87, 27–45. [Google Scholar] [CrossRef]
- Zupkovitz, G.; Grausenburger, R.; Brunmeir, R.; Senese, S.; Tischler, J.; Jurkin, J.; Rembold, M.; Meunier, D.; Egger, G.; Lagger, S.; et al. The cyclin-dependent kinase inhibitor p21 is a crucial target for histone deacetylase 1 as a regulator of cellular proliferation. Mol. Cell. Biol. 2010, 30, 1171–1181. [Google Scholar] [CrossRef] [Green Version]
- Vecera, J.; Bartova, E.; Krejci, J.; Legartova, S.; Komurkova, D.; Ruda-Kucerova, J.; Stark, T.; Drazanova, E.; Kasparek, T.; Sulcova, A.; et al. HDAC1 and HDAC3 underlie dynamic H3K9 acetylation during embryonic neurogenesis and in schizophrenia-like animals. J. Cell. Physiol. 2018, 233, 530–548. [Google Scholar] [CrossRef]
- Sakaue-Sawano, A.; Ohtawa, K.; Hama, H.; Kawano, M.; Ogawa, M.; Miyawaki, A. Tracing the silhouette of individual cells in S/G2/M phases with fluorescence. Chem. Biol. 2008, 15, 1243–1248. [Google Scholar] [CrossRef] [Green Version]
- Bartova, E.; Pachernik, J.; Harnicarova, A.; Kovarik, A.; Kovarikova, M.; Hofmanova, J.; Skalnikova, M.; Kozubek, M.; Kozubek, S. Nuclear levels and patterns of histone H3 modification and HP1 proteins after inhibition of histone deacetylases. J. Cell Sci. 2005, 118, 5035–5046. [Google Scholar] [CrossRef] [Green Version]
- Cincarova, L.; Lochmanova, G.; Novakova, K.; Sultesova, P.; Konecna, H.; Fajkusova, L.; Fajkus, J.; Zdrahal, Z. A combined approach for the study of histone deacetylase inhibitors. Mol. Biosyst. 2012, 8, 2937–2945. [Google Scholar] [CrossRef]
- Perez-Riverol, Y.; Bai, J.; Bandla, C.; Garcia-Seisdedos, D.; Hewapathirana, S.; Kamatchinathan, S.; Kundu, D.J.; Prakash, A.; Frericks-Zipper, A.; Eisenacher, M.; et al. The PRIDE database resources in 2022: A hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 2022, 50, D543–D552. [Google Scholar] [CrossRef] [PubMed]
- Di Liegro, C.M.; Schiera, G.; Di Liegro, I. H1.0 Linker Histone as an Epigenetic Regulator of Cell Proliferation and Differentiation. Genes 2018, 9, 310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okuwaki, M.; Abe, M.; Hisaoka, M.; Nagata, K. Regulation of Cellular Dynamics and Chromosomal Binding Site Preference of Linker Histones H1.0 and H1.X. Mol. Cell. Biol. 2016, 36, 2681–2696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, Y.; John, S.; Pesavento, J.J.; Schultz-Norton, J.R.; Schiltz, R.L.; Baek, S.; Nardulli, A.M.; Hager, G.L.; Kelleher, N.L.; Mizzen, C.A. Histone H1 phosphorylation is associated with transcription by RNA polymerases I and II. J. Cell. Biol. 2010, 189, 407–415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, D.J.; Martin, C.M.; Sun, Z.; Cagliero, C.; Zhou, Y.N. Nucleolus-like compartmentalization of the transcription machinery in fast-growing bacterial cells. Crit. Rev. Biochem. Mol. Biol. 2017, 52, 96–106. [Google Scholar] [CrossRef] [Green Version]
- Sarg, B.; Helliger, W.; Talasz, H.; Forg, B.; Lindner, H.H. Histone H1 phosphorylation occurs site-specifically during interphase and mitosis: Identification of a novel phosphorylation site on histone H1. J. Biol. Chem. 2006, 281, 6573–6580. [Google Scholar] [CrossRef] [Green Version]
- Green, A.; Sarg, B.; Green, H.; Lonn, A.; Lindner, H.H.; Rundquist, I. Histone H1 interphase phosphorylation becomes largely established in G1 or early S phase and differs in G1 between T-lymphoblastoid cells and normal T cells. Epigenet. Chromatin 2011, 4, 15. [Google Scholar] [CrossRef] [Green Version]
- Talasz, H.; Sarg, B.; Lindner, H.H. Site-specifically phosphorylated forms of H1.5 and H1.2 localized at distinct regions of the nucleus are related to different processes during the cell cycle. Chromosoma 2009, 118, 693–709. [Google Scholar] [CrossRef]
- Starkova, T.Y.; Polyanichko, A.M.; Artamonova, T.O.; Khodorkovskii, M.A.; Kostyleva, E.I.; Chikhirzhina, E.V.; Tomilin, A.N. Post-translational modifications of linker histone H1 variants in mammals. Phys. Biol. 2017, 14, 016005. [Google Scholar] [CrossRef] [PubMed]
- van Noort, V.; Seebacher, J.; Bader, S.; Mohammed, S.; Vonkova, I.; Betts, M.J.; Kuhner, S.; Kumar, R.; Maier, T.; O’Flaherty, M.; et al. Cross-talk between phosphorylation and lysine acetylation in a genome-reduced bacterium. Mol. Syst. Biol. 2012, 8, 571. [Google Scholar] [CrossRef] [PubMed]
- Uhart, M.; Bustos, D.M. Human 14-3-3 paralogs differences uncovered by cross-talk of phosphorylation and lysine acetylation. PLoS ONE 2013, 8, e55703. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Happel, N.; Warneboldt, J.; Hanecke, K.; Haller, F.; Doenecke, D. H1 subtype expression during cell proliferation and growth arrest. Cell Cycle 2009, 8, 2226–2232. [Google Scholar] [CrossRef] [Green Version]
- Sadoni, N.; Langer, S.; Fauth, C.; Bernardi, G.; Cremer, T.; Turner, B.M.; Zink, D. Nuclear organization of mammalian genomes. Polar chromosome territories build up functionally distinct higher order compartments. J. Cell. Biol. 1999, 146, 1211–1226. [Google Scholar] [CrossRef] [Green Version]

| Resolution (µm) | Value (µm) for 63×/1.4 Objective for Wavelengths | ||
|---|---|---|---|
| λex. = 358 nm | λex. = 554 nm | λex. = 649 nm | |
| Lateral resolution | 0.17 | 0.20 | 0.24 |
| Axial resolution | 0.50 | 0.60 | 0.70 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Legartová, S.; Lochmanová, G.; Bártová, E. The Highest Density of Phosphorylated Histone H1 Appeared in Prophase and Prometaphase in Parallel with Reduced H3K9me3, and HDAC1 Depletion Increased H1.2/H1.3 and H1.4 Serine 38 Phosphorylation. Life 2022, 12, 798. https://doi.org/10.3390/life12060798
Legartová S, Lochmanová G, Bártová E. The Highest Density of Phosphorylated Histone H1 Appeared in Prophase and Prometaphase in Parallel with Reduced H3K9me3, and HDAC1 Depletion Increased H1.2/H1.3 and H1.4 Serine 38 Phosphorylation. Life. 2022; 12(6):798. https://doi.org/10.3390/life12060798
Chicago/Turabian StyleLegartová, Soňa, Gabriela Lochmanová, and Eva Bártová. 2022. "The Highest Density of Phosphorylated Histone H1 Appeared in Prophase and Prometaphase in Parallel with Reduced H3K9me3, and HDAC1 Depletion Increased H1.2/H1.3 and H1.4 Serine 38 Phosphorylation" Life 12, no. 6: 798. https://doi.org/10.3390/life12060798
APA StyleLegartová, S., Lochmanová, G., & Bártová, E. (2022). The Highest Density of Phosphorylated Histone H1 Appeared in Prophase and Prometaphase in Parallel with Reduced H3K9me3, and HDAC1 Depletion Increased H1.2/H1.3 and H1.4 Serine 38 Phosphorylation. Life, 12(6), 798. https://doi.org/10.3390/life12060798







