The Endoplasmic Reticulum Cargo Receptor FgErv14 Regulates DON Production, Growth and Virulence in Fusarium graminearum
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fungal Strains and Growth Conditions
2.2. Assessment of Conidia Germination and Perithecial Development
2.3. Plant Infection Assays
2.4. DON Production and Quantitative Real-Time PCR (qRT-PCR) Analysis
2.5. Microscopic Observations
2.6. Statistical Analyses
3. Results
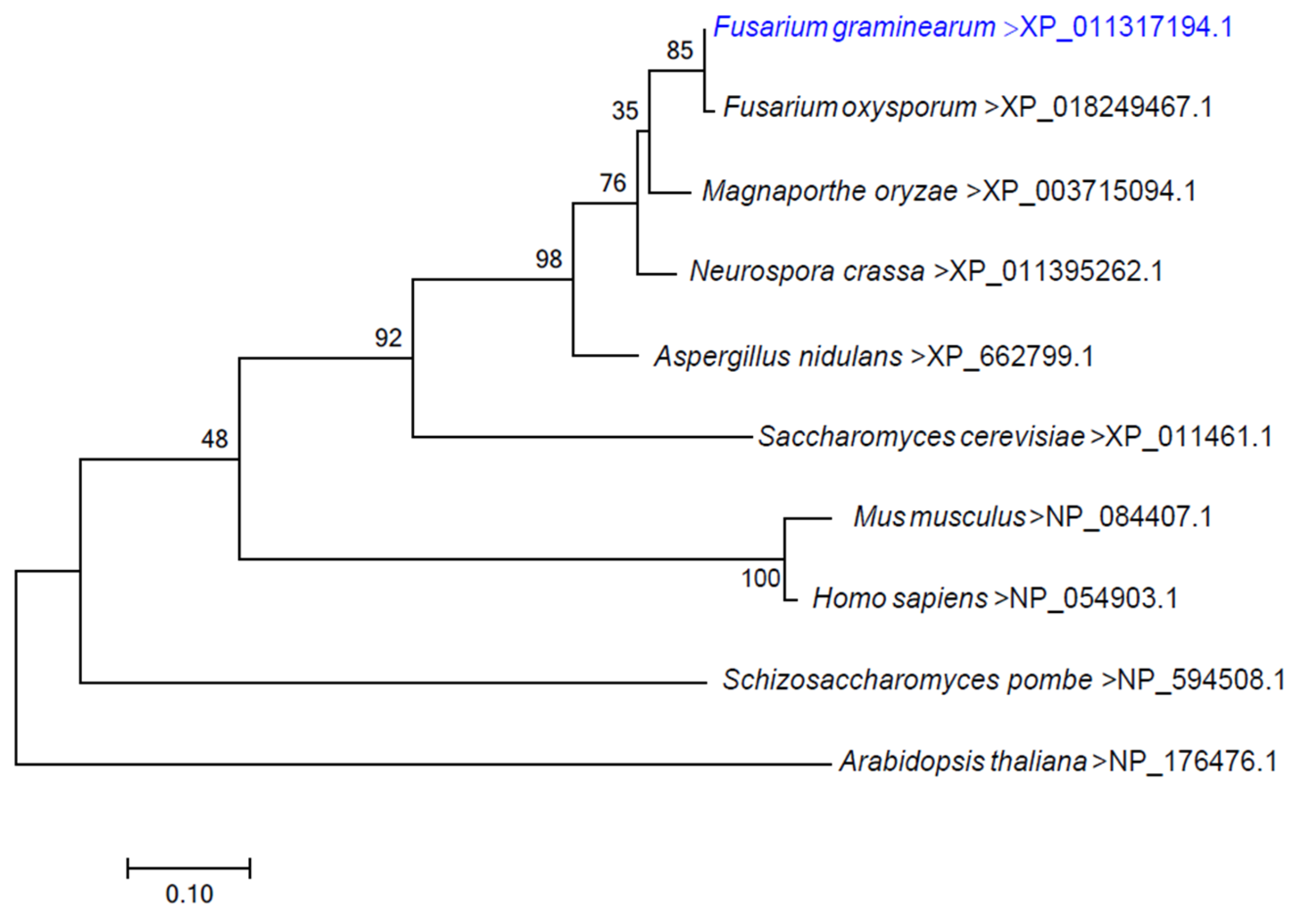
3.1. Identification and Knockout of the FgErv14 Protein in F. graminearum
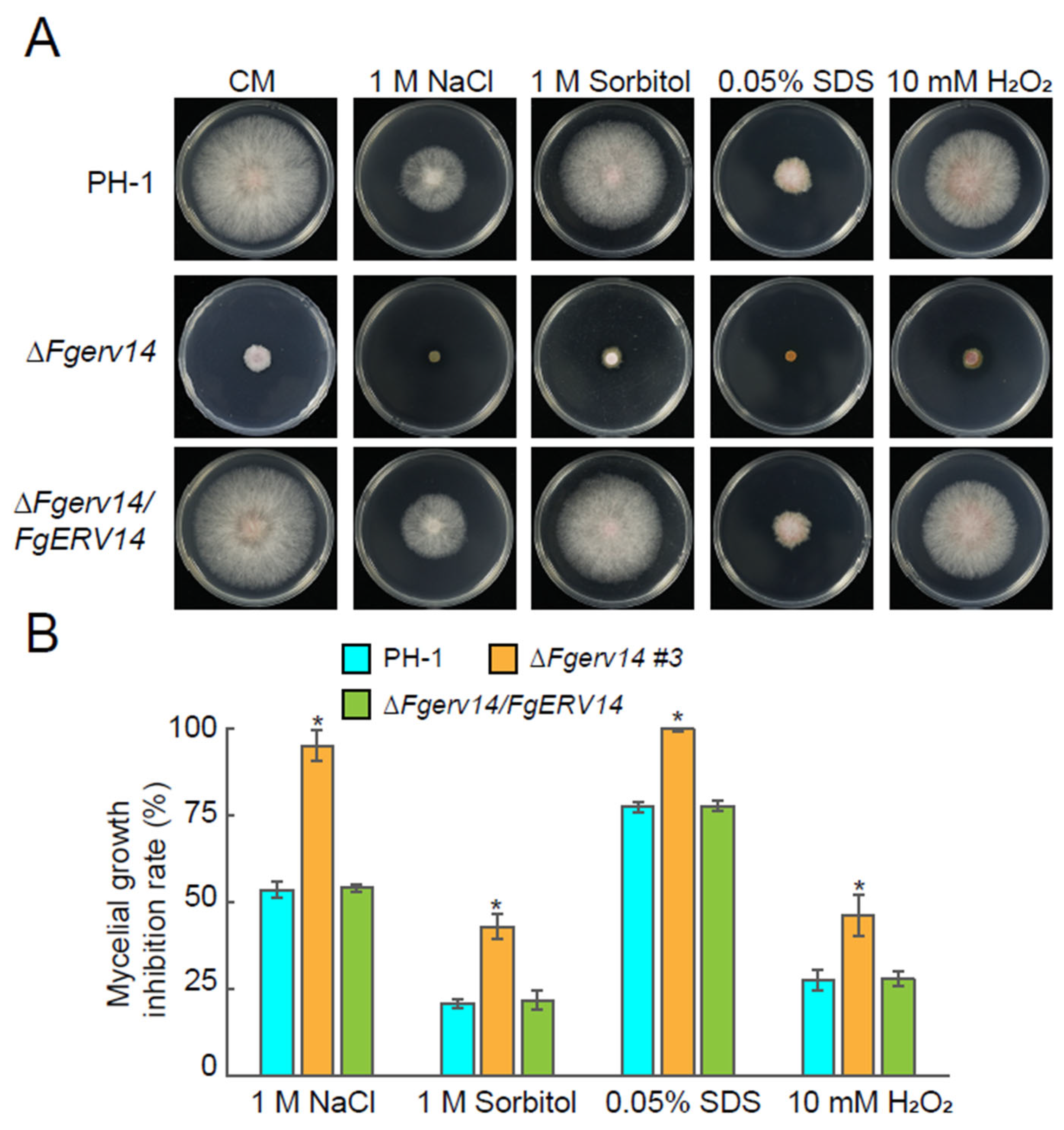
3.2. Involvement of FgErv14 in Vegetative Growth and Stress Resistance in F. graminearum
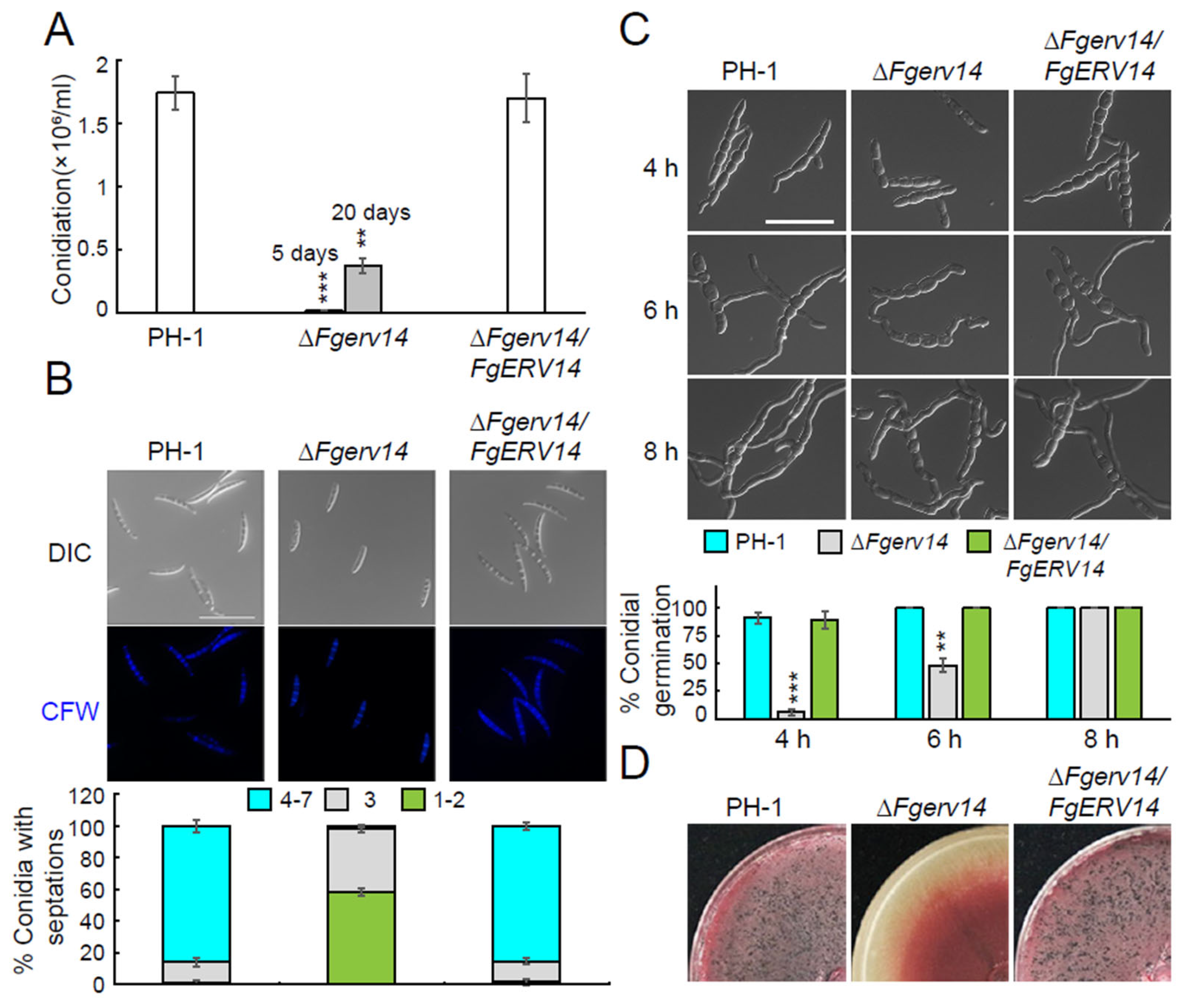
3.3. FgErv14 Controls F. graminearum Asexual and Sexual Reproduction
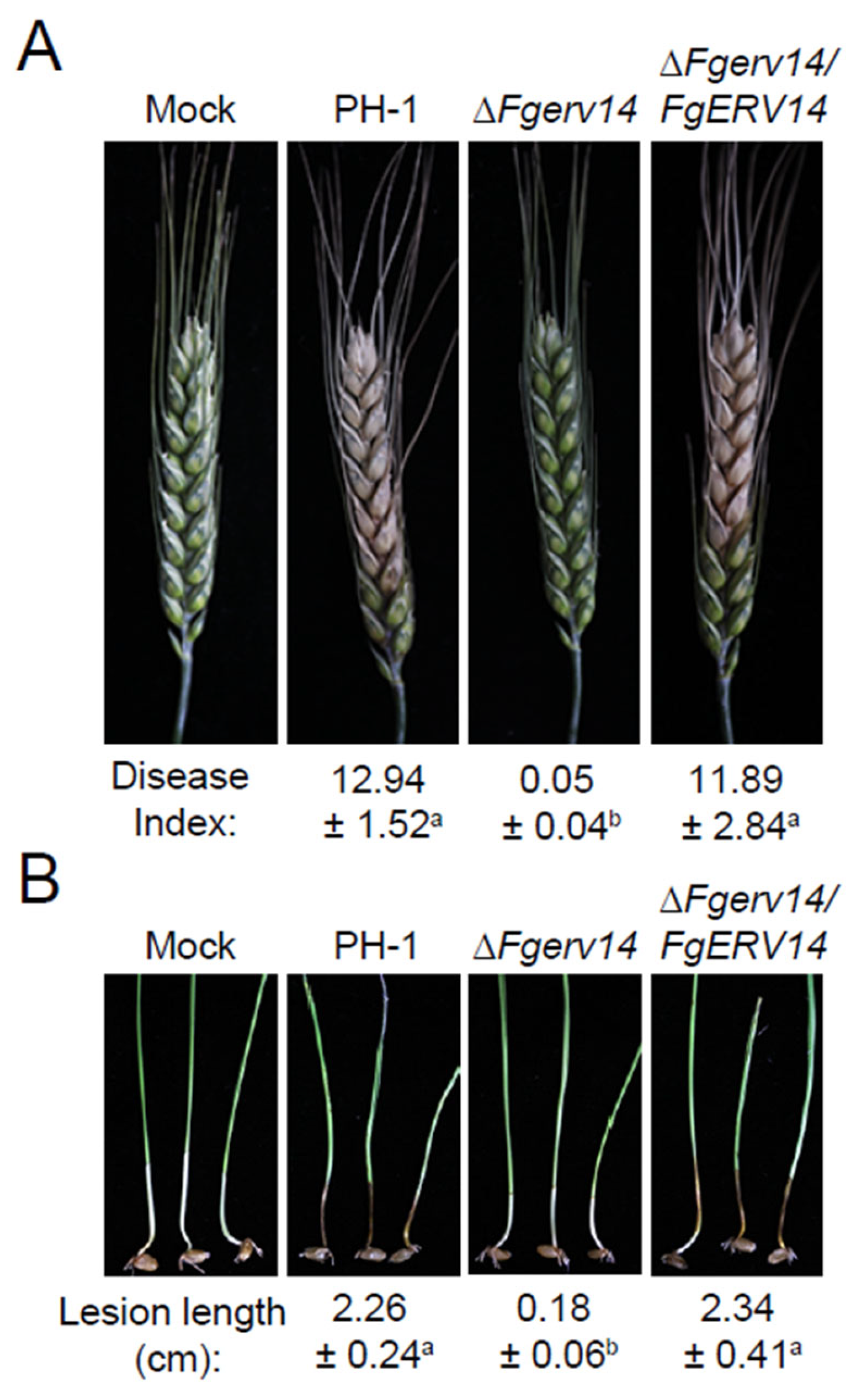
3.4. FgErv14 Is Important for F. graminearum Virulence
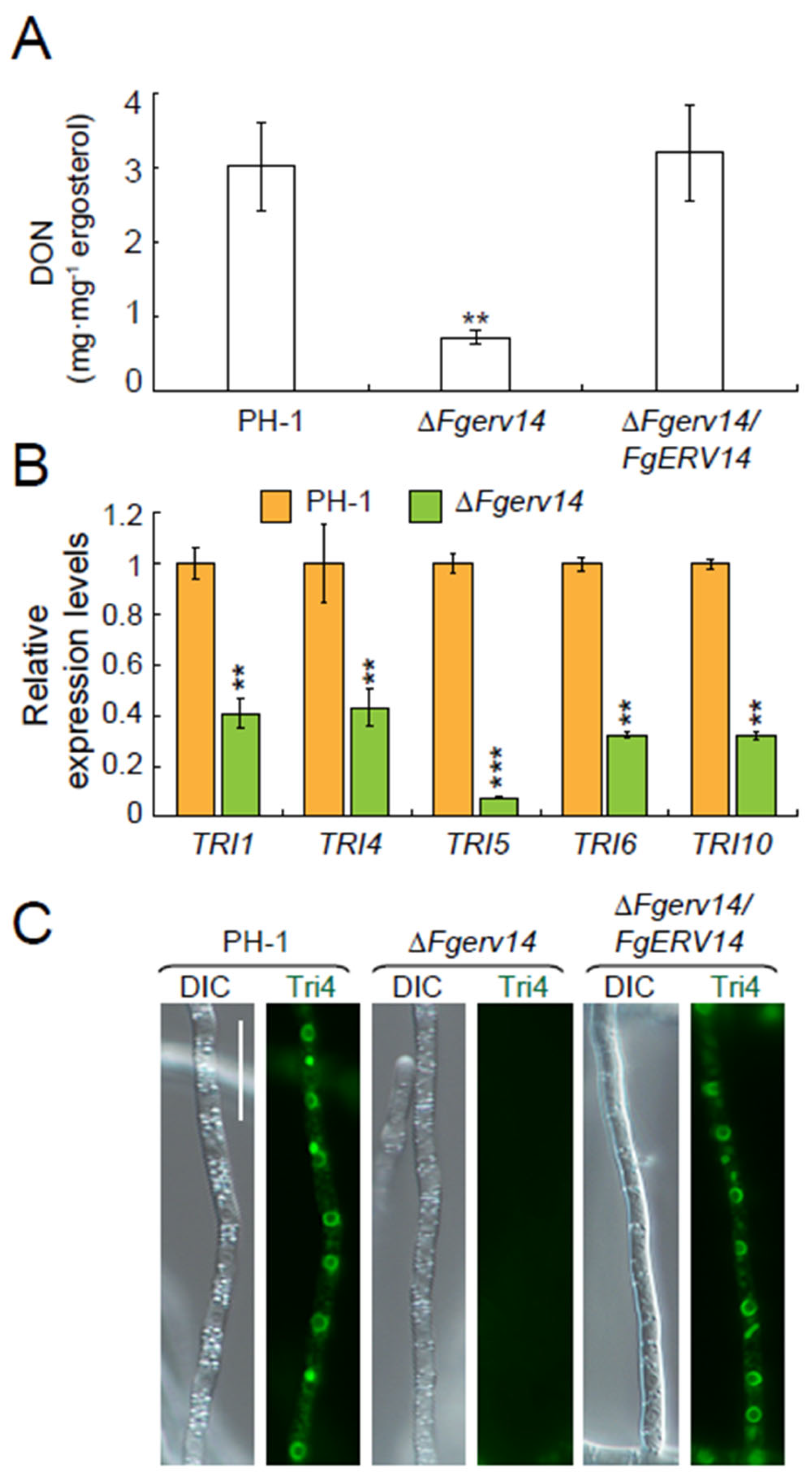
3.5. FgErv14 Is Necessary for the Production of DON in F. graminearum
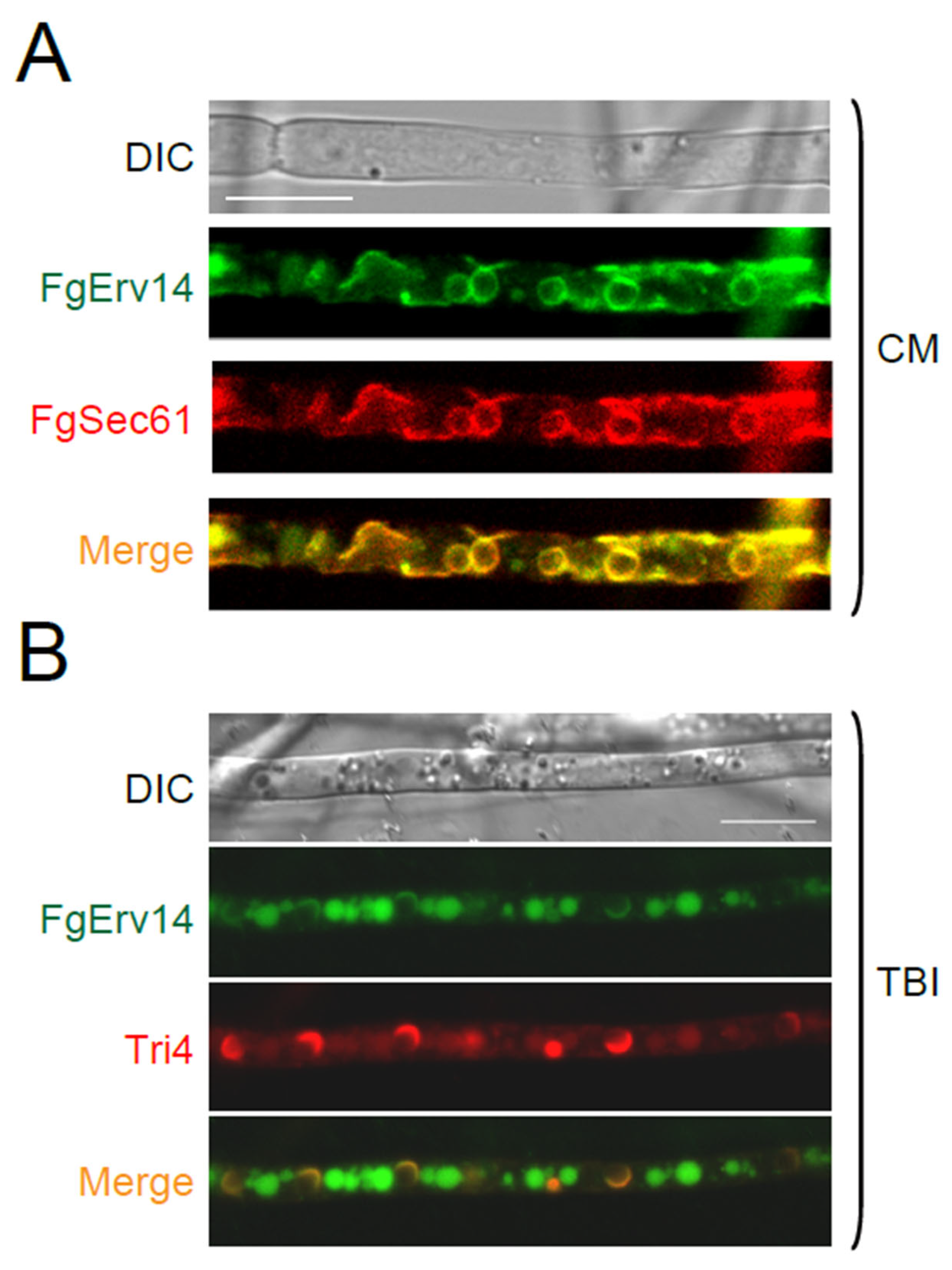
3.6. FgErv14 Is Mainly Localized in the ER and Toxisome in F. graminearum
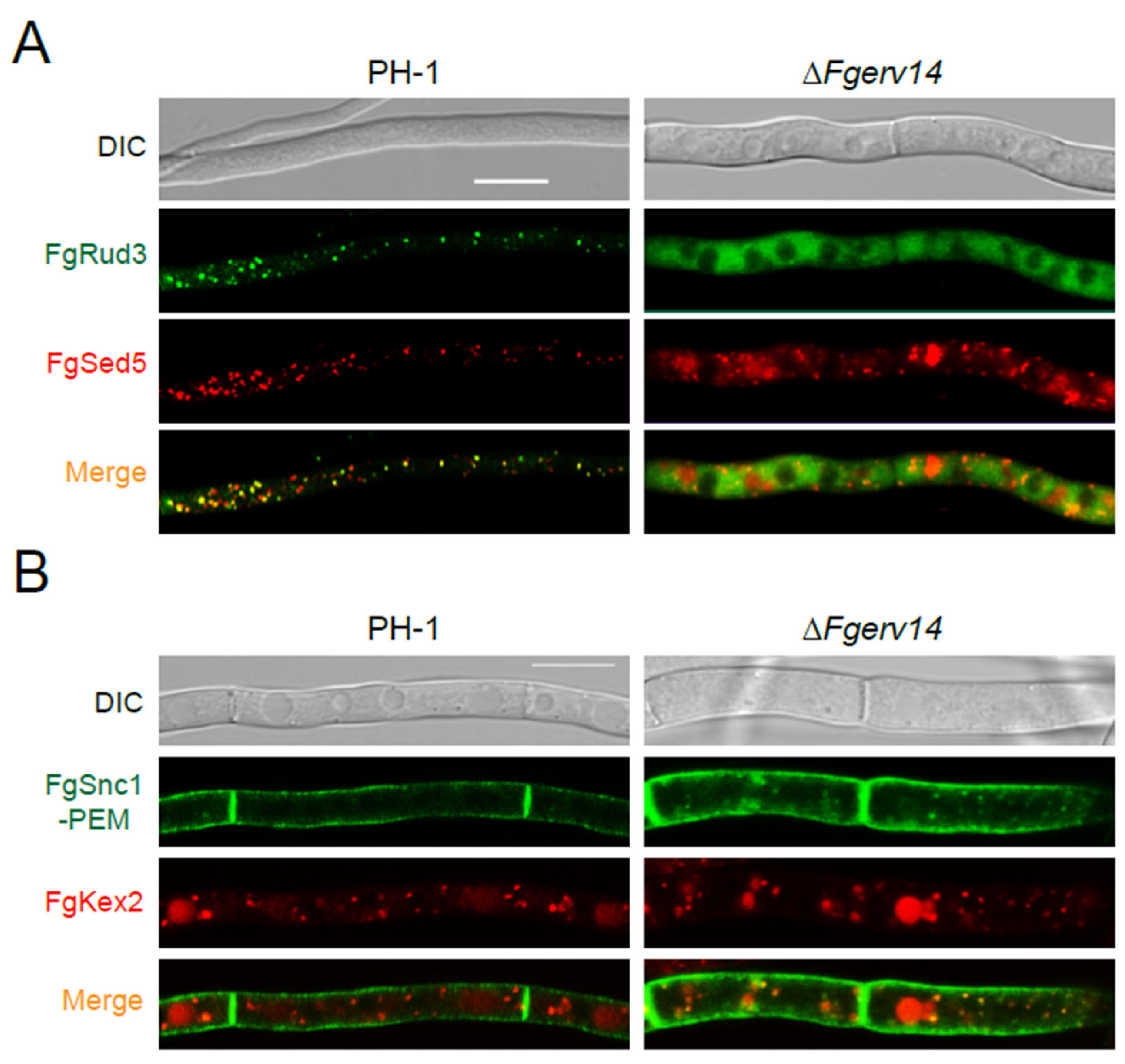
3.7. FgErv14 Is Required for FgRud3 and FgSnc1-PEM Localization in F. graminearum
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kazan, K.; Gardiner, D.M. Transcriptomics of cereal-Fusarium graminearum interactions: What we have learned so far. Mol. Plant Pathol. 2018, 19, 764–778. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boddu, J.; Cho, S.; Kruger, W.M.; Muehlbauer, G.J. Transcriptome analysis of the barley-Fusarium graminearum interaction. Mol. Plant-Microbe Interact. 2006, 19, 407–417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Guo, Y.; Ma, C.; Zhang, D.; Wang, C.; Yang, Q. Transcriptome analysis of maize resistance to Fusarium graminearum. BMC Genom. 2016, 17, 477. [Google Scholar]
- Oghenekaro, A.O.; Oviedo-Ludena, M.A.; Serajazari, M.; Wang, X.; Henriquez, M.A.; Wenner, N.G.; Kuldau, G.A.; Navabi, A.; Kutcher, H.R.; Fernando, W.G.D. Population genetic structure and chemotype diversity of Fusarium graminearum populations from wheat in canada and north eastern united states. Toxins 2021, 13, 180. [Google Scholar] [CrossRef]
- Xu, X.; Nicholson, P. Community ecology of fungal pathogens causing wheat head blight. Annu. Rev. Phytopathol. 2009, 47, 83–103. [Google Scholar] [CrossRef]
- Pestka, J.J. Deoxynivalenol: Mechanisms of action, human exposure, and toxicological relevance. Arch. Toxicol. 2010, 84, 663–679. [Google Scholar] [CrossRef]
- Desjardins, A.E.; Proctor, R.H. Molecular biology of Fusarium mycotoxins. Int. J. Food Microbiol. 2007, 119, 47–50. [Google Scholar] [CrossRef]
- Paul, P.A.; Lipps, P.E.; Hershman, D.E.; McMullen, M.P.; Draper, M.A.; Madden, L.V. Efficacy of triazole-based fungicides for Fusarium head blight and deoxynivalenol control in wheat: A multivariate meta-analysis. Phytopathology 2008, 98, 999–1011. [Google Scholar] [CrossRef] [Green Version]
- Zhou, F.; Li, D.X.; Hu, H.Y.; Song, Y.L.; Fan, Y.C.; Guan, Y.Y.; Song, P.W.; Wei, Q.C.; Yan, H.F.; Li, C.W. Biological characteristics and molecular mechanisms of fludioxonil resistance in Fusarium graminearum in China. Plant Dis. 2020, 104, 2426–2433. [Google Scholar] [CrossRef]
- Bonnighausen, J.; Schauer, N.; Schafer, W.; Bormann, J. Metabolic profiling of wheat rachis node infection by Fusarium graminearum-decoding deoxynivalenol-dependent susceptibility. New Phytol. 2019, 221, 459–469. [Google Scholar] [CrossRef] [Green Version]
- Jansen, C.; von Wettstein, D.; Schafer, W.; Kogel, K.H.; Felk, A.; Maier, F.J. Infection patterns in barley and wheat spikes inoculated with wild-type and trichodiene synthase gene disrupted Fusarium graminearum. Proc. Natl. Acad. Sci. USA 2005, 102, 16892–16897. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malachova, A.; Stockova, L.; Wakker, A.; Varga, E.; Krska, R.; Michlmayr, H.; Adam, G.; Berthiller, F. Critical evaluation of indirect methods for the determination of deoxynivalenol and its conjugated forms in cereals. Anal. Bioanal. Chem. 2015, 407, 6009–6020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mishra, S.; Dixit, S.; Dwivedi, P.D.; Pandey, H.P.; Das, M. Influence of temperature and pH on the degradation of deoxynivalenol (DON) in aqueous medium: Comparative cytotoxicity of DON and degraded product. Food Addit. Contam. Part A Chem. Anal. Control. Expo. Risk Assess. 2014, 31, 121–131. [Google Scholar] [CrossRef]
- Wang, H.; Sun, S.; Ge, W.; Zhao, L.; Hou, B.; Wang, K.; Lyu, Z.; Chen, L.; Xu, S.; Guo, J.; et al. Horizontal gene transfer of Fhb7 from fungus underlies Fusarium head blight resistance in wheat. Science 2020, 368, eaba5435. [Google Scholar] [CrossRef] [PubMed]
- Brown, D.W.; Dyer, R.B.; McCormick, S.P.; Kendra, D.F.; Plattner, R.D. Functional demarcation of the Fusarium core trichothecene gene cluster. Fungal Genet. Biol. 2004, 41, 454–462. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Kistler, H.C.; Ma, Z. Fusarium graminearum trichothecene mycotoxins: Biosynthesis, regulation, and management. Annu. Rev. Phytopathol. 2019, 57, 15–39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boenisch, M.J.; Broz, K.L.; Purvine, S.O.; Chrisler, W.B.; Nicora, C.D.; Connolly, L.R.; Freitag, M.; Baker, S.E.; Kistler, H.C. Structural reorganization of the fungal endoplasmic reticulum upon induction of mycotoxin biosynthesis. Sci. Rep. 2017, 7, 44296. [Google Scholar] [CrossRef] [Green Version]
- Tang, G.; Chen, Y.; Xu, J.R.; Kistler, H.C.; Ma, Z. The fungal myosin I is essential for Fusarium toxisome formation. PLoS Pathog. 2018, 14, e1006827. [Google Scholar] [CrossRef] [Green Version]
- Tokai, T.; Koshino, H.; Takahashi-Ando, N.; Sato, M.; Fujimura, M.; Kimura, M. Fusarium Tri4 encodes a key multifunctional cytochrome P450 monooxygenase for four consecutive oxygenation steps in trichothecene biosynthesis. Biochem. Biophys. Res. Commun. 2007, 353, 412–417. [Google Scholar] [CrossRef]
- McCormick, S.P.; Harris, L.J.; Alexander, N.J.; Ouellet, T.; Saparno, A.; Allard, S.; Desjardins, A.E. Tri1 in Fusarium graminearum encodes a P450 oxygenase. Appl. Environ. Microbiol. 2004, 70, 2044–2051. [Google Scholar] [CrossRef] [Green Version]
- Desjardins, A.E.; Bai, G.; Plattner, R.D.; Proctor, R.H. Analysis of aberrant virulence of Gibberella zeae following transformation-mediated complementation of a trichothecene-deficient (Tri5) mutant. Microbiology 2000, 146, 2059–2068. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Proctor, R.H.; Hohn, T.M.; McCormick, S.P. Restoration of wild-type virulence to Tri5 disruption mutants of Gibberella zeae via gene reversion and mutant complementation. Microbiology 1997, 143, 2583–2591. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nasmith, C.G.; Walkowiak, S.; Wang, L.; Leung, W.W.; Gong, Y.; Johnston, A.; Harris, L.J.; Guttman, D.S.; Subramaniam, R. Tri6 is a global transcription regulator in the phytopathogen Fusarium graminearum. PLoS Pathog. 2011, 7, e1002266. [Google Scholar] [CrossRef] [PubMed]
- Tag, A.G.; Garifullina, G.F.; Peplow, A.W.; Ake, C., Jr.; Phillips, T.D.; Hohn, T.M.; Beremand, M.N. A novel regulatory gene, Tri10, controls trichothecene toxin production and gene expression. Appl. Environ. Microbiol. 2001, 67, 5294–5302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Li, B.; Fang, Q.; Li, Y.; Zheng, X.; Zhang, Z. SNARE protein FgVam7 controls growth, asexual and sexual development, and plant infection in Fusarium graminearum. Mol. Plant Pathol. 2016, 17, 108–119. [Google Scholar] [CrossRef]
- Han, X.; Chen, L.; Li, W.; Zhang, L.; Zhang, L.; Zou, S.; Liang, Y.; Yu, J.; Dong, H. Endocytic FgEde1 regulates virulence and autophagy in Fusarium graminearum. Fungal Genet. Biol. 2020, 141, 103400. [Google Scholar] [CrossRef]
- Wang, C.; Wang, Y.; Zhang, L.; Yin, Z.; Liang, Y.; Chen, L.; Zou, S.; Dong, H. The Golgin Protein RUD3 Regulates Fusarium graminearum Growth and Virulence. Appl. Environ. Microbiol. 2021, 87, e02522-20. [Google Scholar] [CrossRef]
- Zheng, H.; Li, L.; Yu, Z.; Yuan, Y.; Zheng, Q.; Xie, Q.; Li, G.; Abubakar, Y.S.; Zhou, J.; Wang, Z.; et al. FgSpa2 recruits FgMsb3, a Rab8 GAP, to the polarisome to regulate polarized trafficking, growth and pathogenicity in Fusarium graminearum. New Phytol. 2021, 229, 1665–1683. [Google Scholar] [CrossRef]
- Li, B.; Dong, X.; Zhao, R.; Kou, R.; Zheng, X.; Zhang, H. The t-SNARE protein FgPep12, associated with FgVam7, is essential for ascospore discharge and plant infection by trafficking Ca2+ ATPase FgNeo1 between Golgi and endosome/vacuole in Fusarium graminearum. PLoS Pathog. 2019, 15, e1007754. [Google Scholar] [CrossRef]
- Zheng, H.; Zheng, W.; Wu, C.; Yang, J.; Xi, Y.; Xie, Q.; Zhao, X.; Deng, X.; Lu, G.; Li, G.; et al. Rab GTPases are essential for membrane trafficking-dependent growth and pathogenicity in Fusarium graminearum. Environ. Microbiol. 2015, 17, 4580–4599. [Google Scholar] [CrossRef]
- Barlowe, C. Coupled ER to Golgi transport reconstituted with purified cytosolic proteins. J. Cell Biol. 1997, 139, 1097–1108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, X.; Ballew, N.; Barlowe, C. Initial docking of ER-derived vesicles requires Uso1p and Ypt1p but is independent of SNARE proteins. EMBO J. 1998, 17, 2156–2165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, H.; Reinisch, K.; Ferro-Novick, S. Coats, tethers, Rabs, and SNAREs work together to mediate the intracellular destination of a transport vesicle. Dev. Cell 2007, 12, 671–682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, Y.; Zhang, M.; Li, J.; Yang, C.; Abubakar, Y.S.; Chen, X.; Zheng, W.; Wang, Z.; Zheng, H.; Zhou, J. The small GTPase FgRab1 plays indispensable roles in the vegetative growth, vesicle fusion, autophagy and pathogenicity of Fusarium graminearum. Int. J. Mol. Sci. 2022, 23, 895. [Google Scholar] [CrossRef]
- Mancias, J.D.; Goldberg, J. The transport signal on Sec22 for packaging into COPII-coated vesicles is a conformational epitope. Mol. Cell 2007, 26, 403–414. [Google Scholar] [CrossRef]
- Adnan, M.; Fang, W.; Sun, P.; Zheng, Y.; Abubakar, Y.S.; Zhang, J.; Lou, Y.; Zheng, W.; Lu, G.D. R-SNARE FgSec22 is essential for growth, pathogenicity and DON production of Fusarium graminearum. Curr. Genet. 2020, 66, 421–435. [Google Scholar] [CrossRef]
- Xie, C.; Shang, Q.; Mo, C.; Xiao, Y.; Wang, G.; Xie, J.; Jiang, D.; Xiao, X. Early Secretory pathway-associated proteins SsEmp24 and SsErv25 are involved in morphogenesis and pathogenicity in a filamentous phytopathogenic fungus. mBio 2021, 12, e0317321. [Google Scholar] [CrossRef]
- Powers, J.; Barlowe, C. Erv14p directs a transmembrane secretory protein into COPII-coated transport vesicles. Mol. Biol. Cell 2002, 13, 880–891. [Google Scholar] [CrossRef] [Green Version]
- Kuehn, M.J.; Herrmann, J.M.; Schekman, R. COPII-cargo interactions direct protein sorting into ER-derived transport vesicles. Nature 1998, 391, 187–190. [Google Scholar] [CrossRef]
- Muniz, M.; Nuoffer, C.; Hauri, H.P.; Riezman, H. The Emp24 complex recruits a specific cargo molecule into endoplasmic reticulum-derived vesicles. J. Cell Biol. 2000, 148, 925–930. [Google Scholar] [CrossRef] [Green Version]
- Dancourt, J.; Barlowe, C. Protein sorting receptors in the early secretory pathway. Annu. Rev. Biochem. 2010, 79, 777–802. [Google Scholar] [CrossRef] [PubMed]
- Nakanishi, H.; Suda, Y.; Neiman, A.M. Erv14 family cargo receptors are necessary for ER exit during sporulation in Saccharomyces cerevisiae. J. Cell Sci. 2007, 120, 908–916. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herzig, Y.; Sharpe, H.J.; Elbaz, Y.; Munro, S.; Schuldiner, M. A systematic approach to pair secretory cargo receptors with their cargo suggests a mechanism for cargo selection by Erv14. PLoS Biol. 2012, 10, e1001329. [Google Scholar] [CrossRef]
- Pagant, S.; Wu, A.; Edwards, S.; Diehl, F.; Miller, E.A. Sec24 is a coincidence detector that simultaneously binds two signals to drive ER export. Curr. Biol. 2015, 25, 403–412. [Google Scholar] [CrossRef] [Green Version]
- Gillingham, A.K.; Tong, A.H.; Boone, C.; Munro, S. The GTPase Arf1p and the ER to Golgi cargo receptor Erv14p cooperate to recruit the golgin Rud3p to the cis-Golgi. J. Cell Biol. 2004, 167, 281–292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Papouskova, K.; Moravcova, M.; Masrati, G.; Ben-Tal, N.; Sychrova, H.; Zimmermannova, O. C5 conserved region of hydrophilic C-terminal part of Saccharomyces cerevisiae Nha1 antiporter determines its requirement of Erv14 COPII cargo receptor for plasma-membrane targeting. Mol. Microbiol. 2021, 115, 41–57. [Google Scholar] [CrossRef]
- Zimmermannova, O.; Felcmanova, K.; Rosas-Santiago, P.; Papouskova, K.; Pantoja, O.; Sychrova, H. Erv14 cargo receptor participates in regulation of plasma-membrane potential, intracellular pH and potassium homeostasis via its interaction with K+-specific transporters Trk1 and Tok1. BBA-Mol. Cell Res. 2019, 1866, 1376–1388. [Google Scholar] [CrossRef]
- Wang, C.; Wang, Y.; Wang, Y.; Wang, Z.; Zhang, L.; Liang, Y.; Chen, L.; Zou, S.; Dong, H. The ADP-ribosylation factor-like small GTPase FgArl1 participates in growth, pathogenicity and DON production in Fusarium graminearum. Fungal Biol. 2020, 124, 969–980. [Google Scholar] [CrossRef]
- Tang, L.; Yu, X.; Zhang, L.; Zhang, L.; Chen, L.; Zou, S.; Liang, Y.; Yu, J.; Dong, H. Mitochondrial FgEch1 is responsible for conidiation and full virulence in Fusarium graminearum. Curr. Genet. 2020, 66, 361–371. [Google Scholar] [CrossRef]
- Chong, X.; Wang, C.; Wang, Y.; Wang, Y.; Zhang, L.; Liang, Y.; Chen, L.; Zou, S.; Dong, H. The dynamin-lke GTPase FgSey1 plays a critical role in fungal development and virulence in Fusarium graminearum. Appl. Environ. Microbiol. 2020, 86, e02720-19. [Google Scholar] [CrossRef]
- Fan, X.; He, F.; Ding, M.; Geng, C.; Chen, L.; Zou, S.; Liang, Y.; Yu, J.; Dong, H. Thioredoxin reductase is involved in development and pathogenicity in Fusarium graminearum. Front. Microbiol. 2019, 10, 393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cavinder, B.; Sikhakolli, U.; Fellows, K.M.; Trail, F. Sexual development and ascospore discharge in Fusarium graminearum. J. Vis. Exp. 2012, 61, 5407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, M.; Li, J.; Fan, X.; He, F.; Yu, X.; Chen, L.; Zou, S.; Liang, Y.; Yu, J. Aquaporin1 regulates development, secondary metabolism and stress responses in Fusarium graminearum. Curr. Genet. 2018, 64, 1057–1069. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Liu, L.; Li, Y.; Dong, X.; Zhang, H.; Chen, H.; Zheng, X.; Zhang, Z. The FgVps39-FgVam7-FgSso1 complex mediates vesicle trafficking and is important for the development and virulence of Fusarium graminearum. Mol. Plant-Microbe Interact. 2017, 30, 410–422. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Rosas-Santiago, P.; Zimmermannova, O.; Vera-Estrella, R.; Sychrova, H.; Pantoja, O. Erv14 cargo receptor participates in yeast salt tolerance via its interaction with the plasma-membrane Nha1 cation/proton antiporter. Biochim. Biophys. Acta 2016, 1858, 67–74. [Google Scholar] [CrossRef] [Green Version]
- Seong, K.Y.; Zhao, X.; Xu, J.R.; Guldener, U.; Kistler, H.C. Conidial germination in the filamentous fungus Fusarium graminearum. Fungal Genet. Biol. 2008, 45, 389–399. [Google Scholar] [CrossRef]
- Flynn, C.M.; Broz, K.; Jonkers, W.; Schmidt-Dannert, C.; Kistler, H.C. Expression of the Fusarium graminearum terpenome and involvement of the endoplasmic reticulum-derived toxisome. Fungal Genet. Biol. 2019, 124, 78–87. [Google Scholar] [CrossRef]
- Qian, B.; Su, X.; Ye, Z.; Liu, X.; Liu, M.; Shen, D.; Chen, H.; Zhang, H.; Wang, P.; Zhang, Z. MoErv29 promotes apoplastic effector secretion contributing to virulence of the rice blast fungus Magnaporthe oryzae. New Phytol. 2022, 233, 1289–1302. [Google Scholar] [CrossRef]
- Pinar, M.; Pantazopoulou, A.; Arst, H.N., Jr.; Penalva, M.A. Acute inactivation of the Aspergillus nidulans Golgi membrane fusion machinery: Correlation of apical extension arrest and tip swelling with cisternal disorganization. Mol. Microbiol. 2013, 89, 228–248. [Google Scholar] [CrossRef]
- Hernandez-Gonzalez, M.; Penalva, M.A.; Pantazopoulou, A. Conditional inactivation of Aspergillus nidulans sarA(SAR1) uncovers the morphogenetic potential of regulating endoplasmic reticulum (ER) exit. Mol. Microbiol. 2015, 95, 491–508. [Google Scholar] [CrossRef] [PubMed]
- McMullen, M.; Jones, R.; Gallenberg, D. Scab of wheat and barley: A re-emerging disease of devastating impact. Plant Dis. 1997, 81, 1340–1348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guenther, J.C.; Trail, F. The development and differentiation of Gibberella zeae (anamorph: Fusarium graminearum) during colonization of wheat. Mycologia 2005, 97, 229–237. [Google Scholar] [CrossRef] [PubMed]
- Shostak, K.; Bonner, C.; Sproule, A.; Thapa, I.; Shields, S.W.J.; Blackwell, B.; Vierula, J.; Overy, D.; Subramaniam, R. Activation of biosynthetic gene clusters by the global transcriptional regulator TRI6 in Fusarium graminearum. Mol. Microbiol. 2020, 114, 664–680. [Google Scholar] [CrossRef] [PubMed]
- Molnar, O.; Schatzmayr, G.; Fuchs, E.; Prillinger, H. Trichosporon mycotoxinivorans sp. nov., a new yeast species useful in biological detoxification of various mycotoxins. Syst. Appl. Microbiol. 2004, 27, 661–671. [Google Scholar] [CrossRef]
- Chen, L.; Zhang, L.; Mei, X.; Wang, C.; Guo, Z.; Li, L.; Li, B.; Liang, Y.; Zou, S.; Dong, H. The type II phosphoinositide 4-kinase FgLsb6 is important for the development and virulence of Fusarium graminearum. Fungal Genet. Biol. 2020, 144, 103443. [Google Scholar] [CrossRef] [PubMed]
- Castillon, G.A.; Watanabe, R.; Taylor, M.; Schwabe, T.M.; Riezman, H. Concentration of GPI-anchored proteins upon ER exit in yeast. Traffic 2009, 10, 186–200. [Google Scholar] [CrossRef]
- Bravo-Plaza, I.; Hernandez-Gonzalez, M.; Pinar, M.; Diaz, J.F.; Penalva, M.A. Identification of the guanine nucleotide exchange factor for SAR1 in the filamentous fungal model Aspergillus nidulans. BBA-Mol. Cell Res. 2019, 1866, 118551. [Google Scholar] [CrossRef]
- Zheng, H.; Li, L.; Miao, P.; Wu, C.; Chen, X.; Yuan, M.; Fang, T.; Norvienyeku, J.; Li, G.; Zheng, W.; et al. FgSec2A, a guanine nucleotide exchange factor of FgRab8, is important for polarized growth, pathogenicity and deoxynivalenol production in Fusarium graminearum. Environ. Microbiol. 2018, 20, 3378–3392. [Google Scholar] [CrossRef]
- Wang, C.; Zhang, S.; Hou, R.; Zhao, Z.; Zheng, Q.; Xu, Q.; Zheng, D.; Wang, G.; Liu, H.; Gao, X.; et al. Functional analysis of the kinome of the wheat scab fungus Fusarium graminearum. PLoS Pathog. 2011, 7, e1002460. [Google Scholar] [CrossRef] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, F.; Lv, B.; Zhang, X.; Wang, C.; Zhang, L.; Chen, X.; Liang, Y.; Chen, L.; Zou, S.; Dong, H. The Endoplasmic Reticulum Cargo Receptor FgErv14 Regulates DON Production, Growth and Virulence in Fusarium graminearum. Life 2022, 12, 799. https://doi.org/10.3390/life12060799
Sun F, Lv B, Zhang X, Wang C, Zhang L, Chen X, Liang Y, Chen L, Zou S, Dong H. The Endoplasmic Reticulum Cargo Receptor FgErv14 Regulates DON Production, Growth and Virulence in Fusarium graminearum. Life. 2022; 12(6):799. https://doi.org/10.3390/life12060799
Chicago/Turabian StyleSun, Fengjiang, Beibei Lv, Xuemeng Zhang, Chenyu Wang, Liyuan Zhang, Xiaochen Chen, Yuancun Liang, Lei Chen, Shenshen Zou, and Hansong Dong. 2022. "The Endoplasmic Reticulum Cargo Receptor FgErv14 Regulates DON Production, Growth and Virulence in Fusarium graminearum" Life 12, no. 6: 799. https://doi.org/10.3390/life12060799
APA StyleSun, F., Lv, B., Zhang, X., Wang, C., Zhang, L., Chen, X., Liang, Y., Chen, L., Zou, S., & Dong, H. (2022). The Endoplasmic Reticulum Cargo Receptor FgErv14 Regulates DON Production, Growth and Virulence in Fusarium graminearum. Life, 12(6), 799. https://doi.org/10.3390/life12060799






