A Bocage Landscape Restricts the Gene Flow of Pest Vole Populations
Abstract
:1. Introduction
2. Material and Methods
2.1. Study Area
2.2. Specimen Collection
2.3. Graph-Based Landscape Model
2.4. Microsatellite Genotyping
2.5. Genetic Diversity, Structure, and Differentiation
2.6. Effect of the Landscape on Genetic Structure
3. Results
3.1. Genetic Diversity
3.2. Population Structure in A. scherman
3.3. Effect of the Bocage Landscape on Genetic Structure
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gilarranz, L.J.; Bascompte, J. Spatial network structure and metapopulation persistence. J. Theor. Biol. 2012, 297, 11–16. [Google Scholar] [CrossRef] [PubMed]
- Driscoll, D.A.; Banks, S.C.; Barton, P.S.; Lindenmayer, D.B.; Smith, A.L. Conceptual domain of the matrix in fragmented landscapes. Trends Ecol. Evol. 2013, 28, 605–613. [Google Scholar] [CrossRef] [PubMed]
- Chiu, M.C.; Nukazawa, K.; Carvajal, T.; Resh, V.H.; Li, B.; Watanabe, K. Simulation modeling reveals the evolutionary role of landscape shape and species dispersal on genetic variation within a metapopulation. Ecography 2020, 43, 1891–1901. [Google Scholar] [CrossRef]
- Berthier, K.; Piry, S.; Cosson, J.F.; Giraudoux, P.; Foltête, J.C.; Defaut, R.; Truchete, D.; Lambin, X. Dispersal, landscape and travelling waves in cyclic vole populations. Ecol. Lett. 2014, 17, 53–64. [Google Scholar] [CrossRef]
- Craig, V.J.; Klenner, W.; Feller, M.C.; Sullivan, T.P. Population dynamics of meadow voles (Microtus pennsylvanicus) and long-tailed voles (M. longicaudus) and their relationship to downed wood in early successional forest habitats. Mammal Res. 2015, 60, 29–38. [Google Scholar] [CrossRef]
- Wiegand, T.; Revilla, E.; Moloney, K.A. Effects of habitat loss and fragmentation on population dynamics. Conserv. Biol. 2005, 19, 108–121. [Google Scholar] [CrossRef]
- Galpern, P.; Manseau, M.; Fall, A. Patch-based graphs of landscape connectivity: A guide to construction, analysis and application for conservation. Biol. Conserv. 2011, 144, 44–55. [Google Scholar] [CrossRef]
- Gauffre, B.; Mallez, S.; Chapuis, M.P.; Leblois, R.; Litrico, I.; Delaunay, S.; Badenhausser, I. Spatial heterogeneity in landscape structure influences dispersal and genetic structure: Empirical evidence from a grasshopper in an agricultural landscape. Mol. Ecol. 2015, 24, 1713–1728. [Google Scholar] [CrossRef]
- Janova, E.; Heroldova, M. Response of small mammals to variable agricultural landscapes in Central Europe. Mamm. Biol.-Z. Für Säugetierkunde 2016, 81, 488–493. [Google Scholar] [CrossRef]
- Jacob, J.; Hempel, N. Effects of farming practices on spatial behaviour of common voles. J. Ethol. 2003, 21, 45–50. [Google Scholar] [CrossRef]
- Marchi, C.; Andersen, L.W.; Damgaard, C.; Olsen, K.; Jensen, T.S.; Loeschcke, V. Gene flow and population structure of a common agricultural wild species (Microtus agrestis) under different land management regimes. Heredity 2013, 111, 486–494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Foltête, J.C.; Couval, G.; Fontanier, M.; Vuidel, G.; Giraudoux, P. A graph-based approach to defend agro-ecological systems against water vole outbreaks. Ecol. Indic. 2016, 71, 87–98. [Google Scholar] [CrossRef]
- Howell, P.E.; Delgado, M.L.; Scribner, K.T. Landscape genetic analysis of co-distributed white-footed mice (Peromyscus leucopus) and prairie deer mice (Peromyscus maniculatus bairdii) in an agroecosystem. J. Mammal. 2017, 98, 793–803. [Google Scholar] [CrossRef] [Green Version]
- Vera, N.S.; Chiappero, M.B.; Priotto, J.W.; Sommaro, L.V.; Steinmann, A.R.; Gardenal, C.N. Genetic structure of populations of the Pampean grassland mouse, Akodon azarae, in an agroecosystem under intensive management. Mamm. Biol. 2019, 98, 52–60. [Google Scholar] [CrossRef]
- Otero-Jiménez, B.; Li, K.; Tucker, P.K. Landscape drivers of connectivity for a forest rodent in a coffee agroecosystem. Landsc. Ecol. 2020, 35, 1249–1261. [Google Scholar] [CrossRef]
- Hamilton, G.S.; Mather, P.B.; Wilson, J.C. Habitat heterogeneity influences connectivity in a spatially structured pest population. J. Appl. Ecol. 2006, 43, 219–226. [Google Scholar] [CrossRef]
- Gauffre, B.; Estoup, A.; Bretagnolle, V.; Cosson, J.F. Spatial genetic structure of a small rodent in a heterogeneous landscape. Mol. Ecol. 2008, 17, 4619–4629. [Google Scholar] [CrossRef]
- Berthier, K.; Charbonnel, N.; Galan, M.; Chaval, Y.; Cosson, J.F. Migration and recovery of the genetic diversity during the increasing density phase in cyclic vole populations. Mol. Ecol. 2006, 15, 2665–2676. [Google Scholar] [CrossRef]
- Jones, K.E.; Safi, K. Ecology and evolution of mammalian biodiversity. Philos. Trans. R. Soc. Lond. B 2011, 366, 2451–2461. [Google Scholar] [CrossRef] [Green Version]
- Martin, B.G. The role of small ground-foraging mammals in topsoil health and biodiversity: Implications to management and restoration. Ecol. Manag. Restor. 2003, 4, 114–119. [Google Scholar] [CrossRef]
- Schweizer, M.; Excoffier, L.; Heckel, G. Fine-scale genetic structure and dispersal in the common vole (Microtus arvalis). Mol. Ecol. 2007, 16, 2463–2473. [Google Scholar] [CrossRef] [PubMed]
- Herrera, J.M.; Salgueiro, P.A.; Medinas, D.; Costa, P.; Encarnação, C.; Mira, A. Generalities of vertebrate responses to landscape composition and configuration gradients in a highly heterogeneous Mediterranean region. J. Biogeogr. 2016, 43, 1203–1214. [Google Scholar] [CrossRef] [Green Version]
- Weber de Melo, V.; Sheikh Ali, H.; Freise, J.; Kühnert, D.; Essbauer, S.; Mertens, M.; Wanka, K.M.; Drewes, S.; Ulrich, R.G.; Heckel, G. Spatiotemporal dynamics of Puumala hantavirus associated with its rodent host, Myodes glareolus. Evol. Appl. 2015, 8, 545–559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drewes, S.; Sheikh Ali, H.; Saxenhofer, M.; Rosenfeld, U.M.; Binder, F.; Cuypers, F.; Schlegel, M.; Röhrs, S.; Heckel, G.; Ulrich, R.G. Host-associated absence of human Puumala virus infections in northern and eastern Germany. Emerg. Infect. Dis. 2017, 23, 83–86. [Google Scholar] [CrossRef] [Green Version]
- Rodríguez-Pastor, R.; Escudero, R.; Lambin, X.; Vidal, M.D.; Gil, H.; Jado, I.; Luque-Larena, J.J.; Mougeot, F. Zoonotic pathogens in fluctuating common vole (Microtus arvalis) populations: Occurrence and dynamics. Parasitology 2019, 146, 389–398. [Google Scholar] [CrossRef] [Green Version]
- Joannon, A.; Vialatte, A.; Vasseur, C.; Baudry, J.; Thenail, C. Combining studies on crop mosaic dynamics and pest population dynamics to foster biological control. In Landscape Management for Functional Biodiversity; Rossing, W.A.H., Poehling, H.M., Van Helden, M., Eds.; IOBC wprs Bulletin: Zurich, Switzerland, 2008; Volume 34, pp. 45–48. [Google Scholar]
- Veres, A.; Petit, S.; Conord, C.; Lavigne, C. Does landscape composition affect pest abundance and their control by natural enemies? A review. Agric. Ecosyst. Environ. 2013, 166, 110–117. [Google Scholar] [CrossRef]
- Benton, T.G.; Vickery, J.A.; Wilson, J.D. Farmland biodiversity: Is habitat heterogeneity the key? Trends Ecol. Evol. 2003, 18, 182–188. [Google Scholar] [CrossRef]
- Chase, J.M.; Blowes, S.A.; Knight, T.M.; Gerstner, K.; May, F. Ecosystem decay exacerbates biodiversity loss with habitat loss. Nature 2020, 584, 238–243. [Google Scholar] [CrossRef]
- Balmori-de la Puente, A.; Ventura, J.; Miñarro, M.; Somoano, A.; Hey, J.; Castresana, J. Divergence time estimation using ddRAD data and an isolation-with-migration model applied to water vole populations of Arvicola. Sci. Rep. 2022, 12, 4065. [Google Scholar] [CrossRef]
- Delattre, P.; Giraudoux, P.; Baudry, J.; Musard, P.; Toussaint, M.; Truchetete, D.; Stahl, P.; Poule, M.L.; Artois, M.; Damange, J.P.; et al. Land use patterns and types of common vole (Microtus arvalis) population kinetics. Agric. Ecosyst. Environ. 1992, 39, 153–168. [Google Scholar] [CrossRef]
- Giraudoux, P.; Delattre, P.; Habert, M.; Quéré, J.P.; Deblay, S.; Defaut, R.; Duhamel, R.; Moissenet, M.F.; Salvi, D.; Truchetet, D. Population dynamics of fossorial water vole (Arvicola terrestris scherman): A land use and landscape perspective. Agric. Ecosyst. Environ. 1997, 66, 47–60. [Google Scholar] [CrossRef]
- Viel, J.F.; Giraudoux, P.; Abrial, V.; Bresson-Andi, S. Water vole (Arvicola terrestris scherman) density as a risk factor for human alveolar echinococcosis. Am. J. Trop. Med. Hyg. 1999, 61, 559–565. [Google Scholar] [CrossRef] [Green Version]
- Espí, A.; Del Cerro, A.; Somoano, A.; García, V.; Prieto, J.M.; Barandika, J.F.; García-Pérez, A.L. Borrelia burgdorferi sensu lato prevalence and diversity in ticks and small mammals in a Lyme borreliosis endemic Nature Reserve in North-Western Spain. Incidence in surrounding human populations. Enferm. Infecc. Y Microbiol. Clin. 2017, 35, 563–568. (In English) [Google Scholar] [CrossRef] [PubMed]
- Jacob, J.; Imholt, C.; Caminero-Saldaña, C.; Couval, G.; Giraudoux, P.; Herrero-Cófreces, S.; Horváth, G.; Luque-Larena, J.J.; Tkadlec, E.; Wymenga, E. Europe-wide outbreaks of common voles in 2019. J. Pest Sci. 2020, 93, 703–709. [Google Scholar] [CrossRef] [Green Version]
- Somoano, A.; Ventura, J.; Miñarro, M. Continuous breeding of fossorial water voles in northwestern Spain: Potential impact on apple orchards. Folia Zool. 2017, 66, 37–49. [Google Scholar] [CrossRef]
- Halliez, G.; Renault, F.; Vannard, E.; Farny, G.; Lavorel, S.; Giraudoux, P. Historical agricultural changes and the expansion of a water vole population in an Alpine valley. Agric. Ecosyst. Environ. 2015, 212, 198–206. [Google Scholar] [CrossRef]
- Berthier, K.; Galan, M.; Foltete, J.C.; Charbonnel, N.; Cosson, J.F. Genetic structure of the cyclic fossorial water vole (Arvicola terrestris): Landscape and demographic influences. Mol. Ecol. 2005, 14, 2861–2871. [Google Scholar] [CrossRef]
- Somoano, A. The role of the montane water vole (Arvicola scherman) as a crop pest in NW Spain: Since when? Galemys 2020, 32, 61–63. [Google Scholar] [CrossRef]
- Miramontes-Carballada, Á. Los paisajes agrarios de España: La evolución de la nueva horticultura y cultivos especializados frente a la agricultura tradicional en la España Atlántica. Rev. De Geogr. E Ordenam. Do Territ. 2014, 5, 161–180. [Google Scholar]
- Baudry, J.; Bunce, R.G.H.; Burel, F. Hedgerows: An international perspective on their origin, function and management. J. Environ. Manag. 2000, 60, 7–22. [Google Scholar] [CrossRef]
- Le Cœur, D.; Baudry, J.; Burel, F.; Thenail, C. Why and how we should study field boundary biodiversity in an agrarian landscape context. Agric. Ecosyst. Environ. 2002, 89, 23–40. [Google Scholar] [CrossRef]
- Couval, G.; Truchetet, D. Le concept de lutte raisonnée: Combiner des méthodes collectives contre le campagnol terrestre afin de conserver une autonomie furragère. Fourrages 2014, 220, 343–347. [Google Scholar]
- Gauffre, B.; Berthier, K.; Inchausti, P.; Chaval, Y.; Bretagnolle, V.; Cosson, J.F. Short-term variations in gene flow related to cyclic density fluctuations in the common vole. Mol. Ecol. 2014, 23, 3214–3225. [Google Scholar] [CrossRef] [PubMed]
- Garrido-Garduño, T.; Téllez-Valdés, O.; Manel, S.; Vázquez-Domínguez, E. Role of habitat heterogeneity and landscape connectivity in shaping gene flow and spatial population structure of a dominant rodent species in a tropical dry forest. J. Zool. 2015, 298, 293–302. [Google Scholar] [CrossRef]
- Anderson, S.J.; Kierepka, E.M.; Swihart, R.K.; Latch, E.K.; Rhodes, O.E., Jr. Assessing the permeability of landscape features to animal movement: Using genetic structure to infer functional connectivity. PLoS ONE 2015, 10, e0117500. [Google Scholar] [CrossRef] [Green Version]
- Russo, I.R.M.; Sole, C.L.; Barbato, M.; Von Bramann, U.; Bruford, M.W. Landscape determinants of fine-scale genetic structure of a small rodent in a heterogeneous landscape (Hluhluwe-iMfolozi Park, South Africa). Sci. Rep. 2016, 6, 29168. [Google Scholar] [CrossRef] [Green Version]
- Fernández-Ceballos, A. Análisis De La Distribución De Arvicola terrestris a Media Escala. Master’s Thesis, University of Oviedo, Asturias, Spain, 2005. [Google Scholar]
- BOE. Real Decreto 409/2008, de 28 de marzo. Boletín Of. Del Estado 2008, 86, 19217–19219. [Google Scholar]
- Directive 2010/63/UE. Horizontal Legislation on the Protection of Animal Used for Scientific Purposes. Off. J. Eur. Union L 2010, 273, 33–79.
- Hoban, S. An overview of the utility of population simulation software in molecular ecology. Mol. Ecol. 2014, 23, 2383–2401. [Google Scholar] [CrossRef]
- Valcárcel, N.; Villa, G.; Arozarena, A.; Garcia-Asensio, L.; Caballero, M.E.; Porcuna, A.; Domenech, E.; Peces, J.J. SIOSE, a successful test bench towards harmonization and integration of land cover/use information as environmental reference data. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2008, 3, 1159–1164. [Google Scholar]
- Adriaensen, F.; Chardon, J.P.; De Blust, G.; Swinnen, E.; Villalba, S.; Gulinck, H.; Matthysen, E. The application of least-cost modelling as a functional landscape model. Landsc. Urban Plan. 2003, 64, 233–247. [Google Scholar] [CrossRef]
- Sawyer, S.C.; Epps, C.W.; Brashares, J.S. Placing linkages among fragmented habitats: Do least-cost models reflect how animals use landscapes? J. Appl. Ecol. 2011, 48, 668–678. [Google Scholar] [CrossRef]
- Foltête, J.C.; Giraudoux, P. A graph-based approach to investigating the influence of the landscape on population spread processes. Ecol. Indic. 2012, 18, 684–692. [Google Scholar] [CrossRef]
- ESRI. Arcview GIS: The Geographical Information System for Everyone; Environmental Systems Research Institute: New York, NY, USA, 1996. [Google Scholar]
- Fichet-Calvet, E.; Pradier, B.; Quéré, J.P.; Giraudoux, P.; Delattre, P. Landscape composition and vole outbreaks: Evidence from an eight year study of Arvicola terrestris. Ecography 2000, 23, 659–668. [Google Scholar] [CrossRef]
- Stewart, W.A.; Dallas, J.F.; Piertney, S.B.; Marshall, F.; Lambin, X.; Telfer, S. Metapopulation genetic structure in the water vole, Arvicola terrestris, in NE Scotland. Biol. J. Linn. Soc. 1999, 68, 159–171. [Google Scholar] [CrossRef]
- Berthier, K.; Galan, M.; Weber, A.; Loiseau, A.; Cosson, J.F. A multiplex panel of dinucleotide microsatellite markers for the water vole, Arvicola terrestris. Mol. Ecol. Notes 2004, 4, 620–622. [Google Scholar] [CrossRef]
- Glaubitz, J.C. Convert: A user-friendly program to reformat diploid genotypic data for commonly used population genetic software packages. Mol. Ecol. Notes 2004, 4, 309–310. [Google Scholar] [CrossRef]
- Raymond, M.; Rousset, F. GENEPOP (version 1.2): Population genetics software for exact tests and ecumenicism. J. Hered. 1995, 86, 248–249. [Google Scholar] [CrossRef]
- Van Oosterhout, C.; Hutchinson, W.F.; Wills, D.P.; Shipley, P. MICRO-CHECKER: Software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes 2004, 4, 535–538. [Google Scholar] [CrossRef]
- Nei, M. Estimation of average heterozigosity and genetic distance from a small number of individuals. Genetics 1987, 90, 502–503. [Google Scholar]
- Belkhir, K.; Borsa, P.; Chikhi, L.; Raufaste, N.; Bonhomme, F. GENETIX 4.05, Logiciel Sous WindowsTM Pour La Génétique Des Populations. In Laboratoire Génome, Populations, Interactions, CNRS UMR 5000; University of Montpellier II: Montpellier, France, 1996. [Google Scholar]
- Goudet, J. FSTAT (version 1.2): A computer program to calculate F-statistics. J. Hered. 1995, 86, 485–486. [Google Scholar] [CrossRef]
- Piry, S.; Alapetite, A.; Cornuet, J.M.; Paetkau, D.; Baudouin, L.; Estoup, A. GENECLASS2, A software for genetic assignment and first-generation migrant detection. J. Hered. 2004, 95, 536–539. [Google Scholar] [CrossRef] [PubMed]
- Paetkau, D.; Slade, R.; Burden, M.; Estoup, A. Genetic assignment methods for the direct, real-time estimation of migration rate: A simulation-based exploration of accuracy and power. Mol. Ecol. 2004, 13, 55–65. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Earl, D.A. Structure Harvester: A website and program for visualizing Structure output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Guillot, G.; Mortier, F.; Estoup, A. Geneland: A computer package for landscape genetics. Mol. Ecol. Notes 2005, 5, 712–715. [Google Scholar] [CrossRef]
- Meirmans, P.G. The trouble with isolation by distance. Mol. Ecol. 2012, 21, 2839–2846. [Google Scholar] [CrossRef]
- Drummond, C.S.; Hamilton, M.B. Hierarchical components of genetic variation at a species boundary: Population structure in two sympatric varieties of Lupinus microcarpus (Leguminosae). Mol. Ecol. 2007, 16, 753–769. [Google Scholar] [CrossRef]
- Excoffier, L.; Lischer, H.E. Arlequin suite ver 3.5, A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Mantel, N. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967, 27, 209–220. [Google Scholar]
- Bohonak, A.J. IBD (Isolation By Distance): A program for analyses of isolation by distance. J. Hered. 2002, 93, 153–154. [Google Scholar] [CrossRef] [PubMed]
- Rousset, F. Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics 1997, 145, 1219–1228. [Google Scholar] [CrossRef] [PubMed]
- Cushman, S.; Wasserman, T.; Landguth, E.; Shirk, A. Re-evaluating causal modeling with Mantel tests in landscape genetics. Diversity 2013, 5, 51–72. [Google Scholar] [CrossRef] [Green Version]
- Ackiss, A.S.; Bird, C.E.; Akita, Y.; Santos, M.D.; Tachihara, K.; Carpenter, K.E. Genetic patterns in peripheral marine populations of the fusilier fish Caesio cuning within the Kuroshio Current. Ecol. Evol. 2008, 8, 11875–11886. [Google Scholar] [CrossRef] [Green Version]
- Peakall, R.; Smouse, P.E. GENALEX 6, Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Grant, A.H.; Liebgold, E.B. Color-biased dispersal inferred by fine-scale genetic spatial autocorrelation in a color polymorphic salamander. J. Hered. 2017, 108, 588–593. [Google Scholar] [CrossRef]
- Peakall, R.; Ruibal, M.; Lindenmayer, D.B. Spatial autocorrelation analysis offers new insights into gene flow in the Australian bush rat, Rattus fuscipes. Evolution 2003, 57, 1182–1195. [Google Scholar] [CrossRef]
- Guivier, E.; Galan, M.; Chaval, Y.; Xuéreb, A.; Ribas Salvador, A.; Poulle, M.L.; Voutilainen, L.; Henttonen, H.; Charbonnel, N.; Cosson, J.F. Landscape genetics highlights the role of bank vole metapopulation dynamics in the epidemiology of Puumala hantavirus. Mol. Ecol. 2011, 20, 3569–3583. [Google Scholar] [CrossRef]
- Melis, C.; Borg, Å.A.; Jensen, H.; Bjørkvoll, E.; Ringsby, T.H.; Sæther, B.E. Genetic variability and structure of the water vole Arvicola amphibius across four metapopulations in northern Norway. Ecol. Evol. 2013, 3, 770–778. [Google Scholar] [CrossRef]
- Palsbøll, P.J.; Berube, M.; Allendorf, F.W. Identification of management units using population genetic data. Trends Ecol. Evol. 2007, 22, 11–16. [Google Scholar] [CrossRef]
- Piertney, S.B.; Black, A.; Watt, L.; Christie, D.; Poncet, S.; Collins, M.A. Resolving patterns of population genetic and phylogeographic structure to inform control and eradication initiatives for brown rats Rattus norvegicus on South Georgia. J. Appl. Ecol. 2016, 53, 332–339. [Google Scholar] [CrossRef]
- Hanski, I.; Ovaskainen, O. Metapopulation theory for fragmented landscapes. Theor. Popul. Biol. 2003, 64, 119–127. [Google Scholar] [CrossRef]
- Vignieri, S.N. Streams over mountains: Influence of riparian connectivity on gene flow in the Pacific jumping mouse (Zapus trinotatus). Mol. Ecol. 2005, 14, 1925–1937. [Google Scholar] [CrossRef] [PubMed]
- Telfer, S.; Piertney, S.B.; Dallas, J.F.; Stewart, W.A.; Marshall, F.; Gow, J.L.; Lambin, X. Parentage assignment detects frequent and large-scale dispersal in water voles. Mol. Ecol. 2003, 12, 1939–1949. [Google Scholar] [CrossRef] [PubMed]
- Le Galliard, J.F.; Remy, A.; Ims, R.A.; Lambin, X. Patterns and processes of dispersal behaviour in arvicoline rodents. Mol. Ecol. 2012, 21, 505–523. [Google Scholar] [CrossRef]
- Aars, J.; Dallas, J.F.; Piertney, S.B.; Marshall, F.; Gow, J.L.; Telfer, S.; Lambin, X. Widespread gene flow and high genetic variability in populations of water voles Arvicola terrestris in patchy habitats. Mol. Ecol. 2006, 15, 1455–1466. [Google Scholar] [CrossRef] [PubMed]
- Somoano, A.; Miñarro, M.; Ventura, J. Reproductive potential of a vole pest (Arvicola scherman) in Spanish apple orchards. Span. J. Agric. Res. 2016, 14, e1008. [Google Scholar] [CrossRef]
- Peterman, W.E. ResistanceGA: An R package for the optimization of resistance surfaces using genetic algorithms. Methods Ecol. Evol. 2018, 9, 1638–1647. [Google Scholar] [CrossRef] [Green Version]
- Driscoll, D.A.; Whitehead, C.A.; Lazzari, J. Spatial dynamics of the knob-tailed gecko Nephrurus stellatus in a fragmented agricultural landscape. Landsc. Ecol. 2012, 27, 829–841. [Google Scholar] [CrossRef]
- Saucy, F.; Schneiter, B. Juvenile dispersal in the vole, Arvicola terrestris, during rainy nights: A preliminary report. Bull. De La Société Vaud. Des Sci. Nat. 1998, 84, 33–345. [Google Scholar]
- Giraudoux, P.; Delattre, P.; Quéré, J.P.; Damange, J.P. Structure and kinetics of rodent populations, in a region under agricultural land abandonment. Acta Oecologica 1994, 15, 385–400. [Google Scholar]
- Hahne, J.; Jenkins, T.; Halle, S.; Heckel, G. Establishment success and resulting fitness consequences for vole dispersers. Oikos 2011, 120, 95–105. [Google Scholar] [CrossRef]
- SADEI. Asturian Society for Economic and Industrial Studies; SADEI: Asturias, Spain, 2020. [Google Scholar]
- Morilhat, C.; Bernard, N.; Bournais, C.; Meyer, C.; Lamboley, C.; Giraudoux, P. Responses of Arvicola terrestris scherman populations to agricultural practices, and to Talpa europaea abundance in eastern France. Agric. Ecosyst. Environ. 2007, 122, 392–398. [Google Scholar] [CrossRef]
- Somoano, A.; Bastos-Silveira, C.; Ventura, J.; Miñarro, M.; Heckel, G. Bocage landscape restricts the gene flow of pest vole populations. Dryad Dataset 2022. [Google Scholar] [CrossRef]

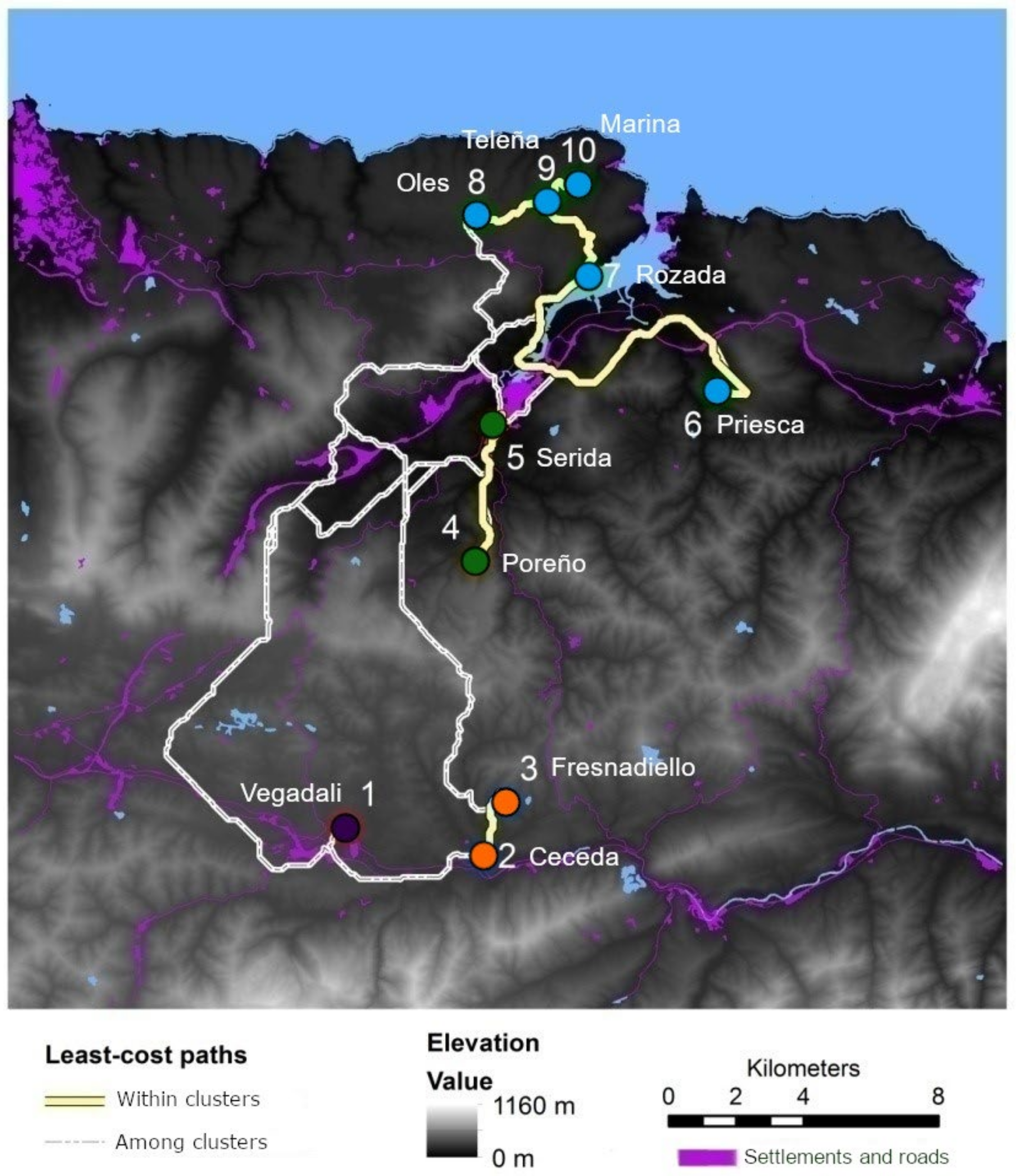
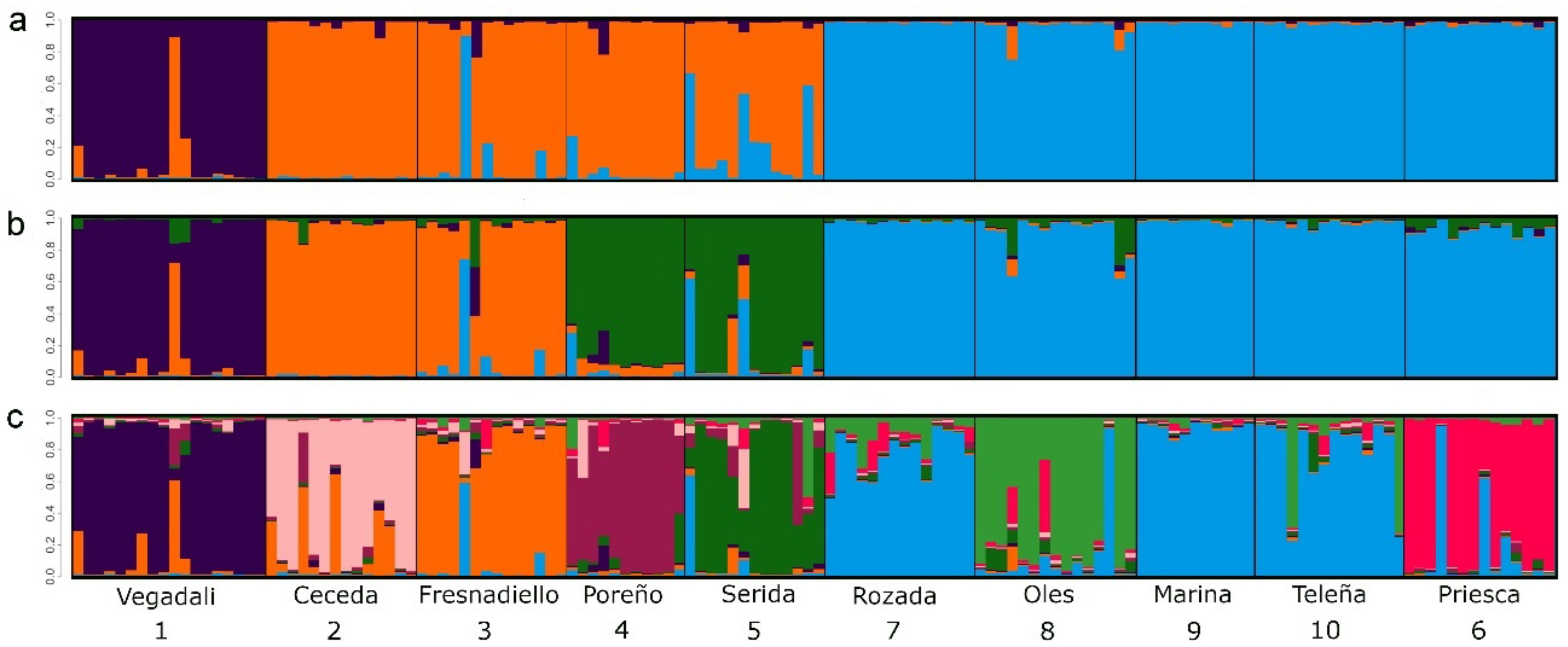
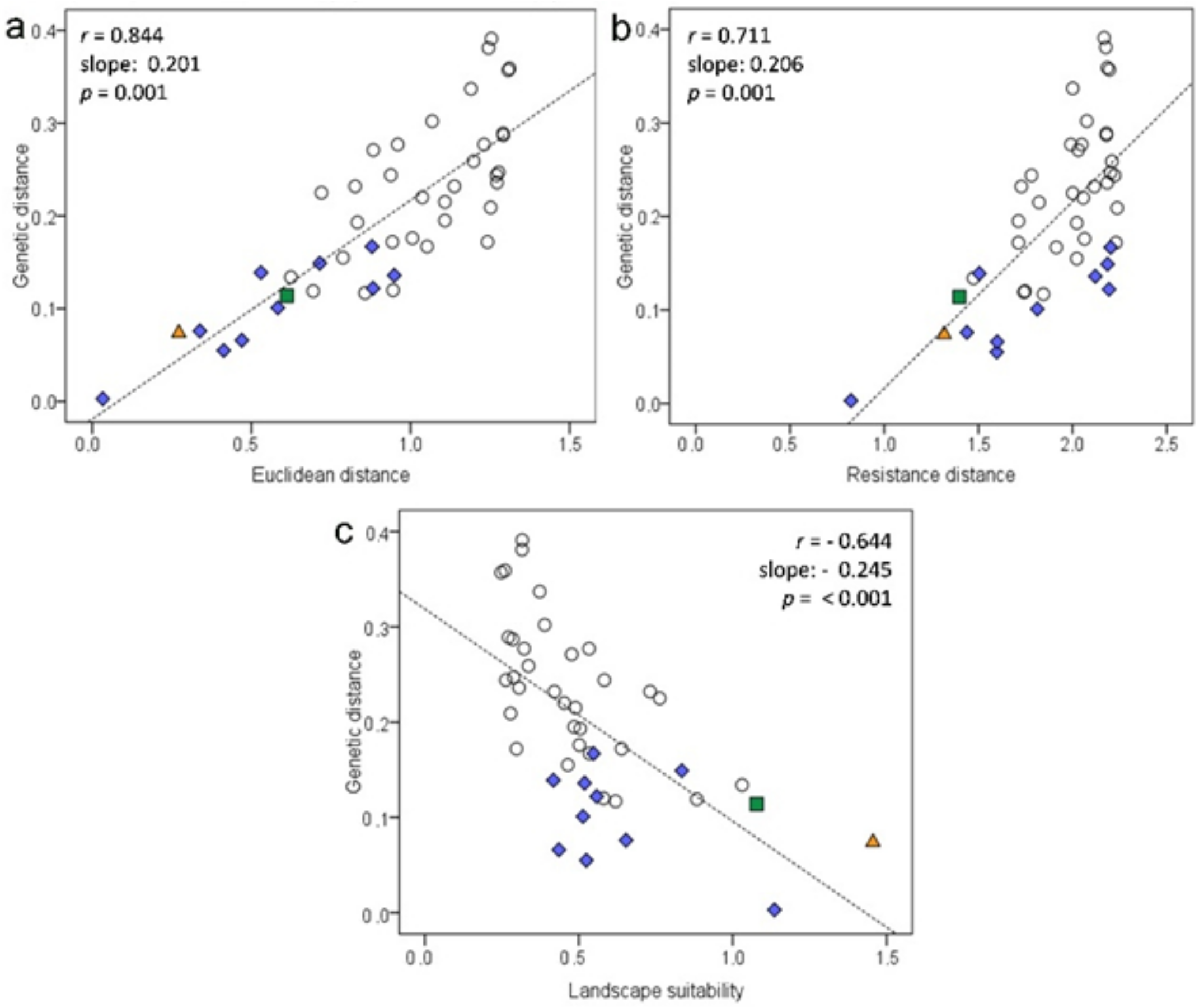
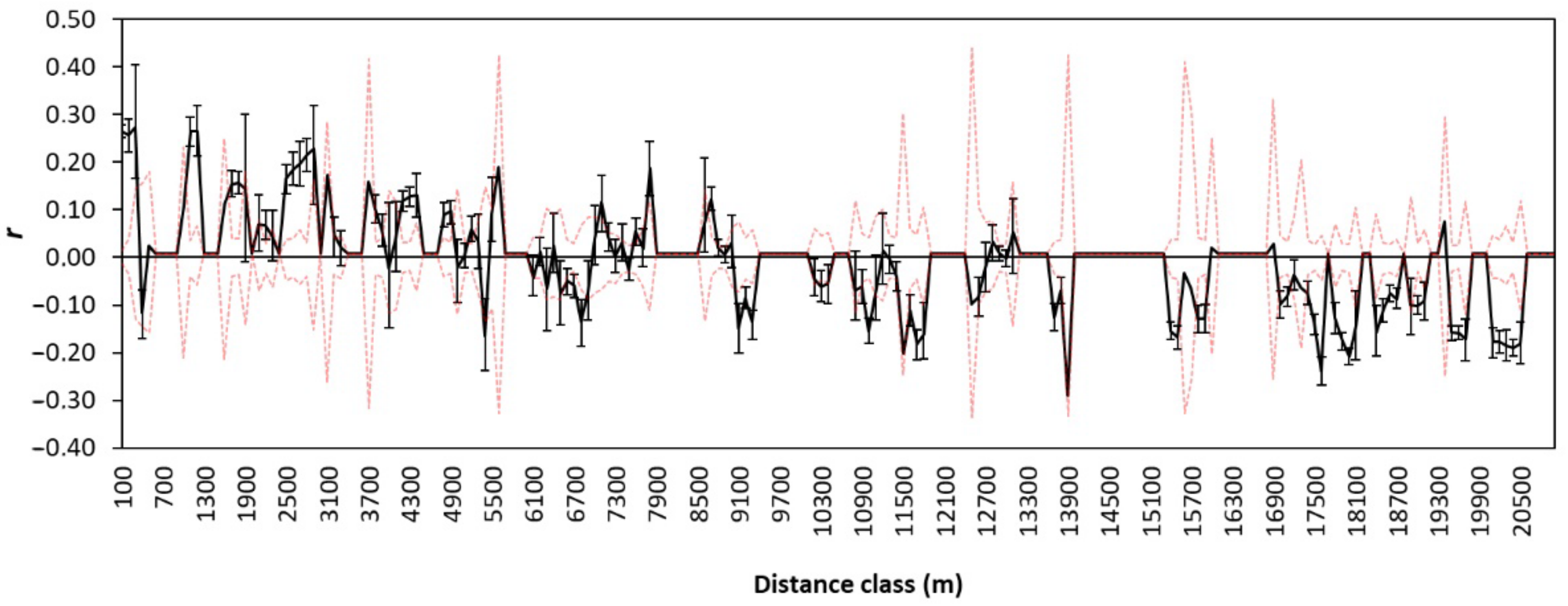
| Orchard/ Deme | Date | UTMx | UTMy | Area (ha) | N | NA | AR | HO | HE | FIS | HWE p-Value | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1-Vegadali | Sep | 2011 | 298049 | 4804471 | 2.31 | 18 | 6.09 | 5.35 | 0.636 | 0.695 | 0.087 | 0.031 |
| 2-Ceceda | Dec | 2012 | 302152 | 4803618 | 5.95 | 14 | 4.82 | 4.53 | 0.604 | 0.629 | 0.042 | 0.434 |
| 3-Fresnadiello | Jun | 2011 | 302809 | 4805229 | 2.84 | 14 | 6.00 | 5.40 | 0.656 | 0.692 | 0.052 | 0.207 |
| 4-Poreño | Jul | 2012 | 301923 | 4812449 | 7.58 | 11 | 4.64 | 4.64 | 0.612 | 0.614 | 0.005 | 0.610 |
| 5-Serida | Jun | 2011 | 302393 | 4816492 | 7.26 | 12 | 4.64 | 4.47 | 0.458 | 0.556 | 0.183 | <0.001 |
| 6-Priesca | Apr | 2011 | 309079 | 4817418 | 2.03 | 14 | 3.55 | 3.44 | 0.591 | 0.554 | −0.071 | 0.582 |
| 7-Rozada | Mar | 2012 | 305277 | 4820835 | 2.99 | 14 | 3.91 | 3.75 | 0.524 | 0.542 | 0.034 | 0.467 |
| 8-Oles | Jun | 2011 | 301922 | 4822568 | 4.55 | 15 | 4.00 | 3.40 | 0.491 | 0.595 | 0.180 | <0.001 |
| 9-Teleña | Jul | 2012 | 304030 | 4823030 | 1.13 | 14 | 3.82 | 3.75 | 0.656 | 0.612 | −0.075 | 0.545 |
| 10-Marina | Jul | 2012 | 304955 | 4823559 | 1.00 | 11 | 3.91 | 3.91 | 0.595 | 0.587 | −0.015 | 0.528 |
| Percentage of Variation | Fixation Indices | p-Value | |||
|---|---|---|---|---|---|
| Between individuals within demes | 3 | 127 | 3.65 | 0.046 | 0.011 |
| 4 | 127 | 3.68 | 0.046 | 0.011 | |
| 8 | 127 | 3.78 | 0.046 | 0.008 | |
| Between demes within clusters | 3 | 7 | 8.95 | 0.100 | <0.001 |
| 4 | 6 | 7.52 | 0.085 | <0.001 | |
| 8 | 2 | 2.84 | 0.033 | <0.001 | |
| Between clusters | 3 | 2 | 10.91 | 0.109 | <0.001 |
| 4 | 3 | 11.81 | 0.118 | <0.001 | |
| 8 | 7 | 14.71 | 0.147 | 0.009 |
| Test | Corrected by | r | p-Value | |
|---|---|---|---|---|
| IBD | Euclidean distance | Resistance distance | 0.630 | 0.001 |
| (SH/TL)/Euclidean distance | 0.733 | 0.002 | ||
| 8-cluster structuring | 0.770 | 0.001 | ||
| IBR | Resistance distance | Euclidean distance | 0.093 | 0.234 |
| (SH/TL)/Euclidean distance | 0.520 | 0.013 | ||
| 8-cluster structuring | 0.568 | 0.004 | ||
| Landscape suitability | (SH/TL)/Euclidean distance | Euclidean distance | −0.151 | 0.175 |
| Resistance distance | −0.246 | 0.052 | ||
| 8-cluster structuring | −0.513 | 1.000 | ||
| Clusters | 8-cluster structuring | Euclidean distance | 0.401 | 0.016 |
| Resistance distance | 0.386 | 0.015 | ||
| (SH/TL)/Euclidean distance | 0.523 | 0.001 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Somoano, A.; Bastos-Silveira, C.; Ventura, J.; Miñarro, M.; Heckel, G. A Bocage Landscape Restricts the Gene Flow of Pest Vole Populations. Life 2022, 12, 800. https://doi.org/10.3390/life12060800
Somoano A, Bastos-Silveira C, Ventura J, Miñarro M, Heckel G. A Bocage Landscape Restricts the Gene Flow of Pest Vole Populations. Life. 2022; 12(6):800. https://doi.org/10.3390/life12060800
Chicago/Turabian StyleSomoano, Aitor, Cristiane Bastos-Silveira, Jacint Ventura, Marcos Miñarro, and Gerald Heckel. 2022. "A Bocage Landscape Restricts the Gene Flow of Pest Vole Populations" Life 12, no. 6: 800. https://doi.org/10.3390/life12060800
APA StyleSomoano, A., Bastos-Silveira, C., Ventura, J., Miñarro, M., & Heckel, G. (2022). A Bocage Landscape Restricts the Gene Flow of Pest Vole Populations. Life, 12(6), 800. https://doi.org/10.3390/life12060800






