Abstract
Seed germination is a critical stage during the life cycle of plants. It is well known that germination is regulated by a series of internal and external factors, especially plant hormones. In Arabidopsis, many germination-related factors have been identified, while in rice, the important crop and monocot model species and the further molecular mechanisms and regulatory networks controlling germination still need to be elucidated. Hormonal signals, especially those of abscisic acid (ABA) and gibberellin (GA), play a dominant role in determining whether a seed germinates or not. The balance between the content and sensitivity of these two hormones is the key to the regulation of germination. In this review, we present the foundational knowledge of ABA and GA pathways obtained from germination research in Arabidopsis. Then, we highlight the current advances in the identification of the regulatory genes involved in ABA- or GA-mediated germination in rice. Furthermore, other plant hormones regulate seed germination, most likely by participating in the ABA or GA pathways. Finally, the results from some regulatory layers, including transcription factors, post-transcriptional regulations, and reactive oxygen species, are also discussed. This review aims to summarize our current understanding of the complex molecular networks involving the key roles of plant hormones in regulating the seed germination of rice.
1. Introduction
Crop seeds are the main source of food and fundamental materials in agricultural production. For improved production, the main agricultural goal is to obtain rapid and uniform germination and seedling emergence once seeds are sown, bringing up the point that germination is considered one of the most critical phases in the plant life cycle [1]. Rice (Oryza sativa L.) is one of the most principal foods in the world. Seed germination has a considerable impact on the yield and quality of rice production. Low seed germination and seedling uniformity lead to a significant reduction in rice production. Seed with rapid and uniform germination is called high-vigor seed, which is essential for vigorous seedling growth, tolerance to adversity, and high yield production. Meanwhile, the inappropriate loss of seed dormancy often results in pre-harvest sprouting, causing substantial losses in the yield and quality of rice [2]. This phenomenon is particularly serious during the production of hybrid rice in southern China due to the long-term rainy weather during harvest season. Aside from that, the emerging rice direct-seeding technology has received much attention because of its low input demand compared with conventional transplantation [3]. However, poor seed germination and seedling emergence in the field are the major constraints of direct-seeding systems. Thus, rice varieties with excellent germination ability (i.e., rapid and uniform germination and seedling emergence in the field context) are required.
Plentiful research and significant progress in understanding seed germination have been made in recent decades. By definition, germination commences with the absorption of water by the quiescent dry seed and is completed with the appearance of the radicle through the surrounding structures [4]. Following the physiological timeline of germination, numerous remarkable reviews have summarized the major physical and metabolic events that occur during germination, including imbibition, resumption of respiration, reserve mobilization, translation, transcription, cellular repair, and radicle extension [1,4,5,6,7]. The emerging area of seed research aims to identify the key regulators controlling germination. In recent years, the regulatory mechanism of germination has been extensively studied in Arabidopsis through molecular genetic studies. Many specific genes correlated with altered seed germination or dormancy have been identified, and the regulatory network, mainly consisting of hormone pathways and environmental factors, has been thoroughly proposed [2,8,9,10,11,12,13,14,15,16]. Unfortunately, the regulatory mechanism of rice, being a model monocotyledon plant and one of the most important crops, has not been explored as much as that in Arabidopsis.
The transition of seeds from a dormant state to germination is the result of the co-regulation of internal and external environmental factors. Environmental factors regulate endogenous components and thus affect the germination of seeds. Hormonal regulation plays a crucial role in this process by regulating the expression of related genes and signal transductions, which might be a highly conserved mechanism among seed plants [8]. ABA (abscisic acid) and GAs (gibberellins) are known to be the primary phytohormones that antagonistically regulate seed dormancy and germination. ABA positively regulates dormancy induction and maintenance, while GAs promote seed germination [17]. Changes in the balance of a seed’s ABA and GA levels and sensitivity influence the dormancy or germination state. That aside, nearly all other classes of plant hormones, including ET (ethylene), BRs (brassinosteroids), auxin, JA (jasmonic acid), SA (salicylic acid), CTKs (cytokinins), and SLs (strigolactones), regulate seed dormancy and germination, most likely by participating in the ABA or GA pathways [2,18]. Overall, hormonal regulation constitutes a central regulatory mechanism underlying the maintenance of seed dormancy and the onset of germination.
Recently, the regulation of rice germination by hormones has drawn much more attention, and several examples of innovative progress have been made. Rice seed germination possesses conserved features compared with Arabidopsis but also has distinct characteristics of its own. In this review, the established knowledge from Arabidopsis serves as a “backbone”, with the latest developments in rice research integrated. This review attempts to comprehensively illustrate the regulatory layers controlling the seed germination of rice. It will guide rice production and new variety breeding in agricultural practice.
2. ABA: The Core Factor in the Maintenance of Seed Dormancy and Inhibition of Seed Germination
2.1. ABA Metabolism Influences Rice Seed Dormancy and Germination
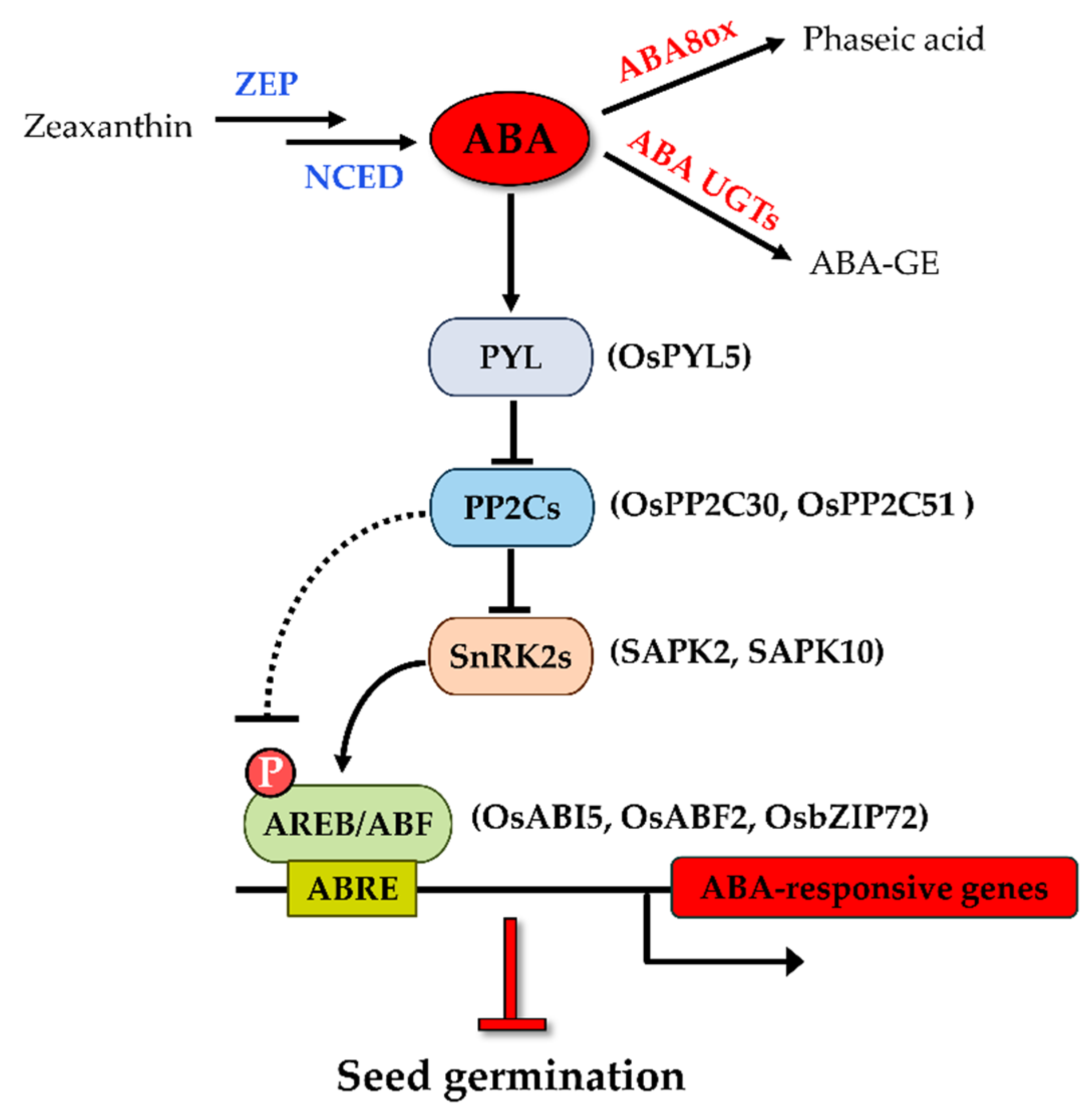
The level of cellular ABA is determined by the dynamic balance of biosynthesis and catabolism of the hormone [19]. Zeaxanthin epoxidase (ZEP) catalyzes the conversion of zeaxanthin to violaxanthin in the upstream of the ABA biosynthetic pathway (Figure 1). In rice, a strong viviparous mutant impaired in ZEP named Osaba1 has been isolated [20]. Nine-cis-epoxycarotenoid dioxygenase (NCED), cleaving the cis-isomers of violaxanthin and neoxanthin to xanthoxin, has been reported to be the rate-limiting step of ABA biosynthesis [21]. Changes in endogenous ABA levels are closely related to the expression of NCED transcripts. Seeds overexpressing AtNCED6 had increased ABA levels during imbibition and were sufficient to prevent germination [22]. ABA catabolism largely relies on the hydroxylation of ABA at the 8′ position under the catalytic action of ABA 8′-hydroxylase (ABA8ox), which is encoded by CYP707A genes [19]. The most abundant ABA hydroxylated catabolite is phaseic acid (PA). A mutation of the CYP707A2 genes in Arabidopsis showed an increase in ABA levels in dry and imbibed seeds and thus decreased germination potential [23]. ABA can also be inactivated by conjugation with glucose to form ABA glucosyl ester (ABA-GE) [19,24]. Several ABA glucosyl-transferases (UGTs) are capable of catalyzing this reaction. ABA-GE can be cleaved by β-glucosidases to form active ABA [19,24]. One of the MYB transcription factor members was recently reported to mediate the deconjugation of ABA-GE to ABA at a very early imbibition time and optimize seed germination in rice [25].
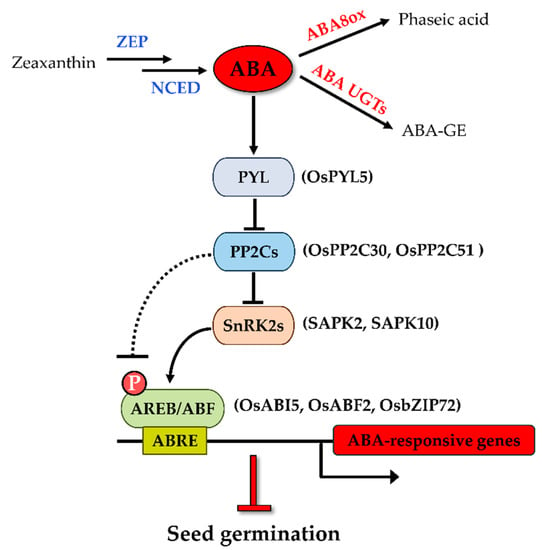
Figure 1.
Key components of ABA metabolism and signaling pathway in rice seed germination.
The expressions of the NCED and ABA8ox genes play significant roles in the control of the ABA level and therefore seed dormancy and germination. There are five members of the NCED gene family in rice. The expression pattern of the OsNCED transcripts was often used as a marker of the ABA level, despite their function during seed germination being far from investigative. The promoted germination or reduced dormancy phenotype was often accompanied by downregulation of all OsNCEDs but OsNCED1 [26,27,28,29,30,31,32]. Similarly, it can be inferred that a reduction in the expression of the catabolic genes OsABA8ox1, OsABA8ox2, and OsABA8ox3 inhibits seed germination [33,34].
ABA is necessary for seed maturation and the maintenance of dormancy. In Arabidopsis, there are two peaks of ABA accumulation that occur during the middle and late phases of seed maturation, of which the latter peak plays a key role in inducing and maintaining seed dormancy and thus preventing vivipary [2,35]. However, a study on rice implicated that the accumulation of ABA occurs at the earlier stage of seed development and then declines and remains low at the late stage [36]. The ABA level of a deep dormant cultivar was higher during the early and middle stages of seed development (10–20 DAP) than that observed in the low dormancy cultivars [36]. A similar result showed that the ABA accumulation peak appears at 10 DAP in hybrid rice seeds [37], suggesting the distinct mechanism of seed dormancy induction between rice and Arabidopsis. Aside from the induction of dormancy in developing seeds, ABA is involved in maintaining dormancy during imbibition [19]. Germination is determined by the decreased endogenous ABA level in the imbibing seed, which results from both the suppression of de novo synthesis and the activation of catabolism [35,38]. ABA Levels decline from imbibition onward, which is conserved in rice, as demonstrated by Liu et al. [36]. Furthermore, previous studies indicated that the reduction of the ABA level during rice imbibition is mainly related to the increased expression of OsABA8ox genes rather than the downregulation of biosynthetic OsNCED genes [33,39].
2.2. ABA Signaling Negatively Regulates Rice Seed Germination
In addition to ABA metabolism, the ABA signaling-dependent pathway also affects seed dormancy and germination. There were five ABA insensitive mutants of Arabidopsis isolated by the screen of ABA-resistant germination [40]. Subsequent research revealed that these mutants are impaired in genes participating in ABA signal transductions, and they are known as ABI1 (ABA Insensitive 1), ABI2, ABI3, ABI4, and ABI5. During seed germination, mutants of these genes are insensitive to ABA (enhanced germination and seedling growth compared with the wild type) and exhibit a reduced seed dormancy phenotype. In the past decade, great efforts in the elucidation of the core ABA-signaling pathway have been made. ABA signaling consists of three main components: ABA receptor PYR/PYL/RCAR (hereafter referred to as PYL) proteins, protein phosphatase 2C (PP2C, the negative regulators), and SNF1-related protein kinase 2 (SnRK2, the positive regulators). The ABA signaling cascade is initiated from the binding of ABA to PYLs, leading to the form of a complex with PP2Cs, thereby suppressing PP2C-mediated dephosphorylation of SnRK2s [12,41]. The ABI1 and ABI2 genes mentioned above belong to group A of the PP2C family, which negatively regulates ABA responses in Arabidopsis seed [42,43]. The activated form of SnRK2s subsequently phosphorylates ABRE-binding protein/ABRE-binding factor (AREB/ABF) transcription factors (TFs), which in turn activates the transcription of ABA-responsive genes [44]. Among the downstream transcription factors, ABI5, a member of the bZIP type transcription factor family, plays a central role in activating the ABA response in seeds [24]. Aside from that, two other TFs, ABI3 (B3 domain TF) and ABI4 (AP2 domain TF), have been reported to function together with ABI5 to induce the ABA-responsive genes. Both TFs act upstream of ABI5, and they are positive regulators of the expression of ABI5 during seed germination [45,46].
ABA signaling and its role in regulating seed dormancy and germination have been intensively studied in Arabidopsis [24]. The established knowledge provided great support for better understanding the ABA regulatory role of seed germination in rice. Thirteen rice PYL orthologues were identified based on amino acid sequence analysis of the AtPYLs, and three members were functionally investigated [47,48]. Transgenic plants overexpressing OsPYL3, OsPYL5, and OsPYL9 were found to be hypersensitive to ABA during rice seed germination, indicating they are positive regulators of ABA signaling in germination. ABA signaling is mediated by physical protein–protein interaction between the PYLs and clade A PP2Cs. It was found that OsPYL5 interacts with OsPP2C30, a homolog of AHG3 (ABA hypersensitive at germination 3), which is a PP2CA known to function specifically in the seed germination of Arabidopsis [49], in an ABA-dependent manner [47]. Other OsPP2CAs bind to OsPYLs in diverse fashions and with different intensities [48]. Among the PP2CA genes, OsPP2C51 is predominantly expressed in seeds and positively regulates seed germination in rice [50]. It is worth noting that OsPYL5 is the target ABA receptor that interacts with OsPP2C51 [50], implying that OsPYL5 is the major PYL for transducing ABA signaling in seed germination. In rice, the downstream SnRKs are designated as Osmotic Stress/ABA-Activated Protein Kinases (SAPKs) [51]. It seems like SAPK2 may predominantly mediate ABA signaling relative to the other SAPKs because SAPK2 interacts physically with OsPP2C30 and OsPP2C51 [47,50], although this is not the ABA-activated protein kinase [51]. A recent study thoroughly investigated the role of SAPK10 in rice seed germination [52]. The SAPK10 overexpression lines showed delayed seed germination and hypersensitivity to ABA. Auto-phosphorylation of SAPK10 on Serine 177 is pivotal to its function, which enables it to activate the downstream TFs. bZIPs are typical AREB or ABF TFs that can be phosphorylated by SnRK2, and they constitute the core downstream component in ABA signaling. ABI5 is a seed-specific bZIP TF-activating ABA signal transduction, and it negatively regulates seed germination in Arabidopsis [53]. Several ABA-regulated ABI5 homologs have been identified in rice such as TRAB1 [54] and OsbZIP10 [55], and their functions regarding germination were well studied. The G-box motifs (containing the core ACGT motif and known as ABREs) bound by bZIP factors are highly enriched in the upstream regions of the ABI5-regulated genes [24]. OsABI5 (also known as OREB1 or OsbZIP10) could bind to a G-box element and transactivate downstream gene expression. A germination assay showed that OsABI5 has a similar function to AtABI5 in the ABA signal pathway [55]. Subsequent studies demonstrated that OsABI5 could be phosphorylated by SAPK2 [47,50] or deactivated by dephosphorylation of the OsPP2C51 detour at SAPK2 [50]. As a group A bZIP factor, OsABF2 (OsbZIP46) binds to ABREs, and the Osabf2 mutant showed a significantly decreased sensitivity to high levels of ABA at the germination stage [56]. OsbZIP72 was found to interact with SAPK10 after a yeast-two-hybrid screen from rice seed [52]. The bzip72 mutant showed no change in germination as well as its closest homolog trab1/osbzip66 mutant did, while its overexpression lines showed enhanced sensitivity to ABA, suggesting OsbZIP72 negatively regulates seed germination. The above research indicates that AREBs, ABFs, and ABI5 are the major positive downstream regulators of ABA signaling during rice seed germination. That aside, as mentioned above, ABI3 and ABI4 are also critical to seed germination because they are positive regulators of ABI5. Their physiological functions in rice will be discussed in the following section. Collectively, these observations provide a core ABA signaling pathway in rice seed germination, consisting of PYL-PP2Cs-SAPKs-bZIPs (Figure 1). Any modification of components in this ABA response pathway can result in changes in the germination potential.
3. GA Promotes Rice Seed Germination by Acting Antagonistically to ABA
3.1. GA Metabolism Influences Rice Seed Germination
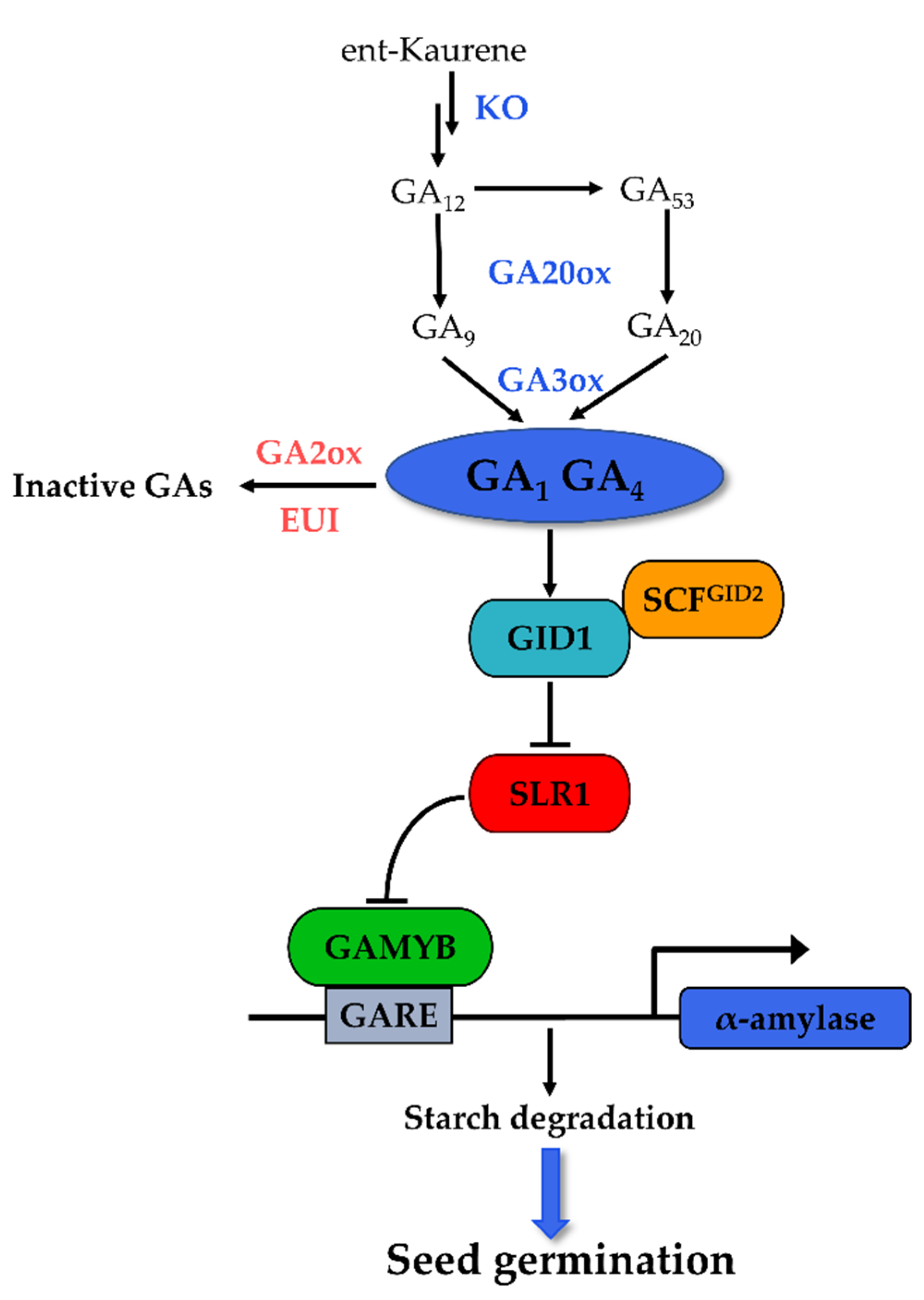
GA plays an antagonistic role to ABA in the regulation of seed germination. The positive regulation of GA in seed germination is determined by the balance between its biosynthesis and inactivation [57]. Bioactive GA is formed in terminal reactions mainly catalyzed by GA 20-oxidase (GA20ox) and GA 3-oxidase (GA3ox), while its inactivation is controlled primarily by GA 2-oxidase (GA2ox) (Figure 2). The endogenous bioactive GA contents and seed germination are correlated with the expression levels of these enzyme-encoding genes in rice. The expression of OsGA3ox2 in an embryo is important for α-amylase induction during rice seed germination [58], while that of GA-inactivating genes (GA2ox) is repressed during the imbibition of non-dormant rice [39]. The Seed Dormancy1-2 (qSD1-2) locus contains a GA synthesis gene OsGA20ox2, and the loss of function mutation in OsGA20ox2 leads to reduced seed GA levels and enhanced dormancy [59]. Likewise, OsGA2ox3 was identified as a candidate gene for controlling seed germination [60]. In a germination-defactive1 (gd1) mutant, the expressions of GA biosynthesis genes OsGA20ox1, OsGA20ox2, and OsGA3ox2 were suppressed, while that of the inactivation gene OsGA2ox3 was dramatically upregulated, resulting in decreased endogenous GA4 content and the inhibition of seed germination [61]. Another key enzyme, ent-kaurene oxidase (KO), is involved in the early steps of GA biosynthesis, converting ent-kaurene to ent-kaurenoic acid [57]. The OsKO1 gene might function mainly at the seed germination and young seedling stages. The defect in this gene leads to a decreasing GA content and shows both delayed germination and the semi-dwarfism phenotype [62]. In addition, OsLOL1 promoted seed germination through the upregulation of OsKO2 [63], suggesting the important role of OsKOs during rice germination.
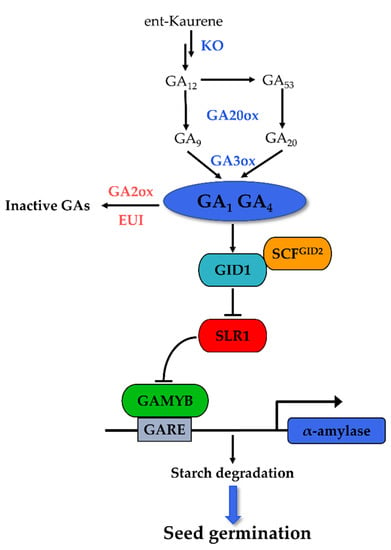
Figure 2.
Key components of GA metabolism and signaling pathway in rice seed germination.
3.2. GA Signaling in the Regulation of Rice Seed Germination
The central GA signaling components include the soluble GA receptor GIBBERELLIN INSENSITIVE DWARF1 (GID1, a positive regulator), the DELLA proteins (negative regulator), and the F-box proteins GID2 (positive regulator, SLEEPY 1 (SLY1) in Arabidopsis) [64]. Bioactive GA is perceived by its receptor GID1, leading to the formation of the GA-GID1-DELLA complex [65]. This complex associates with the F-box protein, the central component of SCFGID2 E3 ubiquitin ligase, resulting in DELLA degradation via the ubiquitin-26S proteasome pathway [64]. The degradation of DELLA proteins activates the downstream GA response TF, typically the GAMYB, and thus mediates the GA signaling effects [66].
As positive regulators in GA signaling, the GID1 and GID2 genes are supposed to promote seed germination, just as found in the observations of Arabidopsis [67,68]. However, the mutation in GID1 and GID2 for the rice does not appear to affect seed germination, although their mutations repress the α-amylase activity, and the expression of GID2 is not associated with the seed germination phenotype [35]. These results suggest that the response to GA signaling in rice seeds may be independent of the GID1 and GID2 levels. The homolog of DELLA proteins in rice was identified as SLENDER RICE1 (SLR1) [69]. There is no direct evidence about the role of SLR1 in germination, but a study on wheat showed that inhibition of the germination of non-dormant seeds is associated with increased expression of RHT1 (DELLA of wheat) [70]. Therefore, it is reasonable that the degradation of SLR1 is essential to rice seed germination.
The molecular mechanisms of downstream transcriptional regulation of GA-responsive genes have been extensively studied, and the response of α-amylase genes in aleurone cells to GA is considered the best-characterized GA response [64]. Many pivotal cis-acting elements and TFs involved in this response were proposed, of which GAMYB plays the central role. GAMYB is a classic MYB TF. In response to a GA signal, it binds to the GA-responsive elements (GAREs) of α-amylase, positively regulating the expression of the gene in the aleurone of rice seeds [71]. Another TF such as DNA binding with one finger (DOF) protein was shown to interact synergistically with GAMYB as a functional unit in the transcription regulation of the GA-responsive RAmy1A gene in rice seed [71], while two WRKY domain TFs, OsWRKY51 and OsWRKY71, worked as a transcriptional repressor of GA signaling by suppression of the OsGAMYB-activated Amy32b expression [72,73,74]. In summary, GA signal cascades during rice seed germination include the following steps. SLR1 binds to GAMYB in the absence of GA, resulting in the inhibition of α-amylase. When GA is present, the formation of a GA-GID1-SLR1 complex is promoted, which facilitates the SCFGID2-mediated degradation of SLR1 through the ubiquitin-proteasome pathway. The released GAMYB from SLR1 then binds to the GARE element in the promoter of α-amylase and activates their expression, consequently leading to the hydrolysis of starchy endosperm reserves (Figure 2).
4. ABA and GA Balance Controls Seed Germination in Rice
ABA or GA does not act solely in controlling seed germination; the dynamic balance between ABA and GA is more critical in this process. What we are concerned about is the crosstalk between these two hormones; that is, ABA and GA reciprocally regulate the metabolism and signal transduction of each other [75].
Dormancy release or germination is often accompanied by decreased ABA levels and increased GA levels, which are contributed by the altered expression patterns of synthetic and catabolic genes. A study indicated that the embryo-imposed dormancy is mainly determined by the ABA/GA ratio at the developmental stage of rice seed. Rice cultivars with a deep dormant phenotype have high ABA/GA ratios and relatively high transcript levels for the key genes in the ABA and GA metabolism pathways [36]. It is well known that ABA suppresses the synthesis of GA and promotes its deactivation, both of which result in a lower GA content in the seeds and lower completion of germination of Arabidopsis. On the contrary, GA also negatively regulates ABA biosynthesis. The expression of ABA biosynthetic genes is increased in GA-deficient mutants, resulting in the accumulation of higher ABA levels [12]. A similar observation is also available for rice. In the OsKO1 and OsKO2 (encoding GA synthetic enzyme) mutants, most of the GA biosynthetic genes and ABA catabolic genes were upregulated, whereas ABA biosynthesis genes were downregulated [62]. Aside from that, trans-genetic lines or mutants generated in rice showing altered germination phenotypes are usually accompanied by a change in expression patterns of synthetic and catabolic genes in the ABA and GA metabolism [76,77]. All results indicate the balance of the ABA and GA levels being crucial to rice seed germination.
The ABA/GA balance is also controlled by mutually antagonistic regulation at the signal transduction layer. Some components of the ABA signal cascade were also found to be regulated by GA. Rice ABA receptor OsPYLs could be degraded by Tiller Enhancer (TE), an activator of the APC/CTE E3 ubiquitin ligase complex [78]. The presence of ABA inhibits APC/CTE activity by phosphorylating TE through activating SnRK2s, while GA can repress the activity of SnRK2s and may promote the degradation of OsPYLs. Thus, the loss-of-function te mutant displays decreased seed sensitivity to ABA and enhanced germination. This work proposed a novel signaling hub mediating the antagonistic action of ABA and GA in plants. Emerging evidence indicates that AP2 domain TFs, one of the important TFs in response to ABA, play key roles in ABA and GA antagonism [79]. In Arabidopsis, ABI4 promotes ABA accumulation through repressing expression of the ABA-inactivating genes CYP707A1 and CYP707A2 but negatively mediates GA biosynthesis during the seed germination processes [80]. Though no direct target of GA metabolism genes by ABI4 has been detected, ABI4 of sorghum can directly bind to the promoter of SbGA2ox3 to activate its expression, thereby causing a reduction in GA and maintaining seed dormancy [81]. As with ABI4, a rice AP2 domain TF, OsAP2-39, regulates germination by modulating the balance between the ABA and GA levels [76]. OsAP2-39 upregulates transcription of the ABA biosynthesis gene OsNCED1 and also enhances expression of the GA-inactivating gene OsEUI (elongated uppermost internode). Furthermore, transcription of the ABA responsive AP2-like (ARAG1) gene was enhanced in a GA-deficient mutant of rice [62]. Taken together, further research on AP2 domain TFs is needed to improve our understanding of the ABA/GA crosstalk, especially in seed germination.
On the other hand, regulators in the GA signaling pathway also influence the regulation of ABA biosynthesis. In addition to the negative role in GA signaling, DELLA proteins act as regulators of GA and ABA crosstalk in germination. DELLA protein RGL2 is a major GA signaling repressor in the germination of Arabidopsis. Inhibition of seed germination by RGL2 is probably achieved by enhanced ABA synthesis and ABI5 activity [82]. When the GA level is low, RGL2 stimulates the expression of XERICO, a gene encoding a RING-H2 zinc finger factor promoting ABA accumulation [83]. In turn, increased endogenous ABA synthesis is necessary to elevate ABI5 at both the RNA and protein levels. The increased ABI5 protein is ultimately responsible for preventing seed germination [82]. Consistently, overexpression of XERICO in rice exhibited hypersensitivity to exogenous ABA during seed germination [84]. The transgenic lines exhibited a significant increase in the endogenous ABA contents and expression levels of OsNCED and OsABI5. Aside from that, as several other studies have demonstrated the regulatory role of DELLA proteins and ABI5 interaction [85,86], a detailed understanding awaits further study for the balance of ABA and GA in rice germination.
5. Other Hormones Involved in the Regulation of Rice Seed Germination
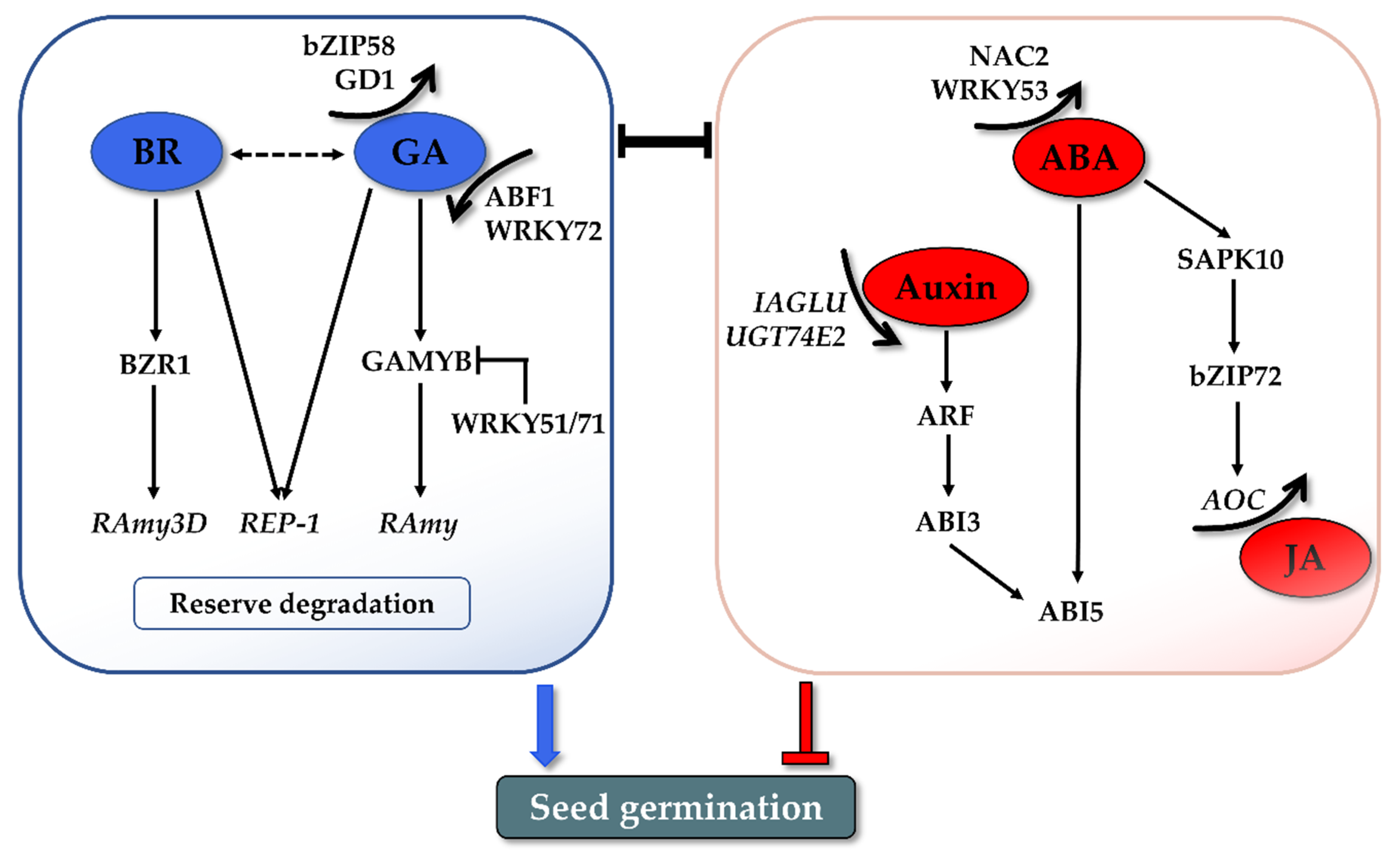
BR was found to promote seed germination. In Arabidopsis, BR can rescue the germination phenotype of severe GA-deficient and GA-insensitive mutants [87]. In addition, germination of both the BR-biosynthetic and BR-insensitive mutants is more strongly inhibited by ABA, suggesting the role of BR in reducing ABA sensitivity during germination [87]. A further study demonstrated that Brassinosteroid Insensitive 2 (BIN2), the key negative regulator in the BR signaling pathway, could phosphorylate and stabilize the ABI5 protein to antagonize the inhibitory roles of ABA on seed germination [88]. All these observations indicated that the positive role of BR is integrated with the balance of ABA/GA in seed germination. The promotion role of BR has also been verified in rice germination. The germination rate was reduced in the presence of BRZ, a BR biosynthesis inhibitor, to mimic BR-deficient germination conditions [89]. A mutation of notched belly grain4 (nbg4), which is an allele of Dwarf 11 encoding cytochrome P450 (CYP724B1), which is involved in BR biosynthesis, also displayed retarded germination [90]. A recent study further investigated the possible mechanism by which GA and BR coordinate seed germination through storage protein mobilization (Figure 3). BR and GA can promote the expression of cysteine proteinase (REP-1) during germination, leading to degradation of the stored glutelin in the endosperm to produce more amino acids, thus facilitating the growth of an embryo [91]. In addition, a novel regulatory module was found independent from the GA pathway [92]. A key downstream TF of the BR signaling cascade, brassinazole-resistant 1 (BZR1), mediates rice seed germination by binding to the α-Amylase 3D (RAmy3D) promoter and activates α-amylase expression and activity. This BZR1-RAmy3D transcriptional module then regulates the degradation of starch in the endosperm, thereby promoting seed germination [92]. More systemic research is still needed to illustrate how BR regulates rice seed germination.
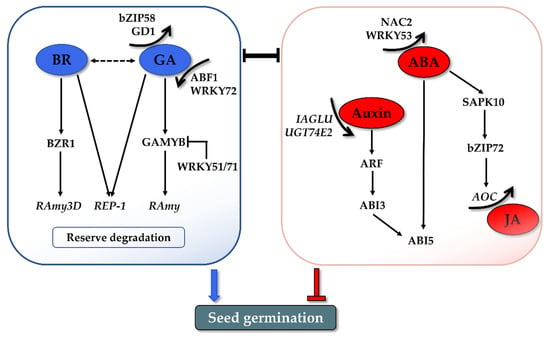
Figure 3.
Preliminary network of hormonal regulation in rice seed germination. GA and BR synergistically promote seed germination through reserve mobilization. Auxin and JA inhibit germination through the crosstalk with ABA. Numerous TFs play crucial roles in connecting upstream hormonal signals and regulating the downstream gene expressions. The interaction of various hormones and TFs influences the balance of ABA and GA, which is the core determinant of seed germination.
Auxin was reported to be involved in maintaining seed dormancy and inhibiting seed germination. Earlier studies showed that exogenous application of the auxin indole-3-acetic acid (IAA) delayed seed germination in wheat and soybean [93,94]. No such direct evidence is available in rice, but studies indicated that the changed endogenous IAA levels influence rice seed germination. Conjugate modification is one of the important processes that tightly controls the active auxin levels, by which the auxin glycosyltransferases catalyze the transfer of activated sugars to auxin [95]. IAA glycosyltransferase (IAGLU) was recently identified in rice and involved in catalyzing IAA glycosylation [96]. The higher levels of free IAA were identified in the germinating seeds of osiaglu mutants with lower germination speeds compared with the wild type. Similarly, the overexpression of UDP-glucosyltransferase 74E2 (UGT74E2), which transfers glucose to indole-3-butyric acid (IBA), could enhance seed germination in rice, which is partially attributed to the lower levels of free IBA [97]. The inhibitory effects of auxin brought out the question of what the mechanism by which auxin controls seed germination is. Emerging data showed that auxin regulates seed dormancy and germination through crosstalk with ABA. The auxin-responsive transcription factors ARF10 and ARF16 act upstream of ABI3 by indirectly promoting its transcription and consequently maintaining seed dormancy and repressing germination [98]. This crosstalk between the auxin and ABA signaling pathways is highly consistent with the observations in the recent studies on rice. Disruption of OsIAGLU elevated both the IAA and ABA contents and continuously induced the expression of OsABIs [96]. Accordingly, both the IBA and ABA levels declined in UGT74E2-overexpressing plants, and their signaling pathways were also affected, including the downregulation of ARF, ABI3, and ABI5 (Figure 3) [97].
JA has been proposed to inhibit rice seed germination at the metabolism and signaling level. The application of Methyl Jasmonate (MeJA) effectively inhibited the germination of rice seeds [99]. In the transgenic rice plants over-accumulating JA, germination was significantly retarded compared with the wild-type [100]. That aside, coi1 mutants of rice, a key component of a JA receptor, exhibited faster germination phenotypes, indicating its negative regulatory role in germination [101]. The inhibitory effect on the germination of JA is usually attributed to its working synergistically with ABA. As a key metabolic intermediate during JA biosynthesis, OPDA acts along with ABA to inhibit seed germination in Arabidopsis [102]. Jasmonate-ZIM domain (JAZ) protein is the repressor of JA signaling. JAZ was revealed to negatively modulate ABA-inhibited seed germination through interaction with ABI5 in bread wheat and Arabidopsis [103]. JAZ3 in bread wheat interacts with ABI5 and represses its transcriptional activation activity, thus resulting in repression of the ABA signal, thereby promoting seed germination [103]. The synergistic effect of ABA and JA in germination inhibition has been recently validated in rice. As described in Section 2.2, the interaction of SAPK10 and bZIP72 constitutes a critical pathway for ABA signaling in rice seed germination [52]. Interestingly, one of the downstream targets activated by bZIP72 is allene oxide cyclase (AOC), which is a JA biosynthesis step-limiting gene. The germination assays in this work also indicated that ABA-inhibited germination is partially based on the activated JA level. In summary, the most compelling evidence of the ABA- and JA-participating pathway in rice seed germination is as follows (Figure 3): exogenous ABA confers auto-phosphorylation of SAPK10, which activates and enhances bZIP72 stability as well as the DNA’s binding ability to the G-box cis-element of the AOC promoter, and the elevated AOC transcription finally increased the endogenous JA level to synergistically inhibit seed germination [52].
6. Transcriptional Regulation of Hormonal Signaling during Rice Seed Germination
Transcription factors play key roles in connecting upstream signals and downstream transcriptional networks, which contribute to the adaptation of plants to the changing environment. To date, several classes of TFs have been demonstrated to be essential for the ABA- or GA-regulated gene expression during rice seed germination: B3, AP2, bZIP, WRKY, and NAC (Table 1).
6.1. TFs Participate in ABA-Regulated Germination
As the ortholog of maize Viviparous1 (VP1) [104], ABI3 represents a classic B3 domain TF member in response to ABA-inhibited germination in Arabidopsis [105]. The positive role of ABI3/VP1 in ABA signaling is achieved by interacting with AREB/ABF/ABI5, which directly binds to ABREs in ABA-regulated genes. ABI3 acts upstream of ABI5 and is essential for ABI5 expression to arrest the growth of germinating embryos [45]. The conserved evidence has also been addressed in the OsVP1 of rice [54]. Oryza sativa Delayed Seed Germination 1 (OsDSG1) encodes a RING finger E3 ligase that negatively regulates ABI3 [106]. The osdsg1 mutant showed a delayed germination phenotype, together with significantly increased transcript levels of ABA-signaling genes such as OsABI3 and OsABI5 [106]. Thus, if the degradation of OsABI3 mediated by OsDSG1 is possibly needed in germinating rice seeds, then the ABA signal cascade is blocked via the OsABI3–OsABI5 pathway, and finally the ABA-inhibited genes (such as those genes encoding hydrolytic enzymes) reactivate, resulting in the promotion of germination.
As an AP2 domain-containing TF, ABI4 positively regulates the primary seed dormancy in Arabidopsis by directly binding to the promoters of CYP707A1 and CYP707A2, the ABA catabolism enzyme-encoding genes, subsequently promoting ABA accumulation [80]. The specific role of the ABI4 homolog in rice seed germination has not been detected thus far. Within the AP2 domain family, ABI4 is most closely related to the Drought Response Element Binding (DREB) subfamily [24]. A DREB-like gene, ABA responsive AP2-like gene (ARAG1), was demonstrated to play a role in rice seed germination [107]. Knockdown of ARAG1 conferred hypersensitivity to ABA inhibition during seed germination and seedling growth, suggesting its negative regulation in the ABA signaling pathway [107]. Furthermore, a transcriptomic study highlighted that the AP2 domain containing TFs, especially some OsDREBs enriched in the initial imbibition of rice seed germination [108]. Since AP2 domain TFs were considered to play key roles in seed dormancy regulation [2], further information is needed about the regulation role of the AP2 family in rice seed dormancy and germination.
The ABA signaling pathway mainly relies on the bZIP transcription factors. Among the large bZIP family members, the AREB/ABF/ABI5 subfamily binds to the ABREs, and OsABI5 (OsbZIP10) plays a key role in suppressing ABA-mediated seed germination (Section 2.2). In addition, several other bZIP members are also involved in the ABA-mediated regulation of seed dormancy and germination in rice. MOTHER OF FT AND TFL 2 (OsMFT2) was found to be a negative regulator of rice seed germination [109]. It delayed germination via interacting with three bZIPs (OsbZIP23, 66, and 72) and enhancing their binding to ABREs of Rab16A, a typical ABA-responsive gene. Notably, OsbZIP23 overexpression could restore the PHS phenotype of osmft2 knockout lines [109]. OsbZIP75 directly binds to the promoter of DELAY OF GERMINATION 1 (DOG1, a major seed dormancy controlling gene) and promotes the accumulation of OsDOG1L-3 [110]. High expression of OsDOG1L-3 then induces the expression of ABA synthesis genes and increased endogenous ABA contents, leading to enhancing the ABA signaling, and finally, seed germination is inhibited [110]. Generally, OsbZIPs negatively regulate rice germination, but a recent study demonstrated that OsbZIP09 positively promotes seed germination [111]. Further evidence indicated that OsbZIP09 regulates seed germination via interacting with ABA signaling to coordinate the expression of common downstream target genes (i.e., by suppressing the expression of OsLEAs (negative regulators of germination) and enhancing the expression of OsLOX2 (a positive regulator of germination)) [111].
WRKY TFs are key regulators of many plant processes, including seed dormancy and seed germination [112]. The involvement of WRKY TFs in seed germination or dormancy is determined by their response to ABA. As ABA-inducible genes, many WRKY TFs are involved in the suppression of germination by ABA. For example, seeds of the AtWRKY2 knockout mutant showed an increased delay in germination in the presence of ABA, and the ABA-induced WRKY2 accumulation during germination requires the participation of ABI3 and ABI5 [113]. In rice, OsWRKY72 and OsWRKY77 enhanced transcription from the promoter of the ABA-responsive gene HVA22 under ABA treatment [114]. OsWRKY53 directly binds to the promoters of OsABA8ox1 and OsABA8ox2 (ABA catabolic genes) to repress their expression, leading to the accumulation of ABA and furthering the inhibition of seed germination [115]. However, WRKY TFs have functional differences in regulating seed germination, because several WRKY proteins were found to promote seed germination by negatively modulating ABA-responsive genes [116]. Likewise, the knockout and RNAi lines of OsWRKY29 had enhanced seed dormancy, whereas its overexpression lines showed significantly increased germination [29]. Further assays showed that OsWRKY29 could bind to the promoters of OsABF1 (ABI5-type TF) and OsVP1 (ABI3-type TF) to inhibit their expression, thus resulting in a weakened ABA response and promoting seed germination [29].
NAC (NAM-ATAFCUC) TFs involve in various events during plant development [117], while their effects on seed germination in rice remain largely unknown. A recent study reported that a rice NAC family member, OsNAC2, negatively regulates seed germination [30]. Despite the binding ability to the promoter of ethylene biosynthetic genes, OsNAC2 probably inhibits germination through the ABA pathway instead of the ethylene and GA pathways. Ethylene is generally recognized to promote seed germination with an antagonistic function to ABA [2]. The results from OsNAC2 research indicate that ABA may play a more advantageous role in the regulation of seed germination in rice. Overall, further investigation is warranted to depict the interactions between ethylene and the upstream transcriptional regulation of NACs during seed germination.
6.2. TFs Participate in GA-Regulated Germination
In addition to ABI3, the ABI3/VP1-related subfamily of B3 domain TFs includes a family of VP1/ABI3-like (VAL) factors and two members of the leafy cotyledon class of regulators that control embryo maturation: Leafy Cotyledon1 (LEC2) and FUSCA3 (FUS3) [24]. These members play a central role in controlling the developmental regulation from embryogenesis to germination. LEC2, FUS3, and ABI3 serve as activators of seed development [118] while VALs repress it [119]. The rice mutant germination-defective1 (gd1) is defective in seed germination and seedling development [61]. GD1 encodes a B3 domain-containing TF with high similarity to VALs in Arabidopsis. In gd1, the inhibited germination is due to the decreased endogenous GA4 level [61]. These data indicate that the VAL subfamily participates directly in regulating GA homeostasis and further regulates rice seed germination and seedling development.
The G-box cis-element exists not only in ABA-inducible genes but also in the promoter of many genes in response to developmental and environmental signals, including GA biosynthesis gene OsKO2. OsbZIP58 binds the G-box cis-element of the OsKO2 promoter and activates its expression, which promotes GA biosynthesis and seed germination [63]. Another piece of evidence of bZIP involvement in GA regulating germination has also been proposed recently. OsABF1, a member of the bZIP TFs, participates in many abiotic stress responses, including salt stress and drought stress, in the ABA-dependent manner [120,121]. However, OsABF1 also functions as a key repressor of GA synthesis in seed germination [122]. Overexpression of OsABF1 showed a typical GA-deficient phenotype with semi-dwarf and retarded seed germination. Further, OsABF1 directly binds to the G-box in the promoter of semi-dwarf 1 (SD1, which encodes GA20ox2 in GA biosynthesis) and suppresses its transcription, thus reducing the endogenous GA level in the seeds [122]. Regarding the roles in ABA signaling and GA biosynthesis suppression, the ABF subfamily of bZIPs is possibly the key hub in mediating the antagonism of GA and ABA.
WRKY genes were suggested to regulate the ABA and GA signaling crosstalk in seed germination. OsWRKY71 acts as a transcriptional repressor of the GA signaling by blocking the activation of the Amy32b promoter by OsGAMYB [72]. Further studies indicate that OsWRKY71 works with another OsWRKY51 to mediate crosstalk between the GA and ABA. The ABA-inducible and GA-repressible OsWRKY51 and OsWRKY71 TFs form a heterotetramer that binds the Amy32b promoter and prevents the activation of the promoter by OsGAMYB [73]. Moreover, in the mutant of Ideal Plant Architecture 1 (IPA1) plants, seed germination and early seedling growth were retarded, together with the significantly upregulated expression of OsWRKY51 and OsWRKY71. IPA1 was further found to directly bind to and activate the expression of OsWRKY51 and OsWRKY71, which would interfere with the binding affinity of GA-induced OsGAMYB to inhibit the expression of a-amylase genes [74]. Aside from that, OsWRKY72 was recently identified as a negative regulator in rice germination through the “LRK1-OsKO2”; pathway in the aleurone layer [123]. WRKY72 directly targets LRK1 (a leucine-rich repeat receptor-like kinase) and activates its transcription, suppressing the ent-kaurene oxidase OsKO2 and thereby resulting in a reduced endogenous GA level and inhibited germination.

Table 1.
Transcription factors involved in hormonal signaling during rice seed germination.
Table 1.
Transcription factors involved in hormonal signaling during rice seed germination.
| TF Class | Gene | Function Description | Role in Germination | Reference |
|---|---|---|---|---|
| bZIP | ABI5/OsbZIP10 | Positively activates expression of ABA-responsive genes in seeds | Negative | [55] |
| ABF2/OsbZIP46 | Positively regulates ABA signaling, and germination is less sensitive to ABA in mutant | Negative | [56] | |
| OsbZIP72 | Phosphorylated by SAPK10 and activates AOC transcription | Negative | [52] | |
| TRAB1/OsbZIP66 | Interacts with VP1 and functions redundantly with OsbZIP72 in ABA signaling | Negative | [52,54] | |
| OsbZIP75 | Interacts with DOG1 to increase endogenous ABA contents and ABA signaling | Negative | [110] | |
| OsbZIP09 | Interacts with ABA signaling to coordinate the expression of common downstream target genes | Positive | [111] | |
| OsbZIP58 | Activates OsKO2 expression to promote GA biosynthesis | Positive | [63] | |
| ABF1 | Suppresses the transcription of SD1, thus reducing the endogenous GA level | Negative | [122] | |
| B3 | ABI3 | Acts upstream of ABI5 to positively regulate ABA signaling | Negative | [54,106] |
| GD1 | Similar to VALs in Arabidopsis, and mutant inhibits germination by decreasing endogenous GA4 level | Positive | [61] | |
| AP2 | OsAP2-39 | Promotes the transcription of OsNCED1 and OsEUI, thus influencing balance of ABA and GA | Negative | [76] |
| ARAG1 | Germination is hypersensitive to ABA in mutant | Positive | [107] | |
| OsDREBs | Enrich in the initial imbibition of seed germination | Not mentioned | [108] | |
| WRKY | OsWRKY72 | Enhances transcription of ABA-responsive gene and also suppresses OsKO2 expression to reduce endogenous GA level | Negative | [114,123] |
| OsWRKY53 | Represses the expression of OsABA8ox1 and OsABA8ox2 to promote ABA accumulation | Negative | [115] | |
| OsWRKY29 | Inhibits the expression of OsABF1 and OsVP1 | Positive | [29] | |
| OsWRKY71 | Interferes with OsGAMYB to block GA signaling | Negative | [72,73,74] | |
| OsWRKY51 | Functions in blocking GA signaling with OsWRKY71 | Negative | [73,74] | |
| NAC | OsNAC2 | Overexpression line delays germination by modulating the expression of OsNCED3, OsZEP1, and OsABA8ox1 | Negative | [30] |
7. Post-Transcriptional Regulation of Seed Germination in Rice
Hormones mainly act as integrators between environmental cues and molecular signals for the transcriptional regulation of gene expression in seed germination. While many other layers of gene expression exist, these include mRNA splicing, epigenetic processing, translation, and degradation [16]. Alternative splicing of mRNA generates multiple transcripts from a single gene and leads to different mRNA variants. The unusual processing patterns in the OsVP1 gene alter preharvest sprouting among rice varieties has been shown [124], with similar observations available in peas and wheat [125,126]. Epigenetic regulation involves histone protein modifications, represented by histone acetylation [127]. Deacetylation of histone is catalyzed by histone deacetylases (HDACs, which remove acetyl groups from histones), which is correlated with transcriptional repression [128]. The overexpression of HDA705, a member of HDACs in rice, delayed seed germination with downregulated expression of GA biosynthetic genes and the upregulation of ABA biosynthetic genes [129]. However, it is still unclear whether HDACs repress their expression through histone deacetylation. Protein ubiquitination is an important protein modification that negatively affects the stability of proteins. Ubiquitination of ABI3 plays a key role in germination [106]. Changes in ubiquitination were detected in 1171 proteins during rice seed germination, which especially occurred within the first 12 h [130].
8. Regulation of Rice Seed Germination by Reactive Oxygen Species
Reactive oxygen species (ROS) consist of the superoxide anion radical (O2−), hydrogen peroxide (H2O2), and the hydroxyl radical (•OH) [131]. Although ROS have long been considered toxic molecules by which ROS accumulation imposes oxidative stress on seed viability during desiccation or deterioration, increasing evidence shows that ROS function as signaling molecules to promote seed germination [131,132]. H2O2 is reported to mediate the regulation of ABA catabolism and GA biosynthesis during germination in Arabidopsis [133]. Links among ABA, ROS, and GA provide a possible explanation for the antagonism between ABA and GA in controlling seed germination [134]. The reduced ROS production by ABA in imbibed rice seeds also led to an inhibition of ascorbic acid (ASC) production. As a substrate in GA biosynthesis, insufficient ASC levels suppressed GA accumulation, which was indicated by the inhibited expression of GA biosynthesis genes [134]. These data suggest ROS and ASC are involved in the ABA-induced inhibition of GA biosynthesis. Furthermore, the ABA and H2O2 interaction during seed germination was investigated [135]. In the transgenic lines showing a delayed germination phenotype, the endogenous ABA levels were significantly higher, and the H2O2 levels significantly lower. Aside from that, exogenous H2O2 treatment can mostly recover the inhibited germination by ABA [135]. Moreover, it has recently been proposed that the improved seed vigor of OsCDP3.10 might be through enhancing H2O2 accumulation in early germination [136]. All of these studies confirmed that seed germination is enhanced through ABA catabolism and H2O2 production. PER1 is a seed-specific peroxiredoxin antioxidant for scavenging ROS [137]. PER1A in rice positively regulates seed vigor under the control of the ABA signaling pathway [138]. Despite these reports, the molecular bases underlying ROS-mediated germination in rice seeds are still elusive.
9. Conclusions and Future Directions
The complex molecular networks that underlie successful seed germination involve the integration of environmental cues and hormonal signals. Utilizing forward and reverse genetic approaches in Arabidopsis, considerable progress has been made in the dissection of molecular mechanisms underlying the hormonal regulation in seed dormancy and germination. ABA is considered the main factor influencing seed dormancy and inhibition of germination, while GA acts as an ABA antagonist, promoting the germination of rice seeds. The metabolic and signaling pathways along which ABA and GA act are determinants, as well as the effect of the balance of ABA and GA on the germination of rice seeds. Some other hormones (brassinosteroids, auxins, and jasmonic acid) contribute to the regulation of rice seed germination, most likely by participating in the ABA and GA pathways. Although several key factors that regulate this important event have been identified in rice, a systematic network of hormone-regulated germination is still lacking, and many important questions remain to be answered.
The research on rice has presented the specific role of plant hormones in seed germination, including ABA, GA, BR, auxin, and JA. However, the involvement of other hormones, such as ET, SA, CTKs, or SLs, is still largely unknown. It will be interesting to explore their interactions with the ABA or GA pathways during germination in rice or even whether they can regulate germination independently like BR does [92]. Genetic studies screening for abnormal responses to these hormones can be employed based on their physiological phenotypes. Aside from that, many key components have been discovered to be involved in metabolism and the signaling pathways for these hormones in Arabidopsis. Functional characterization of their homologs in rice would greatly extend the complex molecular networks of hormones controlling seed germination. The emerging discoveries in Arabidopsis highlighted new layers in the regulation of seed dormancy and germination, including hormone transport in seeds and the epigenetic control of germination [10,14,16]. The knowledge of temporal and tissue-specific regulation of hormone metabolism and signaling during rice seed germination is still lacking. Epigenetic mechanisms can modulate the expression of genes related to germination. More details about chromatin modification, small RNA regulation, or post-translational modifications of hormone-related genes in rice germination are needed.
The main objective of seed dormancy or germination research is to guide rice production and new cultivar breeding in agricultural practice (i.e., we need to breed cultivars with an excellent germination ability to promote direct seeding practices). We can also develop effective seed treatment approaches to benefit seed germination and seedling emergence under adverse environments. Indeed, a high germination ability or strong seed vigor has not been selected as an important breeding trait because of the transplanting program in traditional rice production. Usually, the hybrid rice, including japonica and indica hybrid rice types, possesses stronger seed vigor during the germination stage than the inbred rice under various conditions. The loci or genes for seed germination in hybrid rice represent valuable resources for molecular breeding, and the mechanisms of seed germination enhancement need to be explored. The identification and application of the elite alleles of germination-controlling genes in agricultural conditions will still be challenging for a long time in the future.
Author Contributions
Writing—original draft preparation, D.G. and F.H.; writing—review, editing, and financial support, X.Z.; collection of information and data, J.L., C.Z., Y.W., S.T. and C.S. All authors reviewed the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by Research on breeding technology and variety breeding of high quality japonica hybrid rice (21-110-3-01) and the fund for the National Science Foundation of Tianjin (19JCYBJC29500).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Rajjou, L.; Duval, M.; Gallardo, K.; Catusse, J.; Bally, J.; Job, C.; Job, D. Seed germination and vigor. Annu. Rev. Plant Biol. 2012, 63, 507–533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shu, K.; Liu, X.D.; Xie, Q.; He, Z.H. Two Faces of One Seed: Hormonal Regulation of Dormancy and Germination. Mol. Plant 2016, 9, 34–45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farooq, M.; Siddique, K.H.; Rehman, H.; Aziz, T.; Lee, D.-J.; Wahid, A. Rice direct seeding: Experiences, challenges and opportunities. Soil Till. Res. 2011, 111, 87–98. [Google Scholar] [CrossRef]
- Nonogaki, H.; Bassel, G.W.; Bewley, J.D. Germination—Still a mystery. Plant Sci. 2010, 179, 574–581. [Google Scholar] [CrossRef]
- Bewley, J.D. Seed germination and dormancy. Plant Cell 1997, 9, 1055. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weitbrecht, K.; Müller, K.; Leubner-Metzger, G. First off the mark: Early seed germination. J. Exp. Bot. 2011, 62, 3289–3309. [Google Scholar] [CrossRef] [Green Version]
- Han, C.; Yang, P. Studies on the molecular mechanisms of seed germination. Proteomics 2015, 15, 1671–1679. [Google Scholar] [CrossRef]
- Nonogaki, H. Seed dormancy and germination—Emerging mechanisms and new hypotheses. Front. Plant Sci. 2014, 5, 233. [Google Scholar] [CrossRef] [Green Version]
- Shu, K.; Meng, Y.J.; Shuai, H.W.; Liu, W.G.; Du, J.B.; Liu, J.; Yang, W.Y. Dormancy and germination: How does the crop seed decide? Plant Biol. 2015, 17, 1104–1112. [Google Scholar] [CrossRef]
- Nonogaki, H. Seed Biology Updates—Highlights and New Discoveries in Seed Dormancy and Germination Research. Front. Plant Sci. 2017, 8, 524. [Google Scholar] [CrossRef] [Green Version]
- Penfield, S. Seed dormancy and germination. Curr. Biol. 2017, 27, R874–R878. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, A.; Chen, Z. The pivotal role of abscisic acid signaling during transition from seed maturation to germination. Plant Cell Rep. 2017, 36, 689–703. [Google Scholar] [CrossRef] [PubMed]
- Nonogaki, H. Seed germination and dormancy: The classic story, new puzzles, and evolution. J. Integr. Plant Biol. 2019, 61, 541–563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carrera-Castaño, G.; Calleja-Cabrera, J.; Pernas, M.; Gómez, L.; Oñate-Sánchez, L. An updated overview on the regulation of seed germination. Plants 2020, 9, 703. [Google Scholar] [CrossRef] [PubMed]
- Nonogaki, H. A repressor complex silencing ABA signaling in seeds? J. Exp. Bot. 2020, 71, 2847–2853. [Google Scholar] [CrossRef]
- Tognacca, R.S.; Botto, J.F. Post-transcriptional regulation of seed dormancy and germination: Current understanding and future directions. Plant Commun. 2021, 2, 100169. [Google Scholar] [CrossRef]
- Finkelstein, R.; Reeves, W.; Ariizumi, T.; Steber, C. Molecular aspects of seed dormancy. Annu. Rev. Plant Biol. 2008, 59, 387–415. [Google Scholar] [CrossRef] [Green Version]
- Kucera, B.; Cohn, M.A.; Leubner-Metzger, G. Plant hormone interactions during seed dormancy release and germination. Seed Sci. Res. 2005, 15, 281–307. [Google Scholar] [CrossRef]
- Nambara, E.; Marion-Poll, A. Abscisic acid biosynthesis and catabolism. Annu. Rev. Plant Biol. 2005, 56, 165–185. [Google Scholar] [CrossRef] [Green Version]
- Agrawal, G.K.; Yamazaki, M.; Kobayashi, M.; Hirochika, R.; Miyao, A.; Hirochika, H. Screening of the rice viviparous mutants generated by endogenous retrotransposon Tos17 insertion. Tagging of a zeaxanthin epoxidase gene and a novel OsTATC gene. Plant Physiol. 2001, 125, 1248–1257. [Google Scholar] [CrossRef] [Green Version]
- Schwartz, S.H.; Qin, X.; Zeevaart, J.A. Elucidation of the indirect pathway of abscisic acid biosynthesis by mutants, genes, and enzymes. Plant Physiol. 2003, 131, 1591–1601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martínez-Andújar, C.; Ordiz, M.I.; Huang, Z.; Nonogaki, M.; Beachy, R.N.; Nonogaki, H. Induction of 9-cis-epoxycarotenoid dioxygenase in Arabidopsis thaliana seeds enhances seed dormancy. Proc. Natl. Acad. Sci. USA 2011, 108, 17225–17229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okamoto, M.; Kuwahara, A.; Seo, M.; Kushiro, T.; Asami, T.; Hirai, N.; Kamiya, Y.; Koshiba, T.; Nambara, E. CYP707A1 and CYP707A2, which encode abscisic acid 8′-hydroxylases, are indispensable for proper control of seed dormancy and germination in Arabidopsis. Plant Physiol. 2006, 141, 97–107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Finkelstein, R. Abscisic acid synthesis and response. Arab. Book/Am. Soc. Plant Biol. 2013, 11, e0166. [Google Scholar] [CrossRef] [Green Version]
- Sun, L.; Yuan, Z.; Wang, D.; Li, J.; Shi, J.; Hu, Y.; Yu, J.; Chen, X.; Chen, S.; Liang, W.; et al. Carbon Starved Anther modulates sugar and ABA metabolism to protect rice seed germination and seedling fitness. Plant Physiol. 2021, 187, 2405–2418. [Google Scholar] [CrossRef]
- Song, S.; Dai, X.; Zhang, W.H. A rice F-box gene, OsFbx352, is involved in glucose-delayed seed germination in rice. J. Exp. Bot. 2012, 63, 5559–5568. [Google Scholar] [CrossRef] [Green Version]
- Hu, Q.; Lin, C.; Guan, Y.; Sheteiwy, M.S.; Hu, W.; Hu, J. Inhibitory effect of eugenol on seed germination and pre-harvest sprouting of hybrid rice (Oryza sativa L.). Sci. Rep. 2017, 7, 5295. [Google Scholar] [CrossRef]
- Liu, J.; Hasanuzzaman, M.; Wen, H.; Zhang, J.; Peng, T.; Sun, H.; Zhao, Q. High temperature and drought stress cause abscisic acid and reactive oxygen species accumulation and suppress seed germination growth in rice. Protoplasma 2019, 256, 1217–1227. [Google Scholar] [CrossRef]
- Zhou, C.; Lin, Q.; Lan, J.; Zhang, T.; Liu, X.; Miao, R.; Mou, C.; Nguyen, T.; Wang, J.; Zhang, X.; et al. WRKY Transcription Factor OsWRKY29 Represses Seed Dormancy in Rice by Weakening Abscisic Acid Response. Front. Plant Sci. 2020, 11, 691. [Google Scholar] [CrossRef]
- Yu, J.; Mao, C.; Zhong, Q.; Yao, X.; Li, P.; Liu, C.; Ming, F. OsNAC2 Is Involved in Multiple Hormonal Pathways to Mediate Germination of Rice Seeds and Establishment of Seedling. Front. Plant Sci. 2021, 12, 699303. [Google Scholar] [CrossRef]
- Yuan, Z.; Fan, K.; Wang, Y.; Tian, L.; Zhang, C.; Sun, W.; He, H.; Yu, S. OsGRETCHENHAGEN3-2 modulates rice seed storability via accumulation of abscisic acid and protective substances. Plant Physiol. 2021, 186, 469–482. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Chen, S.; Liu, K.; Chen, Y.; Cheng, Y.; Zeng, P.; Zhu, P.; Xie, T.; Chen, S.; Zhang, H. OsHIPL1, a hedgehog-interacting protein-like 1 protein, increases seed vigor in rice. Plant Biotechnol. J. 2022, 20, 1346–1362. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.; Ye, N.; Zhang, J. Glucose-induced delay of seed germination in rice is mediated by the suppression of ABA catabolism rather than an enhancement of ABA biosynthesis. Plant Cell Physiol. 2009, 50, 644–651. [Google Scholar] [CrossRef]
- Ye, N.; Li, H.; Zhu, G.; Liu, Y.; Liu, R.; Xu, W.; Jing, Y.; Peng, X.; Zhang, J. Copper suppresses abscisic acid catabolism and catalase activity, and inhibits seed germination of rice. Plant Cell Physiol. 2014, 55, 2008–2016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tuan, P.A.; Kumar, R.; Rehal, P.K.; Toora, P.K.; Ayele, B.T. Molecular Mechanisms Underlying Abscisic Acid/Gibberellin Balance in the Control of Seed Dormancy and Germination in Cereals. Front. Plant Sci. 2018, 9, 668. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Fang, J.; Xu, F.; Chu, J.; Yan, C.; Schläppi, M.R.; Wang, Y.; Chu, C. Expression patterns of ABA and GA metabolism genes and hormone levels during rice seed development and imbibition: A comparison of dormant and non-dormant rice cultivars. J. Genet. Genom. 2014, 41, 327–338. [Google Scholar] [CrossRef]
- Zhu, L.W.; Cao, D.D.; Hu, Q.J.; Guan, Y.J.; Hu, W.M.; Nawaz, A.; Hu, J. Physiological changes and sHSPs genes relative transcription in relation to the acquisition of seed germination during maturation of hybrid rice seed. J. Sci. Food Agric. 2016, 96, 1764–1771. [Google Scholar] [CrossRef]
- Kushiro, T.; Okamoto, M.; Nakabayashi, K.; Yamagishi, K.; Kitamura, S.; Asami, T.; Hirai, N.; Koshiba, T.; Kamiya, Y.; Nambara, E. The Arabidopsis cytochrome P450 CYP707A encodes ABA 8′-hydroxylases: Key enzymes in ABA catabolism. EMBO J. 2004, 23, 1647–1656. [Google Scholar] [CrossRef] [Green Version]
- Du, W.; Cheng, J.; Cheng, Y.; Wang, L.; He, Y.; Wang, Z.; Zhang, H. Physiological characteristics and related gene expression of after-ripening on seed dormancy release in rice. Plant Biol. 2015, 17, 1156–1164. [Google Scholar] [CrossRef]
- Koornneef, M.; Reuling, G.; Karssen, C.M. The isolation and characterization of abscisic acid-insensitive mutants of Arabidopsis thaliana. Physiol. Plantarum 1984, 61, 377–383. [Google Scholar] [CrossRef]
- Zhu, J.-K. Abiotic stress signaling and responses in plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leung, J.; Merlot, S.; Giraudat, J. The Arabidopsis ABSCISIC ACID-INSENSITIVE2 (ABI2) and ABI1 genes encode homologous protein phosphatases 2C involved in abscisic acid signal transduction. Plant Cell 1997, 9, 759–771. [Google Scholar] [PubMed] [Green Version]
- Nishimura, N.; Yoshida, T.; Kitahata, N.; Asami, T.; Shinozaki, K.; Hirayama, T. ABA-Hypersensitive Germination1 encodes a protein phosphatase 2C, an essential component of abscisic acid signaling in Arabidopsis seed. Plant J. 2007, 50, 935–949. [Google Scholar] [CrossRef] [PubMed]
- Ng, L.M.; Melcher, K.; Teh, B.T.; Xu, H.E. Abscisic acid perception and signaling: Structural mechanisms and applications. Acta Pharmacol. Sin. 2014, 35, 567–584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lopez-Molina, L.; Mongrand, S.; McLachlin, D.T.; Chait, B.T.; Chua, N.H. ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. Plant J. 2002, 32, 317–328. [Google Scholar] [CrossRef] [Green Version]
- Bossi, F.; Cordoba, E.; Dupre, P.; Mendoza, M.S.; Roman, C.S.; Leon, P. The Arabidopsis ABA-INSENSITIVE (ABI) 4 factor acts as a central transcription activator of the expression of its own gene, and for the induction of ABI5 and SBE2.2 genes during sugar signaling. Plant J. 2009, 59, 359–374. [Google Scholar] [CrossRef]
- Kim, H.; Hwang, H.; Hong, J.W.; Lee, Y.N.; Ahn, I.P.; Yoon, I.S.; Yoo, S.D.; Lee, S.; Lee, S.C.; Kim, B.G. A rice orthologue of the ABA receptor, OsPYL/RCAR5, is a positive regulator of the ABA signal transduction pathway in seed germination and early seedling growth. J. Exp. Bot. 2012, 63, 1013–1024. [Google Scholar] [CrossRef] [Green Version]
- Tian, X.; Wang, Z.; Li, X.; Lv, T.; Liu, H.; Wang, L.; Niu, H.; Bu, Q. Characterization and functional analysis of pyrabactin resistance-like abscisic acid receptor family in rice. Rice 2015, 8, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, T.; Nishimura, N.; Kitahata, N.; Kuromori, T.; Ito, T.; Asami, T.; Shinozaki, K.; Hirayama, T. ABA-hypersensitive germination3 encodes a protein phosphatase 2C (AtPP2CA) that strongly regulates abscisic acid signaling during germination among Arabidopsis protein phosphatase 2Cs. Plant Physiol. 2006, 140, 115–126. [Google Scholar] [CrossRef] [Green Version]
- Bhatnagar, N.; Min, M.K.; Choi, E.H.; Kim, N.; Moon, S.J.; Yoon, I.; Kwon, T.; Jung, K.H.; Kim, B.G. The protein phosphatase 2C clade A protein OsPP2C51 positively regulates seed germination by directly inactivating OsbZIP10. Plant Mol. Biol. 2017, 93, 389–401. [Google Scholar] [CrossRef]
- Kobayashi, Y.; Yamamoto, S.; Minami, H.; Kagaya, Y.; Hattori, T. Differential activation of the rice sucrose nonfermenting1-related protein kinase2 family by hyperosmotic stress and abscisic acid. Plant Cell 2004, 16, 1163–1177. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Hou, Y.; Qiu, J.; Wang, H.; Wang, S.; Tang, L.; Tong, X.; Zhang, J. Abscisic acid promotes jasmonic acid biosynthesis via a ‘SAPK10-bZIP72-AOC’ pathway to synergistically inhibit seed germination in rice (Oryza sativa). New Phytol. 2020, 228, 1336–1353. [Google Scholar] [CrossRef] [PubMed]
- Finkelstein, R.R.; Lynch, T.J. The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 2000, 12, 599–609. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hobo, T.; Kowyama, Y.; Hattori, T. A bZIP factor, TRAB1, interacts with VP1 and mediates abscisic acid-induced transcription. Proc. Natl. Acad. Sci. USA 1999, 96, 15348–15353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zou, M.; Guan, Y.; Ren, H.; Zhang, F.; Chen, F. A bZIP transcription factor, OsABI5, is involved in rice fertility and stress tolerance. Plant Mol. Biol. 2008, 66, 675–683. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.A.; Cho, J.-I.; Han, M.; Ahn, C.-H.; Jeon, J.-S.; An, G.; Park, P.B. The ABRE-binding bZIP transcription factor OsABF2 is a positive regulator of abiotic stress and ABA signaling in rice. J. Plant Physiol. 2010, 167, 1512–1520. [Google Scholar] [CrossRef]
- Yamaguchi, S. Gibberellin metabolism and its regulation. Annu. Rev. Plant Biol. 2008, 59, 225–251. [Google Scholar] [CrossRef]
- Kaneko, M.; Itoh, H.; Ueguchi-Tanaka, M.; Ashikari, M.; Matsuoka, M. The α-amylase induction in endosperm during rice seed germination is caused by gibberellin synthesized in epithelium. Plant Physiol. 2002, 128, 1264–1270. [Google Scholar] [CrossRef] [Green Version]
- Ye, H.; Feng, J.; Zhang, L.; Zhang, J.; Mispan, M.S.; Cao, Z.; Beighley, D.H.; Yang, J.; Gu, X.-Y. Map-based cloning of seed dormancy1-2 identified a gibberellin synthesis gene regulating the development of endosperm-imposed dormancy in rice. Plant Physiol. 2015, 169, 2152–2165. [Google Scholar]
- Magwa, R.A.; Zhao, H.; Xing, Y. Genome-wide association mapping revealed a diverse genetic basis of seed dormancy across subpopulations in rice (Oryza sativa L.). BMC Genet. 2016, 17, 28. [Google Scholar] [CrossRef] [Green Version]
- Guo, X.; Hou, X.; Fang, J.; Wei, P.; Xu, B.; Chen, M.; Feng, Y.; Chu, C. The rice GERMINATION DEFECTIVE 1, encoding a B3 domain transcriptional repressor, regulates seed germination and seedling development by integrating GA and carbohydrate metabolism. Plant J. 2013, 75, 403–416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Li, M.; He, D.; Wang, K.; Yang, P. Mutations on ent-kaurene oxidase 1 encoding gene attenuate its enzyme activity of catalyzing the reaction from ent-kaurene to ent-kaurenoic acid and lead to delayed germination in rice. PLoS Genet. 2020, 16, e1008562. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Zhu, C.; Pang, J.; Zhang, X.; Yang, C.; Xia, G.; Tian, Y.; He, C. OsLOL1, a C2C2-type zinc finger protein, interacts with OsbZIP58 to promote seed germination through the modulation of gibberellin biosynthesis in Oryza sativa. Plant J. 2014, 80, 1118–1130. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.-P.; Gubler, F. Molecular mechanism of gibberellin signaling in plants. Annu. Rev. Plant Biol. 2004, 55, 197–223. [Google Scholar] [CrossRef] [Green Version]
- Murase, K.; Hirano, Y.; Sun, T.P.; Hakoshima, T. Gibberellin-induced DELLA recognition by the gibberellin receptor GID1. Nature 2008, 456, 459–463. [Google Scholar] [CrossRef]
- Gubler, F.; Kalla, R.; Roberts, J.K.; Jacobsen, J.V. Gibberellin-regulated expression of a myb gene in barley aleurone cells: Evidence for Myb transactivation of a high-pI alpha-amylase gene promoter. Plant Cell 1995, 7, 1879–1891. [Google Scholar] [CrossRef] [Green Version]
- Voegele, A.; Linkies, A.; Müller, K.; Leubner-Metzger, G. Members of the gibberellin receptor gene family GID1 (GIBBERELLIN INSENSITIVE DWARF1) play distinct roles during Lepidium sativum and Arabidopsis thaliana seed germination. J. Exp. Bot. 2011, 62, 5131–5147. [Google Scholar] [CrossRef] [Green Version]
- Ariizumi, T.; Lawrence, P.K.; Steber, C.M. The role of two F-box proteins, SLEEPY1 and SNEEZY, in Arabidopsis gibberellin signaling. Plant Physiol. 2011, 155, 765–775. [Google Scholar] [CrossRef] [Green Version]
- Ikeda, A.; Ueguchi-Tanaka, M.; Sonoda, Y.; Kitano, H.; Koshioka, M.; Futsuhara, Y.; Matsuoka, M.; Yamaguchi, J. slender rice, a constitutive gibberellin response mutant, is caused by a null mutation of the SLR1 gene, an ortholog of the height-regulating gene GAI/RGA/RHT/D8. Plant Cell 2001, 13, 999–1010. [Google Scholar] [CrossRef] [Green Version]
- Izydorczyk, C.; Nguyen, T.N.; Jo, S.; Son, S.; Tuan, P.A.; Ayele, B.T. Spatiotemporal modulation of abscisic acid and gibberellin metabolism and signalling mediates the effects of suboptimal and supraoptimal temperatures on seed germination in wheat (Triticum aestivum L.). Plant Cell Environ. 2018, 41, 1022–1037. [Google Scholar] [CrossRef]
- Washio, K. Functional dissections between GAMYB and Dof transcription factors suggest a role for protein-protein associations in the gibberellin-mediated expression of the RAmy1A gene in the rice aleurone. Plant Physiol. 2003, 133, 850–863. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.L.; Xie, Z.; Zou, X.; Casaretto, J.; Ho, T.H.; Shen, Q.J. A rice WRKY gene encodes a transcriptional repressor of the gibberellin signaling pathway in aleurone cells. Plant Physiol. 2004, 134, 1500–1513. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, Z.; Zhang, Z.L.; Zou, X.; Yang, G.; Komatsu, S.; Shen, Q.J. Interactions of two abscisic-acid induced WRKY genes in repressing gibberellin signaling in aleurone cells. Plant J. 2006, 46, 231–242. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Zhu, M.; Li, Z.; Jiang, S.; He, Z.; Xu, S.; Chen, X.; Hu, Z.; Zhang, Z. IPA1 Negatively Regulates Early Rice Seedling Development by Interfering with Starch Metabolism via the GA and WRKY Pathways. Int. J. Mol. Sci. 2021, 22, 6605. [Google Scholar] [CrossRef]
- Bewley, J.D.; Bradford, K.J.; Hilhorst, H.W.; Nonogaki, H. Dormancy and the control of germination. In Seeds; Springer: Berlin/Heidelberg, Germany, 2013; pp. 247–297. [Google Scholar]
- Yaish, M.W.; El-Kereamy, A.; Zhu, T.; Beatty, P.H.; Good, A.G.; Bi, Y.M.; Rothstein, S.J. The APETALA-2-like transcription factor OsAP2-39 controls key interactions between abscisic acid and gibberellin in rice. PLoS Genet. 2010, 6, e1001098. [Google Scholar] [CrossRef] [Green Version]
- Yang, B.; Chen, M.; Zhan, C.; Liu, K.; Cheng, Y.; Xie, T.; Zhu, P.; He, Y.; Zeng, P.; Tang, H. Identification of OsPK5 involved in rice glycolytic metabolism and GA/ABA balance for improving seed germination via GWAS. J. Exp. Bot. 2022, 73, 3446–3461. [Google Scholar] [CrossRef]
- Lin, Q.; Wu, F.; Sheng, P.; Zhang, Z.; Zhang, X.; Guo, X.; Wang, J.; Cheng, Z.; Wang, J.; Wang, H. The SnRK2-APC/CTE regulatory module mediates the antagonistic action of gibberellic acid and abscisic acid pathways. Nat. Commun. 2015, 6, 7981. [Google Scholar] [CrossRef] [Green Version]
- Shu, K.; Zhou, W.; Yang, W. APETALA 2-domain-containing transcription factors: Focusing on abscisic acid and gibberellins antagonism. New Phytol. 2018, 217, 977–983. [Google Scholar] [CrossRef]
- Shu, K.; Zhang, H.; Wang, S.; Chen, M.; Wu, Y.; Tang, S.; Liu, C.; Feng, Y.; Cao, X.; Xie, Q. ABI4 regulates primary seed dormancy by regulating the biogenesis of abscisic acid and gibberellins in Arabidopsis. PLoS Genet. 2013, 9, e1003577. [Google Scholar] [CrossRef] [Green Version]
- Cantoro, R.; Crocco, C.D.; Benech-Arnold, R.L.; Rodríguez, M.V. In vitro binding of Sorghum bicolor transcription factors ABI4 and ABI5 to a conserved region of a GA 2-OXIDASE promoter: Possible role of this interaction in the expression of seed dormancy. J. Exp. Bot. 2013, 64, 5721–5735. [Google Scholar] [CrossRef] [Green Version]
- Piskurewicz, U.; Jikumaru, Y.; Kinoshita, N.; Nambara, E.; Kamiya, Y.; Lopez-Molina, L. The gibberellic acid signaling repressor RGL2 inhibits Arabidopsis seed germination by stimulating abscisic acid synthesis and ABI5 activity. Plant Cell 2008, 20, 2729–2745. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ko, J.H.; Yang, S.H.; Han, K.H. Upregulation of an Arabidopsis RING-H2 gene, XERICO, confers drought tolerance through increased abscisic acid biosynthesis. Plant J. 2006, 47, 343–355. [Google Scholar] [CrossRef] [PubMed]
- Zeng, D.-E.; Hou, P.; Xiao, F.; Liu, Y. Overexpression of Arabidopsis XERICO gene confers enhanced drought and salt stress tolerance in rice (Oryza sativa L.). J. Plant Biochem. Biotechnol. 2015, 24, 56–64. [Google Scholar] [CrossRef]
- Lim, S.; Park, J.; Lee, N.; Jeong, J.; Toh, S.; Watanabe, A.; Kim, J.; Kang, H.; Kim, D.H.; Kawakami, N. ABA-INSENSITIVE3, ABA-INSENSITIVE5, and DELLAs interact to activate the expression of SOMNUS and other high-temperature-inducible genes in imbibed seeds in Arabidopsis. Plant Cell 2013, 25, 4863–4878. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Hu, P.; Huang, M.; Tang, Y.; Li, Y.; Li, L.; Hou, X. The NF-YC-RGL2 module integrates GA and ABA signalling to regulate seed germination in Arabidopsis. Nat. Commun. 2016, 7, 12768. [Google Scholar] [CrossRef] [Green Version]
- Steber, C.M.; McCourt, P. A role for brassinosteroids in germination in Arabidopsis. Plant Physiol. 2001, 125, 763–769. [Google Scholar] [CrossRef] [Green Version]
- Hu, Y.; Yu, D. BRASSINOSTEROID INSENSITIVE2 interacts with ABSCISIC ACID INSENSITIVE5 to mediate the antagonism of brassinosteroids to abscisic acid during seed germination in Arabidopsis. Plant Cell 2014, 26, 4394–4408. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.-F.; Xiong, M.; Xu, P.; Huang, L.-C.; Zhang, C.-Q.; Liu, Q.-Q. Dissection of brassinosteroid-regulated proteins in rice embryos during germination by quantitative proteomics. Sci. Rep. 2016, 6, 34583. [Google Scholar] [CrossRef] [Green Version]
- Tong, X.; Wang, Y.; Sun, A.; Bello, B.K.; Ni, S.; Zhang, J. Notched Belly Grain 4, a Novel Allele of Dwarf 11, Regulates Grain Shape and Seed Germination in Rice (Oryza sativa L.). Int. J. Mol. Sci. 2018, 19, 4069. [Google Scholar] [CrossRef] [Green Version]
- Xiong, M.; Chu, L.Y.; Li, Q.F.; Yu, J.W.; Yang, Y.H.; Zhou, P.; Zhou, Y.; Zhang, C.Q.; Fan, X.L.; Zhao, D.S.; et al. Brassinosteroid and gibberellin coordinate rice seed germination and embryo growth by regulating glutelin mobilization. Crop J. 2021, 9, 1039–1048. [Google Scholar] [CrossRef]
- Xiong, M.; Yu, J.; Wang, J.; Gao, Q.; Huang, L.; Chen, C.; Zhang, C.; Fan, X.; Zhao, D.; Liu, Q.Q.; et al. Brassinosteroids regulate rice seed germination through the BZR1-RAmy3D transcriptional module. Plant Physiol. 2022, 189, 402–418. [Google Scholar] [CrossRef] [PubMed]
- Ramaih, S.; Guedira, M.; Paulsen, G.M. Relationship of indoleacetic acid and tryptophan to dormancy and preharvest sprouting of wheat. Funct. Plant Biol. 2003, 30, 939–945. [Google Scholar] [CrossRef] [PubMed]
- Shuai, H.; Meng, Y.; Luo, X.; Chen, F.; Zhou, W.; Dai, Y.; Qi, Y.; Du, J.; Yang, F.; Liu, J. Exogenous auxin represses soybean seed germination through decreasing the gibberellin/abscisic acid (GA/ABA) ratio. Sci. Rep. 2017, 7, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ludwig-Müller, J. Auxin conjugates: Their role for plant development and in the evolution of land plants. J. Exp. Bot. 2011, 62, 1757–1773. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, Y.; Zhao, J.; Yang, B.; Sun, S.; Peng, L.; Wang, Z. Indole-3-acetate beta-glucosyltransferase OsIAGLU regulates seed vigour through mediating crosstalk between auxin and abscisic acid in rice. Plant Biotechnol. J. 2020, 18, 1933–1945. [Google Scholar] [CrossRef] [Green Version]
- Wang, T.; Li, P.; Mu, T.; Dong, G.; Zheng, C.; Jin, S.; Chen, T.; Hou, B.; Li, Y. Overexpression of UGT74E2, an Arabidopsis IBA Glycosyltransferase, Enhances Seed Germination and Modulates Stress Tolerance via ABA Signaling in Rice. Int. J. Mol. Sci. 2020, 21, 7239. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, H.; Zhao, Y.; Feng, Z.; Li, Q.; Yang, H.-Q.; Luan, S.; Li, J.; He, Z.-H. Auxin controls seed dormancy through stimulation of abscisic acid signaling by inducing ARF-mediated ABI3 activation in Arabidopsis. Proc. Natl. Acad. Sci. USA 2013, 110, 15485–15490. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Zhang, S.; Sun, N.; Liu, H.; Zhao, Y.; Liang, Y.; Zhang, L.; Han, Y. Functional diversity of jasmonates in rice. Rice 2015, 8, 5. [Google Scholar] [CrossRef] [Green Version]
- Lee, D.-E.; Back, K. Ectopic expression of MAP kinase inhibits germination and seedling growth in transgenic rice. Plant Growth Regul. 2005, 45, 251–257. [Google Scholar] [CrossRef]
- Yang, D.-L.; Yao, J.; Mei, C.-S.; Tong, X.-H.; Zeng, L.-J.; Li, Q.; Xiao, L.-T.; Sun, T.-P.; Li, J.; Deng, X.-W. Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc. Natl. Acad. Sci. USA 2012, 109, E1192–E1200. [Google Scholar] [CrossRef] [Green Version]
- Dave, A.; Hernández, M.L.; He, Z.; Andriotis, V.M.; Vaistij, F.E.; Larson, T.R.; Graham, I.A. 12-Oxo-phytodienoic acid accumulation during seed development represses seed germination in Arabidopsis. Plant Cell 2011, 23, 583–599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ju, L.; Jing, Y.; Shi, P.; Liu, J.; Chen, J.; Yan, J.; Chu, J.; Chen, K.M.; Sun, J. JAZ proteins modulate seed germination through interaction with ABI 5 in bread wheat and Arabidopsis. New Phytol. 2019, 223, 246–260. [Google Scholar] [CrossRef] [PubMed]
- McCarty, D.R.; Hattori, T.; Carson, C.B.; Vasil, V.; Lazar, M.; Vasil, I.K. The Viviparous-1 developmental gene of maize encodes a novel transcriptional activator. Cell 1991, 66, 895–905. [Google Scholar] [CrossRef]
- Giraudat, J.; Hauge, B.M.; Valon, C.; Smalle, J.; Parcy, F.; Goodman, H.M. Isolation of the Arabidopsis ABI3 gene by positional cloning. Plant Cell 1992, 4, 1251–1261. [Google Scholar] [CrossRef] [PubMed]
- Park, G.G.; Park, J.J.; Yoon, J.; Yu, S.N.; An, G. A RING finger E3 ligase gene, Oryza sativa Delayed Seed Germination 1 (OsDSG1), controls seed germination and stress responses in rice. Plant Mol. Biol. 2010, 74, 467–478. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Hu, Y.; Chong, K.; Wang, T. ARAG1, an ABA-responsive DREB gene, plays a role in seed germination and drought tolerance of rice. Ann. Bot. 2010, 105, 401–409. [Google Scholar] [CrossRef] [Green Version]
- Yongqi, H.; Jia, Z.; Defeng, F.; Zhibo, H.; Jiaming, L.; Yufei, Z.; Jinping, C.; Jifeng, Y.; Zhoufei, W. RNA-Seq study reveals AP2-Domain-Containing signalling regulators involved in initial imbibition of seed germination in rice. Rice Sci. 2020, 27, 302–314. [Google Scholar] [CrossRef]
- Song, S.; Wang, G.; Wu, H.; Fan, X.; Liang, L.; Zhao, H.; Li, S.; Hu, Y.; Liu, H.; Ayaad, M.; et al. OsMFT2 is involved in the regulation of ABA signaling-mediated seed germination through interacting with OsbZIP23/66/72 in rice. Plant J. 2020, 103, 532–546. [Google Scholar] [CrossRef]
- Wang, Q.; Lin, Q.; Wu, T.; Duan, E.; Huang, Y.; Yang, C.; Mou, C.; Lan, J.; Zhou, C.; Xie, K. OsDOG1L-3 regulates seed dormancy through the abscisic acid pathway in rice. Plant Sci. 2020, 298, 110570. [Google Scholar] [CrossRef]
- Zhu, C.C.; Wang, C.X.; Lu, C.Y.; Wang, J.D.; Zhou, Y.; Xiong, M.; Zhang, C.Q.; Liu, Q.Q.; Li, Q.F. Genome-Wide Identification and Expression Analysis of OsbZIP09 Target Genes in Rice Reveal Its Mechanism of Controlling Seed Germination. Int. J. Mol. Sci. 2021, 22, 1661. [Google Scholar] [CrossRef]
- Rushton, D.L.; Tripathi, P.; Rabara, R.C.; Lin, J.; Ringler, P.; Boken, A.K.; Langum, T.J.; Smidt, L.; Boomsma, D.D.; Emme, N.J. WRKY transcription factors: Key components in abscisic acid signalling. Plant Biotechnol. J. 2012, 10, 2–11. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Yu, D. Arabidopsis WRKY2 transcription factor mediates seed germination and postgermination arrest of development by abscisic acid. BMC Plant Biol. 2009, 9, 96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, Z.; Zhang, Z.-L.; Zou, X.; Huang, J.; Ruas, P.; Thompson, D.; Shen, Q.J. Annotations and functional analyses of the rice WRKY gene superfamily reveal positive and negative regulators of abscisic acid signaling in aleurone cells. Plant Physiol. 2005, 137, 176–189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, W.; Li, X.; Wang, S.; Yuan, M. OsWRKY53 Promotes Abscisic Acid Accumulation to Accelerate Leaf Senescence and Inhibit Seed Germination by Downregulating Abscisic Acid Catabolic Genes in Rice. Front. Plant Sci. 2021, 12, 816156. [Google Scholar] [CrossRef]
- Shang, Y.; Yan, L.; Liu, Z.-Q.; Cao, Z.; Mei, C.; Xin, Q.; Wu, F.-Q.; Wang, X.-F.; Du, S.-Y.; Jiang, T. The Mg-chelatase H subunit of Arabidopsis antagonizes a group of WRKY transcription repressors to relieve ABA-responsive genes of inhibition. Plant Cell 2010, 22, 1909–1935. [Google Scholar] [CrossRef] [Green Version]
- Olsen, A.N.; Ernst, H.A.; Leggio, L.L.; Skriver, K. NAC transcription factors: Structurally distinct, functionally diverse. Trends Plant. Sci. 2005, 10, 79–87. [Google Scholar] [CrossRef]
- Kroj, T.; Savino, G.; Valon, C.; Giraudat, J.; Parcy, F. Regulation of storage protein gene expression in Arabidopsis. Development 2003, 130, 6065–6073. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, M.; Wang, H.H.-Y.; McCarty, D.R. Repression of the LEAFY COTYLEDON 1/B3 regulatory network in plant embryo development by VP1/ABSCISIC ACID INSENSITIVE 3-LIKE B3 genes. Plant Physiol. 2007, 143, 902–911. [Google Scholar] [CrossRef] [Green Version]
- Hossain, A.; Lee, Y.; Cho, J.-I.; Ahn, C.-H.; Lee, S.-K.; Jeon, J.-S.; Kang, H.; Lee, C.-H.; An, G.; Park, P.B. The bZIP transcription factor OsABF1 is an ABA responsive element binding factor that enhances abiotic stress signaling in rice. Plant Mol. Biol. 2010, 72, 557–566. [Google Scholar] [CrossRef]
- Zhang, C.; Li, C.; Liu, J.; Lv, Y.; Yu, C.; Li, H.; Zhao, T.; Liu, B. The OsABF1 transcription factor improves drought tolerance by activating the transcription of COR413-TM1 in rice. J. Exp. Bot. 2017, 68, 4695–4707. [Google Scholar] [CrossRef] [Green Version]
- Tang, L.Q.; Xu, H.Y.; Wang, Y.F.; Wang, H.M.; Li, Z.Y.; Liu, X.X.; Shu, Y.Z.; Li, G.; Liu, W.N.; Ying, J.Z.; et al. OsABF1 Represses Gibberellin Biosynthesis to Regulate Plant Height and Seed Germination in Rice (Oryza sativa L.). Int. J. Mol. Sci. 2021, 22, 12220. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.M.; Hou, Y.X.; Wang, S.; Tong, X.H.; Tang, L.Q.; Ajadi, A.A.; Zhang, J.; Wang, Y.F. WRKY72 Negatively Regulates Seed Germination Through Interfering Gibberellin Pathway in Rice. Rice Sci. 2021, 28, 1–5. [Google Scholar] [CrossRef]
- Fan, J.; Niu, X.; Wang, Y.; Ren, G.; Zhuo, T.; Yang, Y.; Lu, B.R.; Liu, Y. Short, direct repeats (SDRs)-mediated post-transcriptional processing of a transcription factor gene OsVP1 in rice (Oryza sativa). J. Exp. Bot. 2007, 58, 3811–3817. [Google Scholar] [CrossRef] [PubMed]
- McKibbin, R.S.; Wilkinson, M.D.; Bailey, P.C.; Flintham, J.E.; Andrew, L.M.; Lazzeri, P.A.; Gale, M.D.; Lenton, J.R.; Holdsworth, M.J. Transcripts of Vp-1 homeologues are misspliced in modern wheat and ancestral species. Proc. Natl. Acad. Sci. USA 2002, 99, 10203–10208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gagete, A.P.; Riera, M.; Franco, L.; Rodrigo, M.I. Functional analysis of the isoforms of an ABI3-like factor of Pisum sativum generated by alternative splicing. J. Exp. Bot. 2009, 60, 1703–1714. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vaissière, T.; Sawan, C.; Herceg, Z. Epigenetic interplay between histone modifications and DNA methylation in gene silencing. Mutat. Res./Rev. Mutat. Res. 2008, 659, 40–48. [Google Scholar] [CrossRef]
- Grandperret, V.; Nicolas-Francès, V.; Wendehenne, D.; Bourque, S. Type-II histone deacetylases: Elusive plant nuclear signal transducers. Plant Cell Environ. 2014, 37, 1259–1269. [Google Scholar] [CrossRef]
- Zhao, J.; Li, M.; Gu, D.; Liu, X.; Zhang, J.; Wu, K.; Zhang, X.; Teixeira da Silva, J.A.; Duan, J. Involvement of rice histone deacetylase HDA705 in seed germination and in response to ABA and abiotic stresses. Biochem. Biophys. Res. Commun. 2016, 470, 439–444. [Google Scholar] [CrossRef]
- He, D.; Li, M.; Damaris, R.N.; Bu, C.; Xue, J.; Yang, P. Quantitative ubiquitylomics approach for characterizing the dynamic change and extensive modulation of ubiquitylation in rice seed germination. Plant J. 2020, 101, 1430–1447. [Google Scholar] [CrossRef]
- Bailly, C. Active oxygen species and antioxidants in seed biology. Seed Sci. Res. 2004, 14, 93–107. [Google Scholar] [CrossRef]
- Katsuya-Gaviria, K.; Caro, E.; Carrillo-Barral, N.; Iglesias-Fernández, R. Reactive oxygen species (ROS) and nucleic acid modifications during seed dormancy. Plants 2020, 9, 679. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Ye, N.; Liu, R.; Chen, M.; Zhang, J. H2O2 mediates the regulation of ABA catabolism and GA biosynthesis in Arabidopsis seed dormancy and germination. J. Exp. Bot. 2010, 61, 2979–2990. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, N.; Zhu, G.; Liu, Y.; Zhang, A.; Li, Y.; Liu, R.; Shi, L.; Jia, L.; Zhang, J. Ascorbic acid and reactive oxygen species are involved in the inhibition of seed germination by abscisic acid in rice seeds. J. Exp. Bot. 2012, 63, 1809–1822. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, D.; Chen, L.; Li, D.; Lv, B.; Chen, Y.; Chen, J.; Yan, X.; Liang, J. OsRACK1 is involved in abscisic acid- and H2O2-mediated signaling to regulate seed germination in rice (Oryza sativa, L.). PLoS ONE 2014, 9, e97120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, L.; Sun, S.; Yang, B.; Zhao, J.; Li, W.; Huang, Z.; Li, Z.; He, Y.; Wang, Z. Genome-wide association study reveals that the cupin domain protein OsCDP3.10 regulates seed vigour in rice. Plant Biotechnol. J. 2022, 20, 485–498. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Huang, K.; Zhou, Y.; Hu, Y.; Xiao, Y.; Chen, T.; Yin, M.; Liu, Y.; Xu, M.; Jiang, X. Degradome sequencing reveals an integrative miRNA-mediated gene interaction network regulating rice seed vigor. BMC Plant Biol. 2022, 22, 269. [Google Scholar]
- Wang, W.Q.; Xu, D.Y.; Sui, Y.P.; Ding, X.H.; Song, X.J. A multiomic study uncovers a bZIP23-PER1A-mediated detoxification pathway to enhance seed vigor in rice. Proc. Natl. Acad. Sci. USA 2022, 119, e2026355119. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).