Mass Spectrometry Imaging Spatial Tissue Analysis toward Personalized Medicine
Abstract
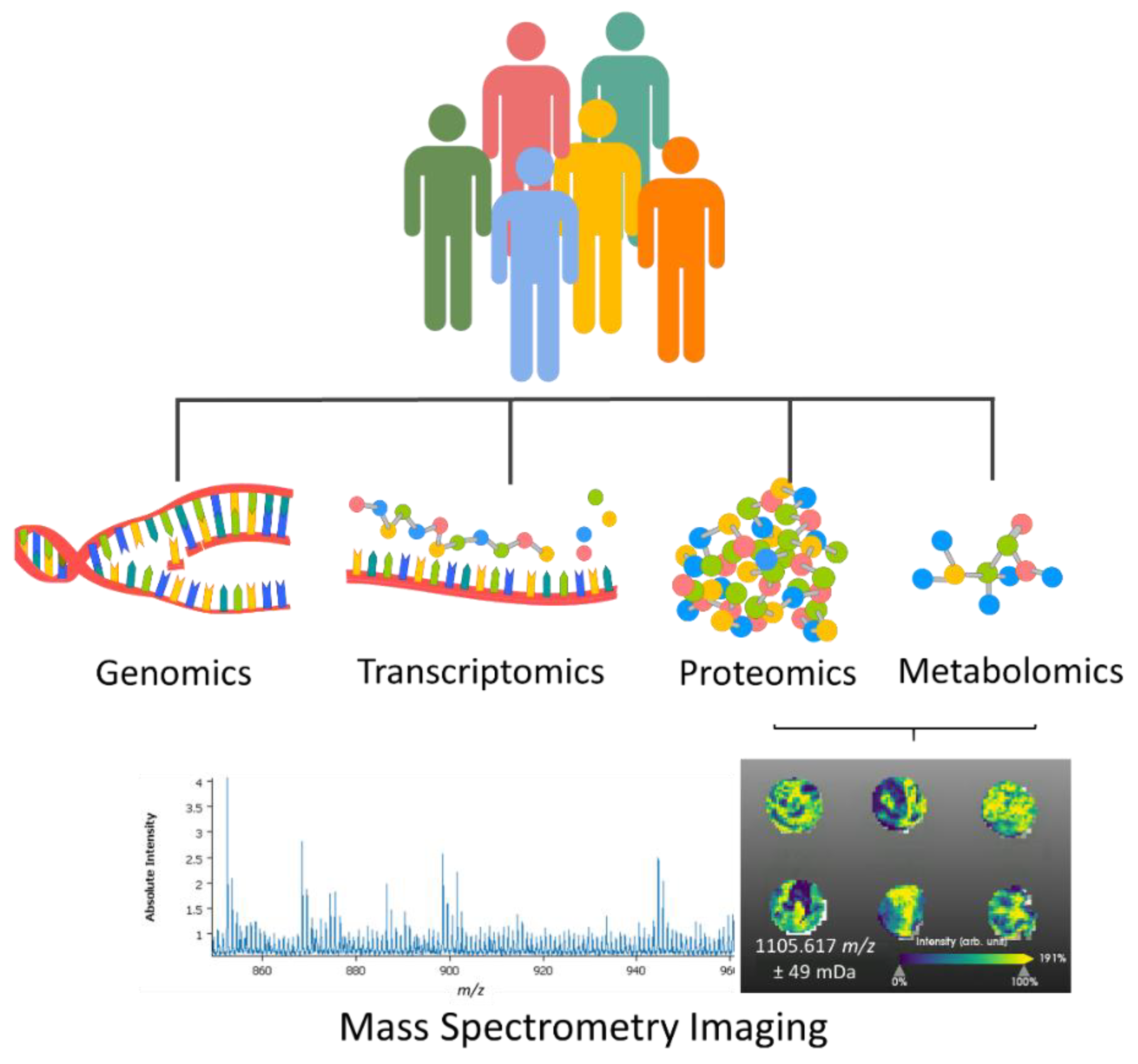
:1. Introduction
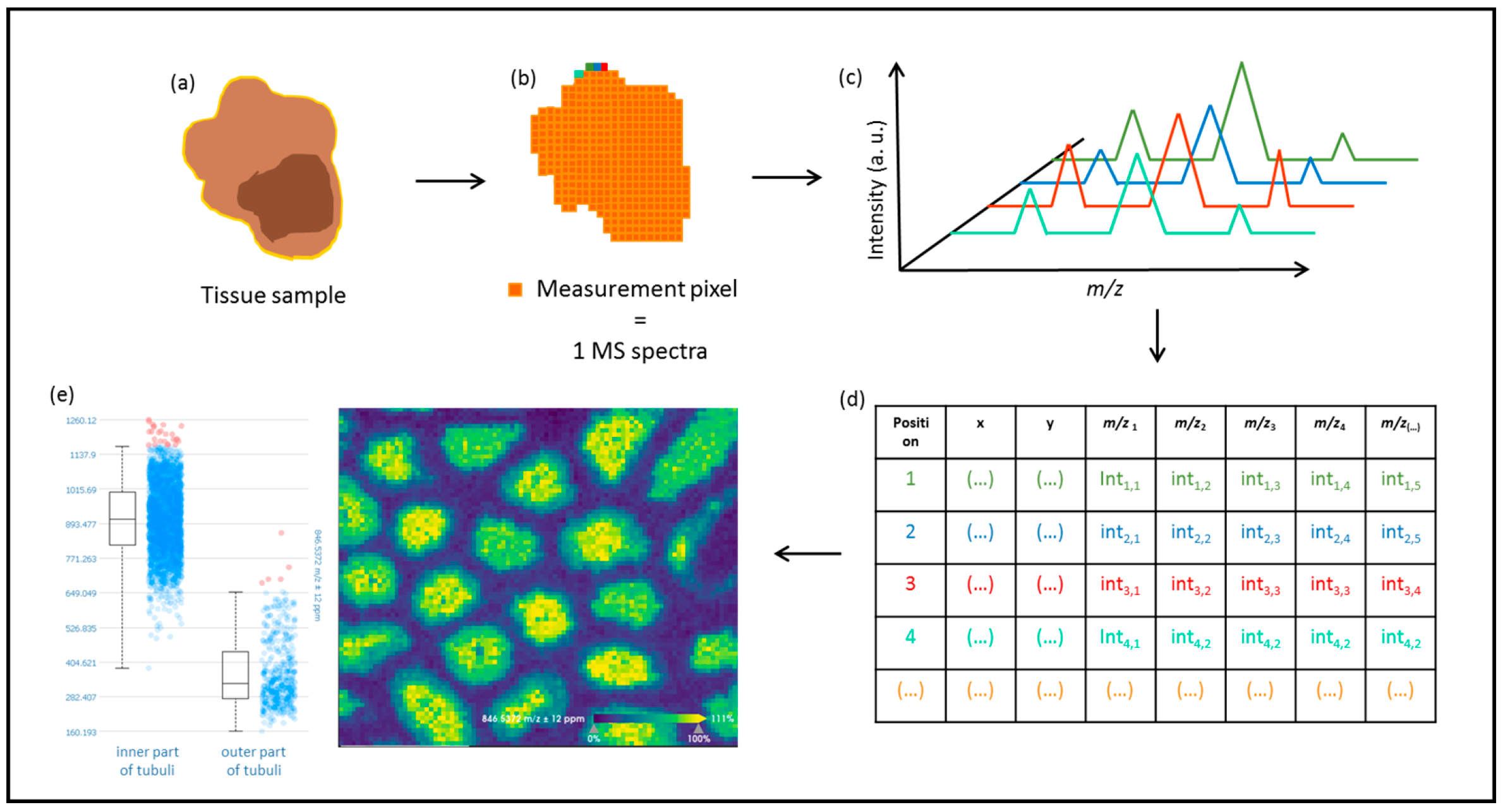
2. Mass Spectrometry Imaging (MSI)
2.1. Mass Spectrometry Imaging for Proteomic Profiling of Cancer
2.2. Prognosis Studies of Tumor Using Mass Spectrometry Imaging
2.3. Mass Spectrometry Imaging in Clinical Diagnosis
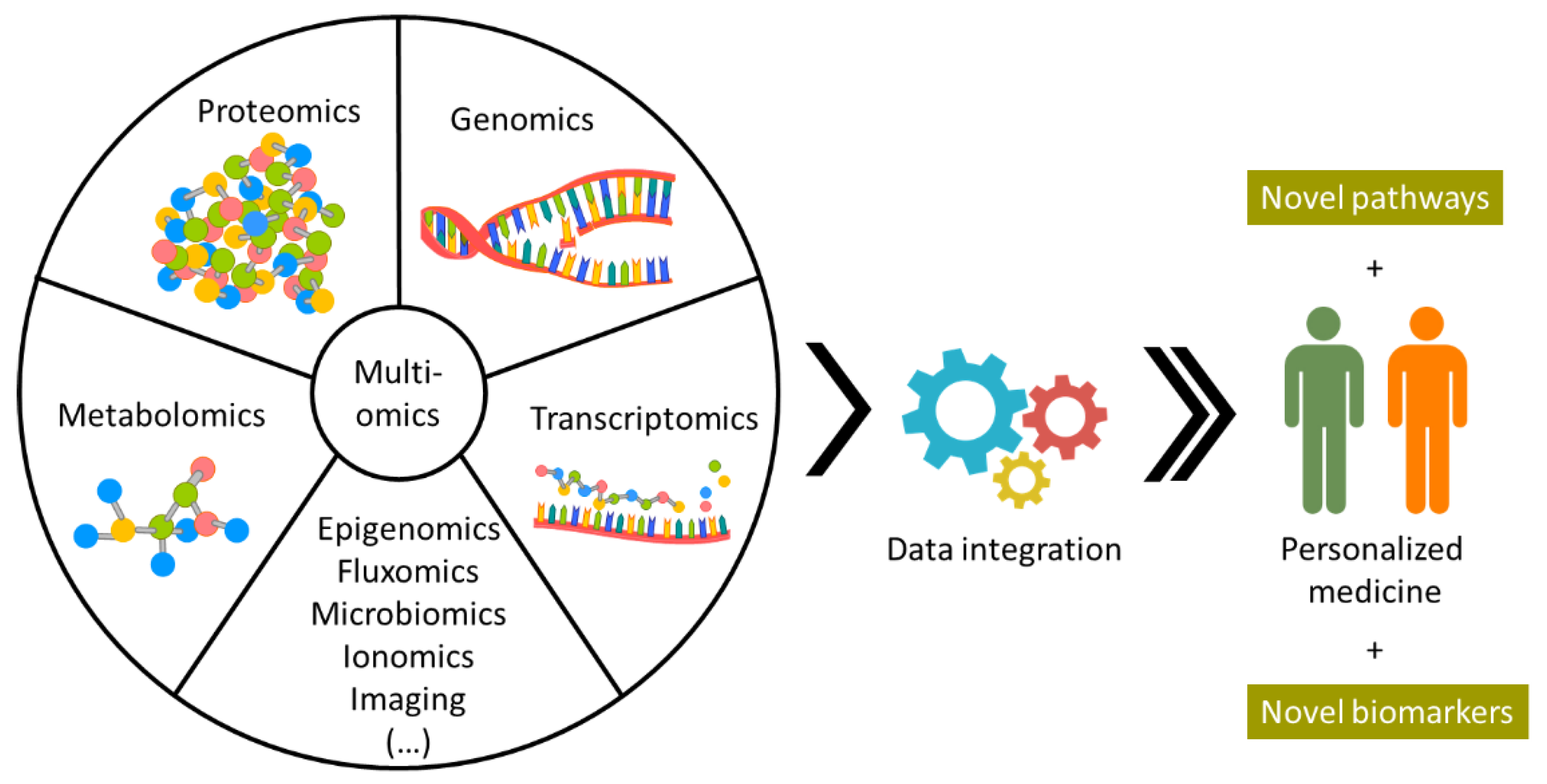
2.4. Mass Spectrometry Imaging for Personalized Medicine
3. Future Perspectives
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Moch, H.; Cubilla, A.L.; Humphrey, P.A.; Reuter, V.E.; Ulbright, T.M. The 2016 WHO Classification of Tumours of the Urinary System and Male Genital Organs—Part A: Renal, Penile, and Testicular Tumours. Eur. Urol. 2016, 70, 93–105. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Beltran, A.; Henriques, V.; Montironi, R.; Cimadamore, A.; Raspollini, M.R.; Cheng, L. Variants and new entities of bladder cancer. Histopathology 2019, 74, 77–96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trpkov, K.; Hes, O. New and emerging renal entities: A perspective post-WHO 2016 classification. Histopathology 2019, 74, 31–59. [Google Scholar] [CrossRef] [Green Version]
- Choi, J.H.; Ro, J.Y. The 2020 WHO Classification of Tumors of Soft Tissue: Selected Changes and New Entities. Adv. Anat. Pathol. 2021, 28, 44–58. [Google Scholar] [CrossRef]
- Hartl, F.U. Protein Misfolding Diseases. Annu. Rev. Biochem. 2017, 86, 21–26. [Google Scholar] [CrossRef] [Green Version]
- Brüning, A.; Jückstock, J. Misfolded proteins: From little villains to little helpers in the fight against cancer. Front. Oncol. 2015, 5, 47. [Google Scholar] [CrossRef]
- Gámez, A.; Yuste-Checa, P.; Brasil, S.; Briso-Montiano, Á.; Desviat, L.R.; Ugarte, M.; Pérez-Cerdá, C.; Pérez, B. Protein misfolding diseases: Prospects of pharmacological treatment. Clin. Genet. 2018, 93, 450–458. [Google Scholar] [CrossRef] [Green Version]
- Wheeler, D.A.; Wang, L. From human genome to cancer genome: The first decade. Genome Res. 2013, 23, 1054. [Google Scholar] [CrossRef] [Green Version]
- Turnbull, C.; Sud, A.; Houlston, R.S. Cancer genetics, precision prevention and a call to action. Nat. Genet. 2018, 50, 1212–1218. [Google Scholar] [CrossRef]
- Sondka, Z.; Bamford, S.; Cole, C.G.; Ward, S.A.; Dunham, I.; Forbes, S.A. The COSMIC Cancer Gene Census: Describing genetic dysfunction across all human cancers. Nat. Rev. Cancer 2018, 18, 696–705. [Google Scholar] [CrossRef]
- Ansari, D.; Torén, W.; Zhou, Q.; Hu, D.; Andersson, R. Proteomic and genomic profiling of pancreatic cancer. Cell Biol. Toxicol. 2019, 35, 333–343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gasparri, R.; Sedda, G.; Noberini, R.; Bonaldi, T.; Spaggiari, L. Clinical Application of Mass Spectrometry-Based Proteomics in Lung Cancer Early Diagnosis. Proteom. Clin. Appl. 2020, 14, e1900138. [Google Scholar] [CrossRef] [PubMed]
- Quail, D.F.; Joyce, J.A. Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 2013, 19, 1423–1437. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Zhuang, X.; Lin, L.; Yu, P.; Wang, Y.; Shi, Y.; Hu, G.; Sun, Y. New horizons in tumor microenvironment biology: Challenges and opportunities. BMC Med. 2015, 13, 45. [Google Scholar] [CrossRef] [Green Version]
- Vaysse, P.-M.; Heeren, R.M.A.; Porta, T.; Balluff, B. Mass spectrometry imaging for clinical research—Latest developments, applications, and current limitations. Analyst 2017, 142, 2690–2712. [Google Scholar] [CrossRef]
- Berghmans, E.; Boonen, K.; Maes, E.; Mertens, I.; Pauwels, P.; Baggerman, G. Implementation of MALDI Mass Spectrometry Imaging in Cancer Proteomics Research: Applications and Challenges. J. Pers. Med. 2020, 10, 54. [Google Scholar] [CrossRef]
- Groseclose, M.R.; Massion, P.P.; Chaurand, P.; Caprioli, R.M. High-throughput proteomic analysis of formalin-fixed paraffin-embedded tissue microarrays using MALDI imaging mass spectrometry. Proteomics 2008, 8, 3715–3724. [Google Scholar] [CrossRef]
- Haag, A.M. Mass analyzers and mass spectrometers. Adv. Exp. Med. Biol. 2016, 919, 157–169. [Google Scholar] [CrossRef]
- Feider, C.L.; Krieger, A.; Dehoog, R.J.; Eberlin, L.S. Ambient Ionization Mass Spectrometry: Recent Developments and Applications. Anal. Chem. 2019, 91, 4266–4290. [Google Scholar] [CrossRef]
- Yoshimura, Y.; Zaima, N. Application of Mass Spectrometry Imaging for Visualizing Food Components. Foods 2020, 9, 575. [Google Scholar] [CrossRef]
- Dinaiz, B.A.B.; Daniel, T.; Antony, S.; Roessner, B.U.; Boughton, B.A.; Thinagaran, D.; Sarabia, Á.D.; Bacic, Á.A.; Roessner, Á.U.; Bacic, A. Mass spectrometry imaging for plant biology: A review. Phytochem. Rev. 2015, 15, 445–488. [Google Scholar] [CrossRef] [Green Version]
- Costa, C.; Jang, M.; De Jesus, J.; Steven, R.T.; Nikula, C.J.; Elia, E.; Bunch, J.; Bellew, A.T.; Watts, J.F.; Hinder, S.; et al. Imaging mass spectrometry: A new way to distinguish dermal contact from administration of cocaine, using a single fingerprint. Analyst 2021, 146, 4010–4021. [Google Scholar] [CrossRef] [PubMed]
- Hinners, P.; O’Neill, K.C.; Lee, Y.J. Revealing Individual Lifestyles through Mass Spectrometry Imaging of Chemical Compounds in Fingerprints. Sci. Rep. 2018, 8, 5149. [Google Scholar] [CrossRef] [Green Version]
- Serratì, S.; de Summa, S.; Pilato, B.; Petriella, D.; Lacalamita, R.; Tommasi, S.; Pinto, R. Next-generation sequencing: Advances and applications in cancer diagnosis. OncoTargets Ther. 2016, 9, 7355. [Google Scholar] [CrossRef] [Green Version]
- Hassan, E.M.; DeRosa, M.C. Recent advances in cancer early detection and diagnosis: Role of nucleic acid based aptasensors. TrAC Trends Anal. Chem. 2020, 124, 115806. [Google Scholar] [CrossRef]
- Fouad, Y.A.; Aanei, C. Revisiting the hallmarks of cancer. Am. J. Cancer Res. 2017, 7, 1016. [Google Scholar] [PubMed]
- Kang, Y.P.; Ward, N.P.; DeNicola, G.M. Recent advances in cancer metabolism: A technological perspective. Exp. Mol. Med. 2018, 50, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crusz, S.M.; Balkwill, F.R. Inflammation and cancer: Advances and new agents. Nat. Rev. Clin. Oncol. 2015, 12, 584–596. [Google Scholar] [CrossRef] [PubMed]
- Pavlova, N.N.; Thompson, C.B. The Emerging Hallmarks of Cancer Metabolism. Cell Metab. 2016, 23, 27–47. [Google Scholar] [CrossRef] [Green Version]
- Mezger, S.T.P.; Mingels, A.M.A.; Bekers, O.; Heeren, R.M.A.; Cillero-Pastor, B. Mass Spectrometry Spatial-Omics on a Single Conductive Slide. Anal. Chem. 2021, 93, 2527–2533. [Google Scholar] [CrossRef]
- Gonçalves, J.P.L.; Bollwein, C.; Schlitter, A.M.; Martin, B.; Märkl, B.; Utpatel, K.; Weichert, W.; Schwamborn, K. The Impact of Histological Annotations for Accurate Tissue Classification Using Mass Spectrometry Imaging. Metabolites 2021, 11, 752. [Google Scholar] [CrossRef] [PubMed]
- Kazdal, D.; Longuespée, R.; Dietz, S.; Casadonte, R.; Schwamborn, K.; Volckmar, A.-L.; Kriegsmann, J.; Kriegsmann, K.; Fresnais, M.; Stenzinger, A.; et al. Digital PCR After MALDI–Mass Spectrometry Imaging to Combine Proteomic Mapping and Identification of Activating Mutations in Pulmonary Adenocarcinoma. Proteom. Clin. Appl. 2019, 13, 1800034. [Google Scholar] [CrossRef] [PubMed]
- Kriegsmann, K.; Longuespée, R.; Hundemer, M.; Zgorzelski, C.; Casadonte, R.; Schwamborn, K.; Weichert, W.; Schirmacher, P.; Harms, A.; Kazdal, D.; et al. Combined Immunohistochemistry after Mass Spectrometry Imaging for Superior Spatial Information. Proteom. Clin. Appl. 2019, 13, 1800035. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arentz, G.; Mittal, P.; Zhang, C.; Ho, Y.Y.; Briggs, M.; Winderbaum, L.; Hoffmann, M.K.; Hoffmann, P. Applications of Mass Spectrometry Imaging to Cancer. In Advances in Cancer Research; Academic Press Inc.: Cambridge, MA, USA, 2017; Volume 134, pp. 27–66. ISBN 9780128052495. [Google Scholar]
- Gonçalves, J.P.L.; Bollwein, C.; Weichert, W.; Schwamborn, K. Implementation of Mass Spectrometry Imaging in Pathology: Advances and Challenges. Clin. Lab. Med. 2021, 41, 173–184. [Google Scholar] [CrossRef]
- Saudemont, P.; Quanico, J.; Robin, Y.M.; Baud, A.; Balog, J.; Fatou, B.; Tierny, D.; Pascal, Q.; Minier, K.; Pottier, M.; et al. Real-Time Molecular Diagnosis of Tumors Using Water-Assisted Laser Desorption/Ionization Mass Spectrometry Technology. Cancer Cell 2018, 34, 840–851.e4. [Google Scholar] [CrossRef] [Green Version]
- Balog, J.; Sasi-Szabó, L.; Kinross, J.; Lewis, M.R.; Muirhead, L.J.; Veselkov, K.; Mirnezami, R.; Dezso, B.; Damjanovich, L.; Darzi, A.; et al. Intraoperative tissue identification using rapid evaporative ionization mass spectrometry. Sci. Transl. Med. 2013, 5, 194ra93. [Google Scholar] [CrossRef]
- St John, E.R.; Balog, J.; McKenzie, J.S.; Rossi, M.; Covington, A.; Muirhead, L.; Bodai, Z.; Rosini, F.; Speller, A.V.M.; Shousha, S.; et al. Rapid evaporative ionisation mass spectrometry of electrosurgical vapours for the identification of breast pathology: Towards an intelligent knife for breast cancer surgery. Breast Cancer Res. 2017, 19, 59. [Google Scholar] [CrossRef]
- Alexander, J.; Gildea, L.; Balog, J.; Speller, A.; McKenzie, J.; Muirhead, L.; Scott, A.; Kontovounisios, C.; Rasheed, S.; Teare, J.; et al. A novel methodology for in vivo endoscopic phenotyping of colorectal cancer based on real-time analysis of the mucosal lipidome: A prospective observational study of the iKnife. Surg. Endosc. 2017, 31, 1361–1370. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Rector, J.; Lin, J.Q.; Young, J.H.; Sans, M.; Katta, N.; Giese, N.; Yu, W.; Nagi, C.; Suliburk, J.; et al. Nondestructive tissue analysis for ex vivo and in vivo cancer diagnosis using a handheld mass spectrometry system. Sci. Transl. Med. 2017, 9, eaan3968. [Google Scholar] [CrossRef] [Green Version]
- Pirro, V.; Jarmusch, A.K.; Alfaro, C.M.; Hattab, E.M.; Cohen-Gadol, A.A.; Cooks, R.G. Utility of neurological smears for intrasurgical brain cancer diagnostics and tumour cell percentage by DESI-MS. Analyst 2017, 142, 449–454. [Google Scholar] [CrossRef]
- Calligaris, D.; Caragacianu, D.; Liu, X.; Norton, I.; Thompson, C.J.; Richardson, A.L.; Golshan, M.; Easterling, M.L.; Santagata, S.; Dillon, D.A.; et al. Application of desorption electrospray ionization mass spectrometry imaging in breast cancer margin analysis. Proc. Natl. Acad. Sci. USA 2014, 111, 15184–15189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Woolman, M.; Ferry, I.; Kuzan-Fischer, C.M.; Wu, M.; Zou, J.; Kiyota, T.; Isik, S.; Dara, D.; Aman, A.; Das, S.; et al. Rapid determination of medulloblastoma subgroup affiliation with mass spectrometry using a handheld picosecond infrared laser desorption probe. Chem. Sci. 2017, 8, 6508–6519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- King, M.E.; Zhang, J.; Lin, J.Q.; Garza, K.Y.; DeHoog, R.J.; Feider, C.L.; Bensussan, A.; Sans, M.; Krieger, A.; Badal, S.; et al. Rapid diagnosis and tumor margin assessment during pancreatic cancer surgery with the MasSpec Pen technology. Proc. Natl. Acad. Sci. USA 2021, 118, e2104411118. [Google Scholar] [CrossRef] [PubMed]
- Hänel, L.; Kwiatkowski, M.; Heikaus, L.; Schlüter, H. Mass spectrometry-based intraoperative tumor diagnostics. Future Sci. OA 2019, 5, FSO373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Margulis, K.; Chiou, A.S.; Aasi, S.Z.; Tibshirani, R.J.; Tang, J.Y.; Zare, R.N. Distinguishing malignant from benign microscopic skin lesions using desorption electrospray ionization mass spectrometry imaging. Proc. Natl. Acad. Sci. USA 2018, 115, 6347–6352. [Google Scholar] [CrossRef] [Green Version]
- Clark, A.R.; Calligaris, D.; Regan, M.S.; Pomeranz Krummel, D.; Agar, J.N.; Kallay, L.; MacDonald, T.; Schniederjan, M.; Santagata, S.; Pomeroy, S.L.; et al. Rapid discrimination of pediatric brain tumors by mass spectrometry imaging. J. Neurooncol. 2018, 140, 269–279. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Li, S.Q.; Lin, J.Q.; Yu, W.; Eberlin, L.S. Mass spectrometry imaging enables discrimination of renal oncocytoma from renal cell cancer subtypes and normal kidney tissues. Cancer Res. 2020, 80, 689–698. [Google Scholar] [CrossRef]
- Bollwein, C.; Gonçalves, J.P.L.; Utpatel, K.; Weichert, W.; Schwamborn, K. MALDI Mass Spectrometry Imaging for the Distinction of Adenocarcinomas of the Pancreas and Biliary Tree. Molecules 2022, 27, 3464. [Google Scholar] [CrossRef]
- Schwamborn, K. The Importance of Histology and Pathology in Mass Spectrometry Imaging; Elsevier: Amsterdam, The Netherlands, 2017; Volume 134, pp. 1–26. [Google Scholar]
- Martin, B.; Gonçalves, J.P.L.; Bollwein, C.; Sommer, F.; Schenkirsch, G.; Jacob, A.; Seibert, A.; Weichert, W.; Märkl, B.; Schwamborn, K. A Mass Spectrometry Imaging Based Approach for Prognosis Prediction in UICC Stage I/II Colon Cancer. Cancers 2021, 13, 5371. [Google Scholar] [CrossRef]
- Mascini, N.E.; Eijkel, G.B.; Ter Brugge, P.; Jonkers, J.; Wesseling, J.; Heeren, R.M.A. The use of mass spectrometry imaging to predict treatment response of patient-derived xenograft models of triple-negative breast cancer. J. Proteome Res. 2015, 14, 1069–1075. [Google Scholar] [CrossRef]
- Sans, M.; Gharpure, K.; Tibshirani, R.; Zhang, J.; Liang, L.; Liu, J.; Young, J.H.; Dood, R.L.; Sood, A.K.; Eberlin, L.S. Metabolic markers and statistical prediction of serous ovarian cancer aggressiveness by ambient ionization mass spectrometry imaging. Cancer Res. 2017, 77, 2903–2913. [Google Scholar] [CrossRef] [Green Version]
- Erlmeier, F.; Sun, N.; Shen, J.; Feuchtinger, A.; Buck, A.; Prade, V.M.; Kunzke, T.; Schraml, P.; Moch, H.; Autenrieth, M.; et al. MALDI Mass Spectrometry Imaging—Prognostic Pathways and Metabolites for Renal Cell Carcinomas. Cancers 2022, 14, 1763. [Google Scholar] [CrossRef] [PubMed]
- Phillips, L.; Gill, A.J.; Baxter, R.C. Novel prognostic markers in triple-negative breast cancer discovered by MALDI-mass spectrometry imaging. Front. Oncol. 2019, 9, 379. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Goldstein, L.; Nesbit, A.; Chen, M.Y. Influence of Hormone Receptor Status on Spinal Metastatic Lesions in Patients with Breast Cancer. World Neurosurg. 2016, 85, 42–48. [Google Scholar] [CrossRef] [Green Version]
- Spurr, I.B.; Birts, C.N.; Cuda, F.; Benkovic, S.J.; Blaydes, J.P.; Tavassoli, A. Targeting Tumour Proliferation with a Small-Molecule Inhibitor of AICAR Transformylase Homodimerization. ChemBioChem 2012, 13, 1628–1634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shepherd, J.H.; Uray, I.P.; Mazumdar, A.; Tsimelzon, A.; Savage, M.; Hilsenbeck, S.G.; Brown, P.H.; Shepherd, J.H.; Uray, I.P.; Mazumdar, A.; et al. The SOX11 transcription factor is a critical regulator of basal-like breast cancer growth, invasion, and basal-like gene expression. Oncotarget 2016, 7, 13106–13121. [Google Scholar] [CrossRef] [PubMed]
- Steurer, S.; Borkowski, C.; Odinga, S.; Buchholz, M.; Koop, C.; Huland, H.; Becker, M.; Witt, M.; Trede, D.; Omidi, M.; et al. MALDI mass spectrometric imaging based identification of clinically relevant signals in prostate cancer using large-scale tissue microarrays. Int. J. Cancer 2013, 133, 920–928. [Google Scholar] [CrossRef]
- Randall, E.C.; Zadra, G.; Chetta, P.; Lopez, B.G.C.; Syamala, S.; Basu, S.S.; Agar, J.N.; Loda, M.; Tempany, C.M.; Fennessy, F.M.; et al. Molecular characterization of prostate cancer with associated Gleason score using mass spectrometry imaging. Mol. Cancer Res. 2019, 17, 1155–1165. [Google Scholar] [CrossRef]
- Wang, X.; Han, J.; Hardie, D.B.; Yang, J.; Pan, J.; Borchers, C.H. Metabolomic profiling of prostate cancer by matrix assisted laser desorption/ionization-Fourier transform ion cyclotron resonance mass spectrometry imaging using Matrix Coating Assisted by an Electric Field (MCAEF). Biochim. Biophys. Acta Proteins Proteom. 2017, 1865, 755–767. [Google Scholar] [CrossRef]
- Sun, N.; Trajkovic-Arsic, M.; Li, F.; Wu, Y.; Münch, C.; Kunzke, T.; Feuchtinger, A.; Steiger, K.; Schlitter, A.M.; Weichert, W.; et al. Native glycan fragments detected by MALDI mass spectrometry imaging are independent prognostic factors in pancreatic ductal adenocarcinoma. EJNMMI Res. 2021, 11, 120. [Google Scholar] [CrossRef]
- González de Vega, R.; Clases, D.; Fernández-Sánchez, M.L.; Eiró, N.; González, L.O.; Vizoso, F.J.; Doble, P.A.; Sanz-Medel, A. MMP-11 as a biomarker for metastatic breast cancer by immunohistochemical-assisted imaging mass spectrometry. Anal. Bioanal. Chem. 2019, 411, 639–646. [Google Scholar] [CrossRef] [PubMed]
- Heijs, B.; Holst, S.; Briaire-De Bruijn, I.H.; Van Pelt, G.W.; De Ru, A.H.; Van Veelen, P.A.; Drake, R.R.; Mehta, A.S.; Mesker, W.E.; Tollenaar, R.A.; et al. Multimodal Mass Spectrometry Imaging of N-Glycans and Proteins from the Same Tissue Section. Anal. Chem. 2016, 88, 7745–7753. [Google Scholar] [CrossRef] [PubMed]
- Desbenoit, N.; Walch, A.; Spengler, B.; Brunelle, A.; Römpp, A. Correlative mass spectrometry imaging, applying time-of-flight secondary ion mass spectrometry and atmospheric pressure matrix-assisted laser desorption/ionization to a single tissue section. Rapid Commun. Mass Spectrom. 2018, 32, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Kaya, I.; Sämfors, S.; Levin, M.; Borén, J.; Fletcher, J.S. Multimodal MALDI Imaging Mass Spectrometry Reveals Spatially Correlated Lipid and Protein Changes in Mouse Heart with Acute Myocardial Infarction. J. Am. Soc. Mass Spectrom. 2020, 31, 2133–2142. [Google Scholar] [CrossRef]
- Black, A.P.; Angel, P.M.; Drake, R.R.; Mehta, A.S. Antibody Panel Based N-Glycan Imaging for N-Glycoprotein Biomarker Discovery. Curr. Protoc. Protein Sci. 2019, 98, 8429–8435. [Google Scholar] [CrossRef]
- Clift, C.L.; Drake, R.R.; Mehta, A.; Angel, P.M. Multiplexed imaging mass spectrometry of the extracellular matrix using serial enzyme digests from formalin-fixed paraffin-embedded tissue sections. Anal. Bioanal. Chem. 2021, 413, 2709–2719. [Google Scholar] [CrossRef]
- Bishop, D.P.; Westerhausen, M.T.; Barthelemy, F.; Lockwood, T.; Cole, N.; Gibbs, E.M.; Crosbie, R.H.; Nelson, S.F.; Miceli, M.C.; Doble, P.A.; et al. Quantitative immuno-mass spectrometry imaging of skeletal muscle dystrophin. Sci. Rep. 2021, 11, 1128. [Google Scholar] [CrossRef]
- Meng, Y.; Gao, C.; Lu, Q.; Ma, S.; Hang, W. Single-Cell Mass Spectrometry Imaging of Multiple Drugs and Nanomaterials at Organelle Level. ACS Nano 2021, 15, 13220–13229. [Google Scholar] [CrossRef]
- Brunner, A.-D.; Thielert, M.; Vasilopoulou, C.; Ammar, C.; Coscia, F.; Mund, A.; Hoerning, O.B.; Bache, N.; Apalategui, A.; Lubeck, M.; et al. Ultra-high sensitivity mass spectrometry quantifies single-cell proteome changes upon perturbation. Mol. Syst. Biol. 2022, 18, e10798. [Google Scholar] [CrossRef]
- Taylor, M.J.; Lukowski, J.K.; Anderton, C.R. Spatially Resolved Mass Spectrometry at the Single Cell: Recent Innovations in Proteomics and Metabolomics. J. Am. Soc. Mass Spectrom. 2021, 32, 872–894. [Google Scholar] [CrossRef]
- Ščupáková, K.; Dewez, F.; Walch, A.K.; Heeren, R.M.A.; Balluff, B.; Dewez, F.; Walch, A.K. Morphometric Cell Classification for Single-Cell MALDI-Mass Spectrometry Imaging. Angew. Chem. 2020, 132, 17600–17603. [Google Scholar] [CrossRef]
- Gilmore, I.S.; Heiles, S.; Pieterse, C.L. Metabolic Imaging at the Single-Cell Scale: Recent Advances in Mass Spectrometry Imaging. Annu. Rev. Anal. Chem. 2019, 12, 201–224. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.; Zhang, Z.; Liu, Y.; Gao, Y.; Gu, J. Recent advances in single-cell analysis by mass spectrometry. Analyst 2019, 144, 824–845. [Google Scholar] [CrossRef] [PubMed]
- Neumann, E.K.; Djambazova, K.V.; Caprioli, R.M.; Spraggins, J.M. Multimodal Imaging Mass Spectrometry: Next Generation Molecular Mapping in Biology and Medicine. J. Am. Soc. Mass Spectrom. 2020, 31, 2401–2415. [Google Scholar] [CrossRef]
- Srivastava, S.C. Paving the Way to Personalized Medicine: Production of Some Promising Theragnostic Radionuclides at Brookhaven National Laboratory. Semin. Nucl. Med. 2012, 42, 151–163. [Google Scholar] [CrossRef]
- Jackson, S.E.; Chester, J.D. Personalised cancer medicine. Int. J. Cancer 2015, 137, 262–266. [Google Scholar] [CrossRef]
- Yagnik, G.; Liu, Z.; Rothschild, K.J.; Lim, M.J. Highly Multiplexed Immunohistochemical MALDI-MS Imaging of Biomarkers in Tissues. J. Am. Soc. Mass Spectrom. 2021, 32, 977–988. [Google Scholar] [CrossRef]
- Robboy, S.J.; Weintraub, S.; Horvath, A.E.; Jensen, B.W.; Alexander, C.B.; Fody, E.P.; Crawford, J.M.; Clark, J.R.; Cantor-Weinberg, J.; Joshi, M.G.; et al. Pathologist workforce in the United States: I. Development of a predictive model to examine factors influencing supply. Arch. Pathol. Lab. Med. 2013, 137, 1723–1732. [Google Scholar] [CrossRef] [Green Version]
- Robboy, S.J.; Gupta, S.; Crawford, J.M.; Cohen, M.B.; Karcher, D.S.; Leonard, D.G.B.; Magnani, B.; Novis, D.A.; Prystowsky, M.B.; Powell, S.Z.; et al. The Pathologist Workforce in the United States: II. An Interactive Modeling Tool for Analyzing Future Qualitative and Quantitative Staffing Demands for Services. Arch. Pathol. Lab. Med. 2015, 139, 1413–1430. [Google Scholar] [CrossRef] [Green Version]
- Märkl, B.; Füzesi, L.; Huss, R.; Bauer, S.; Schaller, T. Number of pathologists in Germany: Comparison with European countries, USA, and Canada. Virchows Arch. 2021, 478, 335–341. [Google Scholar] [CrossRef]
- Buck, A.; Heijs, B.; Beine, B.; Schepers, J.; Cassese, A.; Heeren, R.M.A.; McDonnell, L.A.; Henkel, C.; Walch, A.; Balluff, B. Round robin study of formalin-fixed paraffin-embedded tissues in mass spectrometry imaging. Anal. Bioanal. Chem. 2018, 410, 5969–5980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dekker, T.J.A.; Balluff, B.D.; Jones, E.A.; Schöne, C.D.; Schmitt, M.; Aubele, M.; Kroep, J.R.; Smit, V.T.H.B.M.; Tollenaar, R.A.E.M.; Mesker, W.E.; et al. Multicenter matrix-assisted laser desorption/ionization mass spectrometry imaging (MALDI MSI) identifies proteomic differences in breast-cancer-associated stroma. J. Proteome Res. 2014, 13, 4730–4738. [Google Scholar] [CrossRef] [PubMed]
- Longuespée, R.; Ly, A.; Casadonte, R.; Schwamborn, K.; Kazdal, D.; Zgorzelski, C.; Bollwein, C.; Kriegsmann, K.; Weichert, W.; Kriegsmann, J.; et al. Identification of MALDI Imaging Proteolytic Peptides Using LC-MS/MS-Based Biomarker Discovery Data: A Proof of Concept. Proteom. Clin. Appl. 2019, 13, e1800158. [Google Scholar] [CrossRef] [Green Version]
- Deininger, S.-O.; Bollwein, C.; Casadonte, R.; Wandernoth, P.; Gonçalves, J.P.L.; Kriegsmann, K.; Kriegsmann, M.; Boskamp, T.; Kriegsmann, J.; Weichert, W.; et al. Multicenter Evaluation of Tissue Classification by Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry Imaging. Anal. Chem. 2022, 94, 8194–8201. [Google Scholar] [CrossRef]
- Porcari, A.M.; Zhang, J.; Garza, K.Y.; Rodrigues-Peres, R.M.; Lin, J.Q.; Young, J.H.; Tibshirani, R.; Nagi, C.; Paiva, G.R.; Carter, S.A.; et al. Multicenter Study Using Desorption-Electrospray-Ionization-Mass-Spectrometry Imaging for Breast-Cancer Diagnosis. Anal. Chem. 2018, 90, 11324–11332. [Google Scholar] [CrossRef]
- Lazova, R.; Smoot, K.; Anderson, H.; Powell, M.J.; Rosenberg, A.S.; Rongioletti, F.; Pilloni, L.; D’Hallewin, S.; Gueorguieva, R.; Tantcheva-Poór, I.; et al. Histopathology-guided mass spectrometry differentiates benign nevi from malignant melanoma. J. Cutan. Pathol. 2020, 47, 226–240. [Google Scholar] [CrossRef]
- Chan, A.W.; Tetzlaff, J.M.; Altman, D.G.; Laupacis, A.; Gøtzsche, P.C.; Krleža-Jerić, K.; Hróbjartsson, A.; Mann, H.; Dickersin, K.; Berlin, J.A.; et al. SPIRIT 2013 statement: Defining standard protocol items for clinical trials. Ann. Intern. Med. 2013, 158, 200–207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cruz Rivera, S.; Liu, X.; Chan, A.W.; Denniston, A.K.; Calvert, M.J.; Darzi, A.; Holmes, C.; Yau, C.; Moher, D.; Ashrafian, H.; et al. Guidelines for clinical trial protocols for interventions involving artificial intelligence: The SPIRIT-AI extension. Nat. Med. 2020, 26, 1351–1363. [Google Scholar] [CrossRef]
- Thapa, C.; Camtepe, S. Precision health data: Requirements, challenges and existing techniques for data security and privacy. Comput. Biol. Med. 2021, 129, 104130. [Google Scholar] [CrossRef]
- SPHN. Swiss Personalized Health Network (SPHN). Available online: https://sphn.ch/ (accessed on 30 May 2022).
- Benjamens, S.; Dhunnoo, P.; Meskó, B. The state of artificial intelligence-based FDA-approved medical devices and algorithms: An online database. NPJ Digit. Med. 2020, 3, 118. [Google Scholar] [CrossRef]
- Medical Imaging Cloud AI-Arterys. Available online: https://www.arterys.com/?hsLang=en (accessed on 1 June 2022).
- Computis.org-Home. Available online: http://www.computis.org/index0c51.html?option=com_frontpage&Itemid=1 (accessed on 30 May 2022).
- Robbe, M.F.; Both, J.P.; Prideaux, B.; Klinkert, I.; Picaudd, V.; Schramm, T.; Hester, A.; Guevara, V.; Stoeckli, M.; Roempp, A.; et al. Software tools of the Computis European project to process mass spectrometry images. Eur. J. Mass Spectrom. 2014, 20, 351–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schramm, T.; Hester, A.; Klinkert, I.; Both, J.P.; Heeren, R.M.A.; Brunelle, A.; Laprévote, O.; Desbenoit, N.; Robbe, M.F.; Stoeckli, M.; et al. imzML—A common data format for the flexible exchange and processing of mass spectrometry imaging data. J. Proteom. 2012, 75, 5106–5110. [Google Scholar] [CrossRef] [PubMed]
- Römpp, A.; Schramm, T.; Hester, A.; Klinkert, I.; Both, J.P.; Heeren, R.M.A.; Stöckli, M.; Spengler, B. imzML: Imaging Mass Spectrometry Markup Language: A common data format for mass spectrometry imaging. Methods Mol. Biol. 2011, 696, 205–224. [Google Scholar] [CrossRef]
- Perez-Riverol, Y.; Bai, J.; Bandla, C.; García-Seisdedos, D.; Hewapathirana, S.; Kamatchinathan, S.; Kundu, D.J.; Prakash, A.; Frericks-Zipper, A.; Eisenacher, M.; et al. The PRIDE database resources in 2022: A hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 2022, 50, D543–D552. [Google Scholar] [CrossRef] [PubMed]
- Deutsch, E.W.; Csordas, A.; Sun, Z.; Jarnuczak, A.; Perez-Riverol, Y.; Ternent, T.; Campbell, D.S.; Bernal-Llinares, M.; Okuda, S.; Kawano, S.; et al. The ProteomeXchange consortium in 2017: Supporting the cultural change in proteomics public data deposition. Nucleic Acids Res. 2017, 45, D1100–D1106. [Google Scholar] [CrossRef] [Green Version]
- Alexandrov, T.; Ovchinnikova, K.; Palmer, A.; Kovalev, V.; Tarasov, A.; Stuart, L.; Nigmetzianov, R.; Fay, D.; Contributors, K.M. METASPACE: A community-populated knowledge base of spatial metabolomes in health and disease. bioRxiv 2019, 539478. [Google Scholar] [CrossRef] [Green Version]
- METASPACE Annotation Platform. Available online: https://metaspace2020.eu/ (accessed on 30 May 2022).
- Subramanian, I.; Verma, S.; Kumar, S.; Jere, A.; Anamika, K. Multi-omics Data Integration, Interpretation, and Its Application. Bioinform. Biol. Insights 2020, 14, 7–9. [Google Scholar] [CrossRef] [Green Version]
- Graw, S.; Chappell, K.; Washam, C.L.; Gies, A.; Bird, J.; Robeson, M.S.; Byrum, S.D. Multi-omics data integration considerations and study design for biological systems and disease. Mol. Omi. 2021, 17, 170–185. [Google Scholar] [CrossRef]
- Duan, R.; Gao, L.; Gao, Y.; Hu, Y.; Xu, H.; Huang, M.; Song, K.; Wang, H.; Dong, Y.; Jiang, C.; et al. Evaluation and comparison of multi-omics data integration methods for cancer subtyping. PLoS Comput. Biol. 2021, 17, e1009224. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gonçalves, J.P.L.; Bollwein, C.; Schwamborn, K. Mass Spectrometry Imaging Spatial Tissue Analysis toward Personalized Medicine. Life 2022, 12, 1037. https://doi.org/10.3390/life12071037
Gonçalves JPL, Bollwein C, Schwamborn K. Mass Spectrometry Imaging Spatial Tissue Analysis toward Personalized Medicine. Life. 2022; 12(7):1037. https://doi.org/10.3390/life12071037
Chicago/Turabian StyleGonçalves, Juliana P. L., Christine Bollwein, and Kristina Schwamborn. 2022. "Mass Spectrometry Imaging Spatial Tissue Analysis toward Personalized Medicine" Life 12, no. 7: 1037. https://doi.org/10.3390/life12071037






