Heterologous (Over) Expression of Human SoLute Carrier (SLC) in Yeast: A Well-Recognized Tool for Human Transporter Function/Structure Studies
Abstract
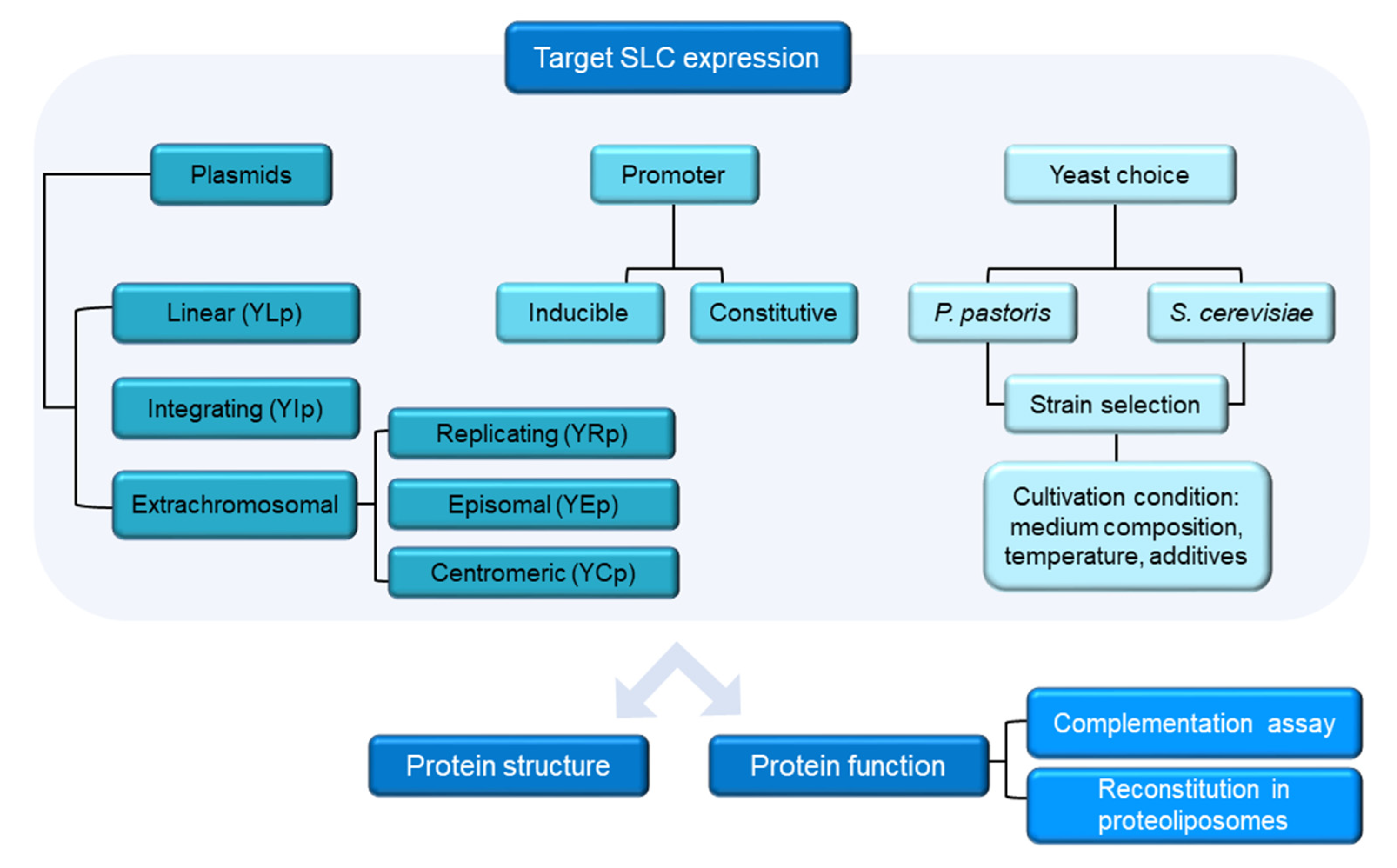
1. Introduction
2. Yeasts as a System for Heterologous Expression of Human SLC Transporters
2.1. The Homologous Recombination
2.2. Vector Choice
2.3. Selection Strategies
2.4. Yeast Transformation Methods
2.5. Saccharomyces Cerevisiae
2.6. Pichia Pastoris
Strategies for Proteins Production in P. pastoris
3. Current Methodology for Purification and Functional Studies of SLCs
3.1. Purification Strategies
3.2. From Complementation Assays to Biochemical Characterization
3.2.1. Sophisticated Complementation Assays
3.2.2. Protein Targeting and Investigation on Specific Organelle Functions/Processes
3.2.3. Study of Pathological SLC Variants
3.2.4. Transporter/Drug Interactions
3.2.5. Biotechnological Implementation
4. Selected Case Studies
4.1. SLC1A5
4.2. SLC2A1, SLC2A2, SLC2A3, SLC2A4, SLC2A5
5. Discussion
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hediger, M.A.; Clemencon, B.; Burrier, R.E.; Bruford, E.A. The ABCs of membrane transporters in health and disease (SLC series): Introduction. Mol. Asp. Med. 2013, 34, 95–107. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhang, G.Q.; Wei, Y.H.; Zhang, J.P.; Zhang, G.R.; Ren, J.X.; Duan, H.G.; Rao, Z.; Wu, X.A. The impact of drug transporters on adverse drug reaction. Eur. J. Drug Metab. Pharmacokinet. 2013, 38, 77–85. [Google Scholar] [CrossRef] [PubMed]
- Yildirim, M.A.; Goh, K.I.; Cusick, M.E.; Barabasi, A.L.; Vidal, M. Drug-target network. Nat. Biotechnol. 2007, 25, 1119–1126. [Google Scholar] [CrossRef] [PubMed]
- Santos, R.; Ursu, O.; Gaulton, A.; Bento, A.P.; Donadi, R.S.; Bologa, C.G.; Karlsson, A.; Al-Lazikani, B.; Hersey, A.; Oprea, T.I.; et al. A comprehensive map of molecular drug targets. Nat. Rev. Drug Discov. 2017, 16, 19–34. [Google Scholar] [CrossRef] [PubMed]
- Scalise, M.; Pochini, L.; Giangregorio, N.; Tonazzi, A.; Indiveri, C. Proteoliposomes as tool for assaying membrane transporter functions and interactions with xenobiotics. Pharmaceutics 2013, 5, 472–497. [Google Scholar] [CrossRef]
- Stein, W.; Litman, T. Channels, Carriers, and Pumps: An Introduction to Membrane Transport, 2nd ed.; Academic Press: Cambridge, MA, USA, 2015. [Google Scholar]
- Pizzagalli, M.D.; Bensimon, A.; Superti-Furga, G. A guide to plasma membrane solute carrier proteins. FEBS J. 2021, 288, 2784–2835. [Google Scholar] [CrossRef]
- Povey, S.; Lovering, R.; Bruford, E.; Wright, M.; Lush, M.; Wain, H. The HUGO Gene Nomenclature Committee (HGNC). Hum. Genet. 2001, 109, 678–680. [Google Scholar] [CrossRef]
- Fredriksson, R.; Nordstrom, K.J.; Stephansson, O.; Hagglund, M.G.; Schioth, H.B. The solute carrier (SLC) complement of the human genome: Phylogenetic classification reveals four major families. FEBS Lett. 2008, 582, 3811–3816. [Google Scholar] [CrossRef]
- Colas, C.; Ung, P.M.; Schlessinger, A. SLC Transporters: Structure, Function, and Drug Discovery. Medchemcomm 2016, 7, 1069–1081. [Google Scholar] [CrossRef]
- Forrest, L.R.; Zhang, Y.W.; Jacobs, M.T.; Gesmonde, J.; Xie, L.; Honig, B.H.; Rudnick, G. Mechanism for alternating access in neurotransmitter transporters. Proc. Natl. Acad. Sci. USA 2008, 105, 10338–10343. [Google Scholar] [CrossRef]
- Bai, X.; Moraes, T.F.; Reithmeier, R.A.F. Structural biology of solute carrier (SLC) membrane transport proteins. Mol. Membr. Biol. 2017, 34, 1–32. [Google Scholar] [CrossRef]
- Cesar-Razquin, A.; Snijder, B.; Frappier-Brinton, T.; Isserlin, R.; Gyimesi, G.; Bai, X.; Reithmeier, R.A.; Hepworth, D.; Hediger, M.A.; Edwards, A.M.; et al. A Call for Systematic Research on Solute Carriers. Cell 2015, 162, 478–487. [Google Scholar] [CrossRef] [PubMed]
- Junge, F.; Schneider, B.; Reckel, S.; Schwarz, D.; Dotsch, V.; Bernhard, F. Large-scale production of functional membrane proteins. Cell Mol. Life Sci. 2008, 65, 1729–1755. [Google Scholar] [CrossRef] [PubMed]
- Koepsell, H. The SLC22 family with transporters of organic cations, anions and zwitterions. Mol. Asp. Med. 2013, 34, 413–435. [Google Scholar] [CrossRef] [PubMed]
- Mueckler, M.; Thorens, B. The SLC2 (GLUT) family of membrane transporters. Mol. Asp. Med. 2013, 34, 121–138. [Google Scholar] [CrossRef]
- Stockbridge, R.B.; Kolmakova-Partensky, L.; Shane, T.; Koide, A.; Koide, S.; Miller, C.; Newstead, S. Crystal structures of a double-barrelled fluoride ion channel. Nature 2015, 525, 548–551. [Google Scholar] [CrossRef]
- Thangaratnarajah, C.; Ruprecht, J.J.; Kunji, E.R. Calcium-induced conformational changes of the regulatory domain of human mitochondrial aspartate/glutamate carriers. Nat. Commun. 2014, 5, 5491. [Google Scholar] [CrossRef]
- Kapoor, K.; Finer-Moore, J.S.; Pedersen, B.P.; Caboni, L.; Waight, A.; Hillig, R.C.; Bringmann, P.; Heisler, I.; Muller, T.; Siebeneicher, H.; et al. Mechanism of inhibition of human glucose transporter GLUT1 is conserved between cytochalasin B and phenylalanine amides. Proc. Natl. Acad. Sci. USA 2016, 113, 4711–4716. [Google Scholar] [CrossRef]
- Garaeva, A.A.; Oostergetel, G.T.; Gati, C.; Guskov, A.; Paulino, C.; Slotboom, D.J. Cryo-EM structure of the human neutral amino acid transporter ASCT2. Nat. Struct. Mol. Biol. 2018, 25, 515–521. [Google Scholar] [CrossRef]
- Deng, D.; Sun, P.; Yan, C.; Ke, M.; Jiang, X.; Xiong, L.; Ren, W.; Hirata, K.; Yamamoto, M.; Fan, S.; et al. Molecular basis of ligand recognition and transport by glucose transporters. Nature 2015, 526, 391–396. [Google Scholar] [CrossRef]
- Yan, R.; Zhao, X.; Lei, J.; Zhou, Q. Structure of the human LAT1-4F2hc heteromeric amino acid transporter complex. Nature 2019, 568, 127–130. [Google Scholar] [CrossRef] [PubMed]
- Galluccio, M.; Console, L.; Pochini, L.; Scalise, M.; Giangregorio, N.; Indiveri, C. Strategies for Successful Over-Expression of Human Membrane Transport Systems Using Bacterial Hosts: Future Perspectives. Int. J. Mol. Sci 2022, 23, 3823. [Google Scholar] [CrossRef] [PubMed]
- Geertsma, E.R.; Groeneveld, M.; Slotboom, D.J.; Poolman, B. Quality control of overexpressed membrane proteins. Proc. Natl. Acad. Sci. USA 2008, 105, 5722–5727. [Google Scholar] [CrossRef]
- Schlegel, S.; Hjelm, A.; Baumgarten, T.; Vikstrom, D.; de Gier, J.W. Bacterial-based membrane protein production. Biochim. Biophys. Acta 2014, 1843, 1739–1749. [Google Scholar] [CrossRef]
- Darby, R.A.; Cartwright, S.P.; Dilworth, M.V.; Bill, R.M. Which yeast species shall I choose? Saccharomyces cerevisiae versus Pichia pastoris (review). Methods Mol. Biol. 2012, 866, 11–23. [Google Scholar] [CrossRef]
- Byrne, B. Pichia pastoris as an expression host for membrane protein structural biology. Curr. Opin. Struct. Biol. 2015, 32, 9–17. [Google Scholar] [CrossRef]
- Lee, J.Y.; Chen, H.; Liu, A.; Alba, B.M.; Lim, A.C. Auto-induction of Pichia pastoris AOX1 promoter for membrane protein expression. Protein Expr. Purif. 2017, 137, 7–12. [Google Scholar] [CrossRef] [PubMed]
- Souabni, H.; Ezzine, A.; Bizouarn, T.; Baciou, L. Functional Assembly of Soluble and Membrane Recombinant Proteins of Mammalian NADPH Oxidase Complex. Methods Mol. Biol. 2017, 1635, 27–43. [Google Scholar] [CrossRef]
- Pingitore, P.; Pochini, L.; Scalise, M.; Galluccio, M.; Hedfalk, K.; Indiveri, C. Large scale production of the active human ASCT2 (SLC1A5) transporter in Pichia pastoris—Functional and kinetic asymmetry revealed in proteoliposomes. Biochim. Biophys. Acta 2013, 1828, 2238–2246. [Google Scholar] [CrossRef]
- Claes, K.; Guerfal, M.; Callewaert, N. Membrane protein expression and analysis in yeast. Methods Enzymol. 2015, 556, 123–140. [Google Scholar] [CrossRef]
- Hartmann, L.; Kugler, V.; Wagner, R. Expression of Eukaryotic Membrane Proteins in Pichia pastoris. Methods Mol. Biol. 2016, 1432, 143–162. [Google Scholar] [CrossRef] [PubMed]
- Scalise, M.; Pappacoda, G.; Mazza, T.; Console, L.; Pochini, L.; Indiveri, C. Cysteine 467 of the ASCT2 Amino Acid Transporter Is a Molecular Determinant of the Antiport Mechanism. Int. J. Mol. Sci. 2022, 23, 1127. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, B.P.; Kumar, H.; Waight, A.B.; Risenmay, A.J.; Roe-Zurz, Z.; Chau, B.H.; Schlessinger, A.; Bonomi, M.; Harries, W.; Sali, A.; et al. Crystal structure of a eukaryotic phosphate transporter. Nature 2013, 496, 533–536. [Google Scholar] [CrossRef] [PubMed]
- Cregg, J.M.; Cereghino, J.L.; Shi, J.; Higgins, D.R. Recombinant protein expression in Pichia pastoris. Mol. Biotechnol. 2000, 16, 23–52. [Google Scholar] [CrossRef]
- Bill, R.M. Yeast—A panacea for the structure-function analysis of membrane proteins? Curr. Genet. 2001, 40, 157–171. [Google Scholar] [CrossRef]
- Karathia, H.; Vilaprinyo, E.; Sorribas, A.; Alves, R. Saccharomyces cerevisiae as a model organism: A comparative study. PLoS ONE 2011, 6, e16015. [Google Scholar] [CrossRef]
- Eckart, M.R.; Bussineau, C.M. Quality and authenticity of heterologous proteins synthesized in yeast. Curr. Opin. Biotechnol. 1996, 7, 525–530. [Google Scholar] [CrossRef]
- Gemmill, T.R.; Trimble, R.B. Overview of N- and O-linked oligosaccharide structures found in various yeast species. Biochim. Biophys. Acta 1999, 1426, 227–237. [Google Scholar] [CrossRef]
- Chiba, Y.; Suzuki, M.; Yoshida, S.; Yoshida, A.; Ikenaga, H.; Takeuchi, M.; Jigami, Y.; Ichishima, E. Production of human compatible high mannose-type (Man5GlcNAc2) sugar chains in Saccharomyces cerevisiae. J. Biol. Chem. 1998, 273, 26298–26304. [Google Scholar] [CrossRef]
- Nakanishi-Shindo, Y.; Nakayama, K.; Tanaka, A.; Toda, Y.; Jigami, Y. Structure of the N-linked oligosaccharides that show the complete loss of alpha-1,6-polymannose outer chain from och1, och1 mnn1, and och1 mnn1 alg3 mutants of Saccharomyces cerevisiae. J. Biol. Chem. 1993, 268, 26338–26345. [Google Scholar] [CrossRef]
- Oka, T.; Jigami, Y. Reconstruction of de novo pathway for synthesis of UDP-glucuronic acid and UDP-xylose from intrinsic UDP-glucose in Saccharomyces cerevisiae. FEBS J. 2006, 273, 2645–2657. [Google Scholar] [CrossRef] [PubMed]
- Chigira, Y.; Oka, T.; Okajima, T.; Jigami, Y. Engineering of a mammalian O-glycosylation pathway in the yeast Saccharomyces cerevisiae: Production of O-fucosylated epidermal growth factor domains. Glycobiology 2008, 18, 303–314. [Google Scholar] [CrossRef] [PubMed]
- Wach, A. PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in S. cerevisiae. Yeast 1996, 12, 259–265. [Google Scholar] [CrossRef]
- Longtine, M.S.; McKenzie, A., 3rd; Demarini, D.J.; Shah, N.G.; Wach, A.; Brachat, A.; Philippsen, P.; Pringle, J.R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 1998, 14, 953–961. [Google Scholar] [CrossRef]
- Storici, F.; Resnick, M.A. The delitto perfetto approach to in vivo site-directed mutagenesis and chromosome rearrangements with synthetic oligonucleotides in yeast. Methods Enzymol. 2006, 409, 329–345. [Google Scholar] [CrossRef] [PubMed]
- Guldener, U.; Heck, S.; Fielder, T.; Beinhauer, J.; Hegemann, J.H. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 1996, 24, 2519–2524. [Google Scholar] [CrossRef]
- Rothstein, R. Targeting, disruption, replacement, and allele rescue: Integrative DNA transformation in yeast. Methods Enzymol. 1991, 194, 281–301. [Google Scholar] [CrossRef]
- Nasmyth, K.A.; Reed, S.I. Isolation of genes by complementation in yeast: Molecular cloning of a cell-cycle gene. Proc. Natl. Acad. Sci. USA 1980, 77, 2119–2123. [Google Scholar] [CrossRef]
- Chan, C.S.; Tye, B.K. Autonomously replicating sequences in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 1980, 77, 6329–6333. [Google Scholar] [CrossRef]
- Clarke, L.; Carbon, J. Isolation of a yeast centromere and construction of functional small circular chromosomes. Nature 1980, 287, 504–509. [Google Scholar] [CrossRef]
- Szostak, J.W.; Blackburn, E.H. Cloning yeast telomeres on linear plasmid vectors. Cell 1982, 29, 245–255. [Google Scholar] [CrossRef]
- Herskowitz, I. Functional inactivation of genes by dominant negative mutations. Nature 1987, 329, 219–222. [Google Scholar] [CrossRef] [PubMed]
- Gardner, J.M.; Jaspersen, S.L. Manipulating the yeast genome: Deletion, mutation, and tagging by PCR. Methods Mol. Biol. 2014, 1205, 45–78. [Google Scholar] [CrossRef]
- Orr-Weaver, T.L.; Szostak, J.W.; Rothstein, R.J. Yeast transformation: A model system for the study of recombination. Proc. Natl. Acad. Sci. USA 1981, 78, 6354–6358. [Google Scholar] [CrossRef] [PubMed]
- Oldenburg, K.R.; Vo, K.T.; Michaelis, S.; Paddon, C. Recombination-mediated PCR-directed plasmid construction in vivo in yeast. Nucleic Acids Res. 1997, 25, 451–452. [Google Scholar] [CrossRef] [PubMed]
- Joska, T.M.; Mashruwala, A.; Boyd, J.M.; Belden, W.J. A universal cloning method based on yeast homologous recombination that is simple, efficient, and versatile. J. Microbiol. Methods 2014, 100, 46–51. [Google Scholar] [CrossRef] [PubMed]
- Zehnpfennig, B.; Wiriyasermkul, P.; Carlson, D.A.; Quick, M. Interaction of alpha-Lipoic Acid with the Human Na+/Multivitamin Transporter (hSMVT). J. Biol. Chem. 2015, 290, 16372–16382. [Google Scholar] [CrossRef]
- Sarder, H.A.M.; Li, X.; Funaya, C.; Cordat, E.; Schmitt, M.J.; Becker, B. Saccharomyces cerevisiae: First Steps to a Suitable Model System To Study the Function and Intracellular Transport of Human Kidney Anion Exchanger 1. mSphere 2020, 5, e00802-19. [Google Scholar] [CrossRef]
- Scharff-Poulsen, P.; Pedersen, P.A. Saccharomyces cerevisiae-based platform for rapid production and evaluation of eukaryotic nutrient transporters and transceptors for biochemical studies and crystallography. PLoS ONE 2013, 8, e76851. [Google Scholar] [CrossRef]
- Xiang, M.; Feng, M.; Muend, S.; Rao, R. A human Na+/H+ antiporter sharing evolutionary origins with bacterial NhaA may be a candidate gene for essential hypertension. Proc. Natl. Acad. Sci. USA 2007, 104, 18677–18681. [Google Scholar] [CrossRef]
- Techau, M.E.; Valdez-Taubas, J.; Popoff, J.F.; Francis, R.; Seaman, M.; Blackwell, J.M. Evolution of differences in transport function in Slc11a family members. J. Biol. Chem. 2007, 282, 35646–35656. [Google Scholar] [CrossRef] [PubMed]
- Doring, F.; Walter, J.; Will, J.; Focking, M.; Boll, M.; Amasheh, S.; Clauss, W.; Daniel, H. Delta-aminolevulinic acid transport by intestinal and renal peptide transporters and its physiological and clinical implications. J. Clin. Investig. 1998, 101, 2761–2767. [Google Scholar] [CrossRef] [PubMed]
- Wieczorke, R.; Dlugai, S.; Krampe, S.; Boles, E. Characterisation of mammalian GLUT glucose transporters in a heterologous yeast expression system. Cell Physiol. Biochem. 2003, 13, 123–134. [Google Scholar] [CrossRef] [PubMed]
- Wieczorke, R.; Krampe, S.; Weierstall, T.; Freidel, K.; Hollenberg, C.P.; Boles, E. Concurrent knock-out of at least 20 transporter genes is required to block uptake of hexoses in Saccharomyces cerevisiae. Febs. Lett. 1999, 464, 123–128. [Google Scholar] [CrossRef]
- Schmidl, S.; Tamayo Rojas, S.A.; Iancu, C.V.; Choe, J.Y.; Oreb, M. Functional Expression of the Human Glucose Transporters GLUT2 and GLUT3 in Yeast Offers Novel Screening Systems for GLUT-Targeting Drugs. Front. Mol. Biosci. 2020, 7, 598419. [Google Scholar] [CrossRef]
- Bonar, P.; Casey, J.R. Purification of functional human Cl−/HCO3− exchanger, AE1, over-expressed in Saccharomyces cerevisiae. Protein Expr. Purif. 2010, 74, 106–115. [Google Scholar] [CrossRef]
- Levine, K.B.; Robichaud, T.K.; Hamill, S.; Sultzman, L.A.; Carruthers, A. Properties of the human erythrocyte glucose transport protein are determined by cellular context. Biochemistry 2005, 44, 5606–5616. [Google Scholar] [CrossRef]
- Li, X.; Cordat, E.; Schmitt, M.J.; Becker, B. Boosting endoplasmic reticulum folding capacity reduces unfolded protein response activation and intracellular accumulation of human kidney anion exchanger 1 in Saccharomyces cerevisiae. Yeast 2021, 38, 521–534. [Google Scholar] [CrossRef]
- Abe, F.; Iida, H. Pressure-induced differential regulation of the two tryptophan permeases Tat1 and Tat2 by ubiquitin ligase Rsp5 and its binding proteins, Bul1 and Bul2. Mol. Cell Biol 2003, 23, 7566–7584. [Google Scholar] [CrossRef]
- Huang, Z.; Srinivasan, S.; Zhang, J.; Chen, K.; Li, Y.; Li, W.; Quiocho, F.A.; Pan, X. Discovering thiamine transporters as targets of chloroquine using a novel functional genomics strategy. PLoS Genet. 2012, 8, e1003083. [Google Scholar] [CrossRef]
- Mayr, J.A.; Merkel, O.; Kohlwein, S.D.; Gebhardt, B.R.; Bohles, H.; Fotschl, U.; Koch, J.; Jaksch, M.; Lochmuller, H.; Horvath, R.; et al. Mitochondrial phosphate-carrier deficiency: A novel disorder of oxidative phosphorylation. Am. J. Hum. Genet. 2007, 80, 478–484. [Google Scholar] [CrossRef] [PubMed]
- Boulet, A.; Vest, K.E.; Maynard, M.K.; Gammon, M.G.; Russell, A.C.; Mathews, A.T.; Cole, S.E.; Zhu, X.; Phillips, C.B.; Kwong, J.Q.; et al. The mammalian phosphate carrier SLC25A3 is a mitochondrial copper transporter required for cytochrome c oxidase biogenesis. J. Biol. Chem. 2018, 293, 1887–1896. [Google Scholar] [CrossRef] [PubMed]
- Hatanaka, T.; Takemoto, Y.; Hashimoto, M.; Majima, E.; Shinohara, Y.; Terada, H. Significant expression of functional human type 1 mitochondrial ADP/ATP carrier in yeast mitochondria. Biol. Pharm. Bull. 2001, 24, 595–599. [Google Scholar] [CrossRef]
- De Marcos Lousa, C.; Trezeguet, V.; Dianoux, A.C.; Brandolin, G.; Lauquin, G.J. The human mitochondrial ADP/ATP carriers: Kinetic properties and biogenesis of wild-type and mutant proteins in the yeast S. cerevisiae. Biochemistry 2002, 41, 14412–14420. [Google Scholar] [CrossRef]
- Hinz, W.; Faller, B.; Gruninger, S.; Gazzotti, P.; Chiesi, M. Recombinant human uncoupling protein-3 increases thermogenesis in yeast cells. FEBS Lett. 1999, 448, 57–61. [Google Scholar] [CrossRef]
- Brown, A.M.; Dolan, J.W.; Willi, S.M.; Garvey, W.T.; Argyropoulos, G. Endogenous mutations in human uncoupling protein 3 alter its functional properties. FEBS Lett. 1999, 464, 189–193. [Google Scholar] [CrossRef]
- Cohen, R.; Engelberg, D. Commonly used Saccharomyces cerevisiae strains (e.g., BY4741, W303) are growth sensitive on synthetic complete medium due to poor leucine uptake. FEMS Microbiol. Lett. 2007, 273, 239–243. [Google Scholar] [CrossRef] [PubMed]
- Cavero, S.; Vozza, A.; del Arco, A.; Palmieri, L.; Villa, A.; Blanco, E.; Runswick, M.J.; Walker, J.E.; Cerdan, S.; Palmieri, F.; et al. Identification and metabolic role of the mitochondrial aspartate-glutamate transporter in Saccharomyces cerevisiae. Mol. Microbiol. 2003, 50, 1257–1269. [Google Scholar] [CrossRef]
- Wongkittichote, P.; Tungpradabkul, S.; Wattanasirichaigoon, D.; Jensen, L.T. Prediction of the functional effect of novel SLC25A13 variants using a S. cerevisiae model of AGC2 deficiency. J. Inherit. Metab. Dis. 2013, 36, 821–830. [Google Scholar] [CrossRef]
- Doimo, M.; Lopreiato, R.; Basso, V.; Bortolotto, R.; Tessa, A.; Santorelli, F.M.; Trevisson, E.; Salviati, L. Heterologous Expression in Yeast of Human Ornithine Carriers ORNT1 and ORNT2 and of ORNT1 Alleles Implicated in HHH Syndrome in Humans. JIMD Rep. 2016, 28, 119–126. [Google Scholar] [CrossRef]
- Visser, W.F.; van Roermund, C.W.; Waterham, H.R.; Wanders, R.J. Identification of human PMP34 as a peroxisomal ATP transporter. Biochem. Biophys. Res. Commun. 2002, 299, 494–497. [Google Scholar] [CrossRef]
- Smith, C.P.; Thorsness, P.E. The molecular basis for relative physiological functionality of the ADP/ATP carrier isoforms in Saccharomyces cerevisiae. Genetics 2008, 179, 1285–1299. [Google Scholar] [CrossRef] [PubMed]
- Di Noia, M.A.; Todisco, S.; Cirigliano, A.; Rinaldi, T.; Agrimi, G.; Iacobazzi, V.; Palmieri, F. The human SLC25A33 and SLC25A36 genes of solute carrier family 25 encode two mitochondrial pyrimidine nucleotide transporters. J. Biol. Chem. 2014, 289, 33137–33148. [Google Scholar] [CrossRef] [PubMed]
- Darbani, B. Genome Evolutionary Dynamics Meets Functional Genomics: A Case Story on the Identification of SLC25A44. Int. J. Mol. Sci. 2021, 22, 5669. [Google Scholar] [CrossRef]
- Kory, N.; Uit de Bos, J.; van der Rijt, S.; Jankovic, N.; Gura, M.; Arp, N.; Pena, I.A.; Prakash, G.; Chan, S.H.; Kunchok, T.; et al. MCART1/SLC25A51 is required for mitochondrial NAD transport. Sci. Adv. 2020, 6, eabe5310. [Google Scholar] [CrossRef]
- Luongo, T.S.; Eller, J.M.; Lu, M.J.; Niere, M.; Raith, F.; Perry, C.; Bornstein, M.R.; Oliphint, P.; Wang, L.; McReynolds, M.R.; et al. SLC25A51 is a mammalian mitochondrial NAD(+) transporter. Nature 2020, 588, 174–179. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Zhu, Z.; Nielsen, J.; Siewers, V. Heterologous transporter expression for improved fatty alcohol secretion in yeast. Metab. Eng. 2018, 45, 51–58. [Google Scholar] [CrossRef]
- Vickers, M.F.; Young, J.D.; Baldwin, S.A.; Ellison, M.J.; Cass, C.E. Functional production of mammalian concentrative nucleoside transporters in Saccharomyces cerevisiae. Mol. Membr. Biol. 2001, 18, 73–79. [Google Scholar] [CrossRef]
- Zhang, J.; Smith, K.M.; Tackaberry, T.; Visser, F.; Robins, M.J.; Nielsen, L.P.; Nowak, I.; Karpinski, E.; Baldwin, S.A.; Young, J.D.; et al. Uridine binding and transportability determinants of human concentrative nucleoside transporters. Mol. Pharmacol. 2005, 68, 830–839. [Google Scholar] [CrossRef]
- Lin, H.; Kumanovics, A.; Nelson, J.M.; Warner, D.E.; Ward, D.M.; Kaplan, J. A single amino acid change in the yeast vacuolar metal transporters ZRC1 and COT1 alters their substrate specificity. J. Biol. Chem. 2008, 283, 33865–33873. [Google Scholar] [CrossRef]
- SenGupta, D.J.; Lum, P.Y.; Lai, Y.; Shubochkina, E.; Bakken, A.H.; Schneider, G.; Unadkat, J.D. A single glycine mutation in the equilibrative nucleoside transporter gene, hENT1, alters nucleoside transport activity and sensitivity to nitrobenzylthioinosine. Biochemistry 2002, 41, 1512–1519. [Google Scholar] [CrossRef] [PubMed]
- Boswell-Casteel, R.C.; Johnson, J.M.; Roe-Žurž, Z.; Duggan, K.D.; Schmitz, H.; Hays, F.A. Expression and purification of human and Saccharomyces cerevisiae equilibrative nucleoside transporters. Protein Expr. Purif. 2018, 142, 68–74. [Google Scholar] [CrossRef] [PubMed]
- Endres, C.J.; Sengupta, D.J.; Unadkat, J.D. Mutation of leucine-92 selectively reduces the apparent affinity of inosine, guanosine, NBMPR [S6-(4-nitrobenzyl)-mercaptopurine riboside] and dilazep for the human equilibrative nucleoside transporter, hENT1. Biochem. J. 2004, 380, 131–137. [Google Scholar] [CrossRef] [PubMed]
- Cotrim, C.A.; Jarrott, R.J.; Whitten, A.E.; Choudhury, H.G.; Drew, D.; Martin, J.L. Heterologous Expression and Biochemical Characterization of the Human Zinc Transporter 1 (ZnT1) and Its Soluble C-Terminal Domain. Front. Chem. 2021, 9, 667803. [Google Scholar] [CrossRef]
- Winzeler, E.A.; Shoemaker, D.D.; Astromoff, A.; Liang, H.; Anderson, K.; Andre, B.; Bangham, R.; Benito, R.; Boeke, J.D.; Bussey, H.; et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 1999, 285, 901–906. [Google Scholar] [CrossRef] [PubMed]
- Tuschl, K.; Clayton, P.T.; Gospe, S.M., Jr.; Gulab, S.; Ibrahim, S.; Singhi, P.; Aulakh, R.; Ribeiro, R.T.; Barsottini, O.G.; Zaki, M.S.; et al. Syndrome of hepatic cirrhosis, dystonia, polycythemia, and hypermanganesemia caused by mutations in SLC30A10, a manganese transporter in man. Am. J. Hum. Genet. 2012, 90, 457–466. [Google Scholar] [CrossRef]
- Zhou, B.; Gitschier, J. hCTR1: A human gene for copper uptake identified by complementation in yeast. Proc. Natl. Acad. Sci. USA 1997, 94, 7481–7486. [Google Scholar] [CrossRef]
- Uemura, S.; Mochizuki, T.; Kurosaka, G.; Hashimoto, T.; Masukawa, Y.; Abe, F. Functional analysis of human aromatic amino acid transporter MCT10/TAT1 using the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta Biomembr. 2017, 1859, 2076–2085. [Google Scholar] [CrossRef]
- Sikorski, R.S.; Hieter, P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 1989, 122, 19–27. [Google Scholar] [CrossRef]
- Strahl-Bolsinger, S.; Scheinost, A. Transmembrane topology of pmt1p, a member of an evolutionarily conserved family of protein O-mannosyltransferases. J. Biol. Chem. 1999, 274, 9068–9075. [Google Scholar] [CrossRef]
- Kuberl, A.; Schneider, J.; Thallinger, G.G.; Anderl, I.; Wibberg, D.; Hajek, T.; Jaenicke, S.; Brinkrolf, K.; Goesmann, A.; Szczepanowski, R.; et al. High-quality genome sequence of Pichia pastoris CBS7435. J. Biotechnol. 2011, 154, 312–320. [Google Scholar] [CrossRef] [PubMed]
- Muraoka, M.; Miki, T.; Ishida, N.; Hara, T.; Kawakita, M. Variety of nucleotide sugar transporters with respect to the interaction with nucleoside mono- and diphosphates. J. Biol. Chem. 2007, 282, 24615–24622. [Google Scholar] [CrossRef] [PubMed]
- Nagampalli, R.S.K.; Quesnay, J.E.N.; Adamoski, D.; Islam, Z.; Birch, J.; Sebinelli, H.G.; Girard, R.; Ascencao, C.F.R.; Fala, A.M.; Pauletti, B.A.; et al. Human mitochondrial pyruvate carrier 2 as an autonomous membrane transporter. Sci. Rep. 2018, 8, 3510. [Google Scholar] [CrossRef]
- Stribny, J.; Thines, L.; Deschamps, A.; Goffin, P.; Morsomme, P. The human Golgi protein TMEM165 transports calcium and manganese in yeast and bacterial cells. J. Biol. Chem. 2020, 295, 3865–3874. [Google Scholar] [CrossRef] [PubMed]
- Lundblad, V. Yeast cloning vectors and genes. Curr. Protoc. Mol. Biol. 2001. [Google Scholar] [CrossRef]
- Peng, B.; Williams, T.C.; Henry, M.; Nielsen, L.K.; Vickers, C.E. Controlling heterologous gene expression in yeast cell factories on different carbon substrates and across the diauxic shift: A comparison of yeast promoter activities. Microb. Cell Fact. 2015, 14, 91. [Google Scholar] [CrossRef]
- Ellis, S.B.; Brust, P.F.; Koutz, P.J.; Waters, A.F.; Harpold, M.M.; Gingeras, T.R. Isolation of alcohol oxidase and two other methanol regulatable genes from the yeast Pichia pastoris. Mol. Cell Biol. 1985, 5, 1111–1121. [Google Scholar] [CrossRef]
- West, R.W., Jr.; Yocum, R.R.; Ptashne, M. Saccharomyces cerevisiae GAL1-GAL10 divergent promoter region: Location and function of the upstream activating sequence UASG. Mol. Cell Biol. 1984, 4, 2467–2478. [Google Scholar] [CrossRef]
- Ye, L.; Berden, J.A.; van Dam, K.; Kruckeberg, A.L. Expression and activity of the Hxt7 high-affinity hexose transporter of Saccharomyces cerevisiae. Yeast 2001, 18, 1257–1267. [Google Scholar] [CrossRef]
- Ruohonen, L.; Aalto, M.K.; Keranen, S. Modifications to the ADH1 promoter of Saccharomyces cerevisiae for efficient production of heterologous proteins. J. Biotechnol. 1995, 39, 193–203. [Google Scholar] [CrossRef]
- Musti, A.M.; Zehner, Z.; Bostian, K.A.; Paterson, B.M.; Kramer, R.A. Transcriptional mapping of two yeast genes coding for glyceraldehyde 3-phosphate dehydrogenase isolated by sequence homology with the chicken gene. Gene 1983, 25, 133–143. [Google Scholar] [CrossRef]
- Bitter, G.A.; Egan, K.M. Expression of heterologous genes in Saccharomyces cerevisiae from vectors utilizing the glyceraldehyde-3-phosphate dehydrogenase gene promoter. Gene 1984, 32, 263–274. [Google Scholar] [CrossRef]
- Pedersen, P.A.; Rasmussen, J.H.; Joorgensen, P.L. Expression in high yield of pig alpha 1 beta 1 Na,K-ATPase and inactive mutants D369N and D807N in Saccharomyces cerevisiae. J. Biol. Chem. 1996, 271, 2514–2522. [Google Scholar] [CrossRef] [PubMed]
- Mumberg, D.; Muller, R.; Funk, M. Regulatable promoters of Saccharomyces cerevisiae: Comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 1994, 22, 5767–5768. [Google Scholar] [CrossRef]
- Tuite, M.F.; Dobson, M.J.; Roberts, N.A.; King, R.M.; Burke, D.C.; Kingsman, S.M.; Kingsman, A.J. Regulated high efficiency expression of human interferon-alpha in Saccharomyces cerevisiae. EMBO J. 1982, 1, 603–608. [Google Scholar] [CrossRef]
- Guarente, L.; Lalonde, B.; Gifford, P.; Alani, E. Distinctly regulated tandem upstream activation sites mediate catabolite repression of the CYC1 gene of S. cerevisiae. Cell 1984, 36, 503–511. [Google Scholar] [CrossRef]
- Humphries, A.; Ationu, A.; Wild, B.; Layton, D.M. The consequence of nucleotide substitutions in the triosephosphate isomerase (TPI) gene promoter. Blood Cells Mol. Dis. 1999, 25, 210–217. [Google Scholar] [CrossRef]
- Elgersma, Y.; van den Berg, M.; Tabak, H.F.; Distel, B. An efficient positive selection procedure for the isolation of peroxisomal import and peroxisome assembly mutants of Saccharomyces cerevisiae. Genetics 1993, 135, 731–740. [Google Scholar] [CrossRef]
- Steiner, S.; Philippsen, P. Sequence and promoter analysis of the highly expressed TEF gene of the filamentous fungus Ashbya gossypii. Mol. Gen. Genet. 1994, 242, 263–271. [Google Scholar] [CrossRef]
- Becker, D.M.; Fikes, J.D.; Guarente, L. A cDNA encoding a human CCAAT-binding protein cloned by functional complementation in yeast. Proc. Natl. Acad. Sci. USA 1991, 88, 1968–1972. [Google Scholar] [CrossRef]
- Etcheverry, T. Induced expression using yeast copper metallothionein promoter. Methods Enzymol. 1990, 185, 319–329. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Li, S.; Liu, L. Engineering redox balance through cofactor systems. Trends Biotechnol. 2014, 32, 337–343. [Google Scholar] [CrossRef] [PubMed]
- Na, D.; Kim, T.Y.; Lee, S.Y. Construction and optimization of synthetic pathways in metabolic engineering. Curr. Opin. Microbiol. 2010, 13, 363–370. [Google Scholar] [CrossRef] [PubMed]
- Tschumper, G.; Carbon, J. Sequence of a yeast DNA fragment containing a chromosomal replicator and the TRP1 gene. Gene 1980, 10, 157–166. [Google Scholar] [CrossRef]
- Struhl, K.; Davis, R.W. A physical, genetic and transcriptional map of the cloned his3 gene region of Saccharomyces cerevisiae. J. Mol. Biol. 1980, 136, 309–332. [Google Scholar] [CrossRef]
- Rose, M.; Grisafi, P.; Botstein, D. Structure and function of the yeast URA3 gene: Expression in Escherichia coli. Gene 1984, 29, 113–124. [Google Scholar] [CrossRef]
- Brachmann, C.B.; Davies, A.; Cost, G.J.; Caputo, E.; Li, J.; Hieter, P.; Boeke, J.D. Designer deletion strains derived from Saccharomyces cerevisiae S288C: A useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 1998, 14, 115–132. [Google Scholar] [CrossRef]
- Britton, Z.; Young, C.; Can, O.; McNeely, P.; Naranjo, A.; Robinson, A.S. Membrane Protein Expression in Saccharomyces cerevisiae. In Production of Membrane Proteins: Strategies for Expression and Isolation; John Wiley & Sons: Hoboken, NJ, USA, 2011; pp. 37–73. [Google Scholar] [CrossRef]
- Hinnen, A.; Hicks, J.B.; Fink, G.R. Transformation of yeast. Proc. Natl. Acad. Sci. USA 1978, 75, 1929–1933. [Google Scholar] [CrossRef]
- Armaleo, D.; Ye, G.N.; Klein, T.M.; Shark, K.B.; Sanford, J.C.; Johnston, S.A. Biolistic nuclear transformation of Saccharomyces cerevisiae and other fungi. Curr. Genet. 1990, 17, 97–103. [Google Scholar] [CrossRef]
- Costanzo, M.C.; Fox, T.D. Transformation of yeast by agitation with glass beads. Genetics 1988, 120, 667–670. [Google Scholar] [CrossRef]
- Burgers, P.M.; Percival, K.J. Transformation of yeast spheroplasts without cell fusion. Anal. Biochem. 1987, 163, 391–397. [Google Scholar] [CrossRef]
- Gietz, D.; St Jean, A.; Woods, R.A.; Schiestl, R.H. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992, 20, 1425. [Google Scholar] [CrossRef] [PubMed]
- Gietz, R.D.; Schiestl, R.H.; Willems, A.R.; Woods, R.A. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 1995, 11, 355–360. [Google Scholar] [CrossRef] [PubMed]
- Delorme, E. Transformation of Saccharomyces cerevisiae by electroporation. Appl. Environ. Microbiol. 1989, 55, 2242–2246. [Google Scholar] [CrossRef]
- Thompson, J.R.; Register, E.; Curotto, J.; Kurtz, M.; Kelly, R. An improved protocol for the preparation of yeast cells for transformation by electroporation. Yeast 1998, 14, 565–571. [Google Scholar] [CrossRef]
- Suga, M.; Hatakeyama, T. High-efficiency electroporation by freezing intact yeast cells with addition of calcium. Curr. Genet. 2003, 43, 206–211. [Google Scholar] [CrossRef]
- Kawai, S.; Hashimoto, W.; Murata, K. Transformation of Saccharomyces cerevisiae and other fungi: Methods and possible underlying mechanism. Bioeng. Bugs. 2010, 1, 395–403. [Google Scholar] [CrossRef]
- Ito, H.; Fukuda, Y.; Murata, K.; Kimura, A. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 1983, 153, 163–168. [Google Scholar] [CrossRef]
- Hashimoto, H.; Morikawa, H.; Yamada, Y.; Kimura, A. Novel method for transformation of intact yeast cells by electroinjection of plasmid DNA. Appl. Microbiol. Biotechnol. 1985, 21, 4. [Google Scholar] [CrossRef]
- Becker, D.M.; Guarente, L. High-efficiency transformation of yeast by electroporation. Methods Enzymol. 1991, 194, 182–187. [Google Scholar] [CrossRef]
- Sherman, F. Getting started with yeast. Methods Enzymol. 2002, 350, 3–41. [Google Scholar] [CrossRef] [PubMed]
- Gasser, B.; Mattanovich, D. Antibody production with yeasts and filamentous fungi: On the road to large scale? Biotechnol. Lett. 2007, 29, 201–212. [Google Scholar] [CrossRef] [PubMed]
- Hackel, B.J.; Huang, D.G.; Buboz, J.C.; Wang, X.X.; Shusta, E.V. Production of soluble and active transferrin receptor-targeting single-chain antibody using Saccharomyces cerevisiae. Pharm. Res. Dordr. 2006, 23, 790–797. [Google Scholar] [CrossRef] [PubMed]
- Joubert, O.; Nehme, R.; Bidet, M.; Mus-Veteau, I. Heterologous Expression of Human Membrane Receptors in the Yeast Saccharomyces cerevisiae. Heterologous Expr. Membr. Proteins Methods Protoc. 2010, 601, 87–103. [Google Scholar] [CrossRef]
- Ferndahl, C.; Bonander, N.; Logez, C.; Wagner, R.; Gustafsson, L.; Larsson, C.; Hedfalk, K.; Darby, R.A.; Bill, R.M. Increasing cell biomass in Saccharomyces cerevisiae increases recombinant protein yield: The use of a respiratory strain as a microbial cell factory. Microb. Cell Fact. 2010, 9, 47. [Google Scholar] [CrossRef]
- Walsh, G. Biopharmaceutical benchmarks 2010. Nat. Biotechnol. 2010, 28, 917–924. [Google Scholar] [CrossRef]
- Vorauer-Uhl, K.; Lhota, G. Assessing the Quality of Recombinant Products Made in Yeast. Methods Mol. Biol. 2019, 1923, 361–384. [Google Scholar] [CrossRef]
- Treco, D.A.; Lundblad, V. Preparation of yeast media. Curr. Protoc. Mol. Biol. 2001. [Google Scholar] [CrossRef]
- Pronk, J.T.; Yde Steensma, H.; Van Dijken, J.P. Pyruvate metabolism in Saccharomyces cerevisiae. Yeast 1996, 12, 1607–1633. [Google Scholar] [CrossRef]
- Rozpedowska, E.; Hellborg, L.; Ishchuk, O.P.; Orhan, F.; Galafassi, S.; Merico, A.; Woolfit, M.; Compagno, C.; Piskur, J. Parallel evolution of the make-accumulate-consume strategy in Saccharomyces and Dekkera yeasts. Nat. Commun. 2011, 2, 302. [Google Scholar] [CrossRef]
- Otterstedt, K.; Larsson, C.; Bill, R.M.; Stahlberg, A.; Boles, E.; Hohmann, S.; Gustafsson, L. Switching the mode of metabolism in the yeast Saccharomyces cerevisiae. EMBO Rep. 2004, 5, 532–537. [Google Scholar] [CrossRef] [PubMed]
- Mentel, M.; Chovancikova, P.; Zeman, I.; Polcic, P. Learning from Yeast about Mitochondrial Carriers. Microorganisms 2021, 9, 2044. [Google Scholar] [CrossRef] [PubMed]
- Monne, M.; Vozza, A.; Lasorsa, F.M.; Porcelli, V.; Palmieri, F. Mitochondrial Carriers for Aspartate, Glutamate and Other Amino Acids: A Review. Int. J. Mol. Sci. 2019, 20, 4456. [Google Scholar] [CrossRef] [PubMed]
- Ferramosca, A.; Zara, V. Mitochondrial Carriers and Substrates Transport Network: A Lesson from Saccharomyces cerevisiae. Int. J. Mol. Sci. 2021, 22, 8496. [Google Scholar] [CrossRef] [PubMed]
- Di Rosa, M.C.; Guarino, F.; Conti Nibali, S.; Magri, A.; De Pinto, V. Voltage-Dependent Anion Selective Channel Isoforms in Yeast: Expression, Structure, and Functions. Front. Physiol. 2021, 12, 675708. [Google Scholar] [CrossRef]
- Bonander, N.; Hedfalk, K.; Larsson, C.; Mostad, P.; Chang, C.; Gustafsson, L.; Bill, R.M. Design of improved membrane protein production experiments: Quantitation of the host response. Protein Sci. 2005, 14, 1729–1740. [Google Scholar] [CrossRef]
- Bonander, N.; Darby, R.A.; Grgic, L.; Bora, N.; Wen, J.; Brogna, S.; Poyner, D.R.; O’Neill, M.A.; Bill, R.M. Altering the ribosomal subunit ratio in yeast maximizes recombinant protein yield. Microb. Cell Fact. 2009, 8, 10. [Google Scholar] [CrossRef]
- Bawa, Z.; Bland, C.E.; Bonander, N.; Bora, N.; Cartwright, S.P.; Clare, M.; Conner, M.T.; Darby, R.A.; Dilworth, M.V.; Holmes, W.J.; et al. Understanding the yeast host cell response to recombinant membrane protein production. Biochem. Soc. Trans. 2011, 39, 719–723. [Google Scholar] [CrossRef]
- Kurtzman, C.P. Biotechnological strains of Komagataella (Pichia) pastoris are Komagataella phaffii as determined from multigene sequence analysis. J. Ind. Microbiol. Biotechnol. 2009, 36, 1435–1438. [Google Scholar] [CrossRef]
- Cregg, J.M.; Vedvick, T.S.; Raschke, W.C. Recent advances in the expression of foreign genes in Pichia pastoris. Biotechnology 1993, 11, 905–910. [Google Scholar] [CrossRef]
- Hamilton, S.R.; Gerngross, T.U. Glycosylation engineering in yeast: The advent of fully humanized yeast. Curr. Opin. Biotechnol. 2007, 18, 387–392. [Google Scholar] [CrossRef] [PubMed]
- Emmerstorfer-Augustin, A.; Wriessnegger, T.; Hirz, M.; Zellnig, G.; Pichler, H. Membrane Protein Production in Yeast: Modification of Yeast Membranes for Human Membrane Protein Production. Methods Mol. Biol. 2019, 1923, 265–285. [Google Scholar] [CrossRef] [PubMed]
- Abad, S.; Kitz, K.; Hormann, A.; Schreiner, U.; Hartner, F.S.; Glieder, A. Real-time PCR-based determination of gene copy numbers in Pichia pastoris. Biotechnol. J. 2010, 5, 413–420. [Google Scholar] [CrossRef] [PubMed]
- Andre, N.; Cherouati, N.; Prual, C.; Steffan, T.; Zeder-Lutz, G.; Magnin, T.; Pattus, F.; Michel, H.; Wagner, R.; Reinhart, C. Enhancing functional production of G protein-coupled receptors in Pichia pastoris to levels required for structural studies via a single expression screen. Protein Sci. 2006, 15, 1115–1126. [Google Scholar] [CrossRef]
- Sunga, A.J.; Tolstorukov, I.; Cregg, J.M. Posttransformational vector amplification in the yeast Pichia pastoris. FEMS Yeast Res. 2008, 8, 870–876. [Google Scholar] [CrossRef] [PubMed]
- Suades, A.; Alcaraz, A.; Cruz, E.; Álvarez-Marimon, E.; Whitelegge, J.P.; Manyosa, J.; Cladera, J.; Perálvarez-Marín, A. Structural biology workflow for the expression and characterization of functional human sodium glucose transporter type 1 in Pichia pastoris. Sci. Rep. 2019, 9, 1203. [Google Scholar] [CrossRef] [PubMed]
- Bird, L.E.; Nettleship, J.E.; Jarvinen, V.; Rada, H.; Verma, A.; Owens, R.J. Expression Screening of Integral Membrane Proteins by Fusion to Fluorescent Reporters. Adv. Exp. Med. Biol. 2016, 922, 1–11. [Google Scholar] [CrossRef]
- Brooks, C.L.; Morrison, M.; Lemieux, M.J. Rapid expression screening of eukaryotic membrane proteins in Pichia pastoris. Protein Sci. 2013, 22, 425–433. [Google Scholar] [CrossRef]
- Kastilan, R.; Boes, A.; Spiegel, H.; Voepel, N.; Chudobova, I.; Hellwig, S.; Buyel, J.F.; Reimann, A.; Fischer, R. Improvement of a fermentation process for the production of two PfAMA1-DiCo-based malaria vaccine candidates in Pichia pastoris. Sci. Rep. 2017, 7, 11991. [Google Scholar] [CrossRef]
- Cregg, J.M.; Madden, K.R.; Barringer, K.J.; Thill, G.P.; Stillman, C.A. Functional characterization of the two alcohol oxidase genes from the yeast Pichia pastoris. Mol. Cell Biol. 1989, 9, 1316–1323. [Google Scholar] [CrossRef]
- Bawa, Z.; Routledge, S.J.; Jamshad, M.; Clare, M.; Sarkar, D.; Dickerson, I.; Ganzlin, M.; Poyner, D.R.; Bill, R.M. Functional recombinant protein is present in the pre-induction phases of Pichia pastoris cultures when grown in bioreactors, but not shake-flasks. Microb. Cell Fact. 2014, 13, 127. [Google Scholar] [CrossRef] [PubMed]
- Cos, O.; Ramon, R.; Montesinos, J.L.; Valero, F. Operational strategies, monitoring and control of heterologous protein production in the methylotrophic yeast Pichia pastoris under different promoters: A review. Microb. Cell Fact. 2006, 5, 17. [Google Scholar] [CrossRef] [PubMed]
- Vogl, T.; Sturmberger, L.; Fauland, P.C.; Hyden, P.; Fischer, J.E.; Schmid, C.; Thallinger, G.G.; Geier, M.; Glieder, A. Methanol independent induction in Pichia pastoris by simple derepressed overexpression of single transcription factors. Biotechnol. Bioeng. 2018, 115, 1037–1050. [Google Scholar] [CrossRef] [PubMed]
- Weis, R.; Luiten, R.; Skranc, W.; Schwab, H.; Wubbolts, M.; Glieder, A. Reliable high-throughput screening with Pichia pastoris by limiting yeast cell death phenomena. FEMS Yeast Res. 2004, 5, 179–189. [Google Scholar] [CrossRef] [PubMed]
- Routledge, S.J.; Mikaliunaite, L.; Patel, A.; Clare, M.; Cartwright, S.P.; Bawa, Z.; Wilks, M.D.; Low, F.; Hardy, D.; Rothnie, A.J.; et al. The synthesis of recombinant membrane proteins in yeast for structural studies. Methods 2016, 95, 26–37. [Google Scholar] [CrossRef]
- Vogl, T.; Thallinger, G.G.; Zellnig, G.; Drew, D.; Cregg, J.M.; Glieder, A.; Freigassner, M. Towards improved membrane protein production in Pichia pastoris: General and specific transcriptional response to membrane protein overexpression. New Biotechnol. 2014, 31, 538–552. [Google Scholar] [CrossRef]
- Guyot, L.; Hartmann, L.; Mohammed-Bouteben, S.; Caro, L.; Wagner, R. Preparation of Recombinant Membrane Proteins from Pichia pastoris for Molecular Investigations. Curr. Protoc. Protein Sci. 2020, 100, e104. [Google Scholar] [CrossRef]
- Alisio, A.; Mueckler, M. Purification and characterization of mammalian glucose transporters expressed in Pichia pastoris. Protein Expr. Purif. 2010, 70, 81–87. [Google Scholar] [CrossRef][Green Version]
- Daniels, M.J.; Jagielnicki, M.; Yeager, M. Structure/Function Analysis of human ZnT8 (SLC30A8): A Diabetes Risk Factor and Zinc Transporter. Curr. Res. Struct. Biol. 2020, 2, 144–155. [Google Scholar] [CrossRef]
- Teo, A.C.K.; Lee, S.C.; Pollock, N.L.; Stroud, Z.; Hall, S.; Thakker, A.; Pitt, A.R.; Dafforn, T.R.; Spickett, C.M.; Roper, D.I. Analysis of SMALP co-extracted phospholipids shows distinct membrane environments for three classes of bacterial membrane protein. Sci. Rep. 2019, 9, 1813. [Google Scholar] [CrossRef]
- Parmar, M.; Rawson, S.; Scarff, C.A.; Goldman, A.; Dafforn, T.R.; Muench, S.P.; Postis, V.L.G. Using a SMALP platform to determine a sub-nm single particle cryo-EM membrane protein structure. Biochim. Biophys. Acta Biomembr. 2018, 1860, 378–383. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Benlekbir, S.; Venkatakrishnan, P.; Wang, Y.; Hong, S.; Hosler, J.; Tajkhorshid, E.; Rubinstein, J.L.; Gennis, R.B. Structure of the alternative complex III in a supercomplex with cytochrome oxidase. Nature 2018, 557, 123–126. [Google Scholar] [CrossRef] [PubMed]
- Qiu, W.; Fu, Z.; Xu, G.G.; Grassucci, R.A.; Zhang, Y.; Frank, J.; Hendrickson, W.A.; Guo, Y. Structure and activity of lipid bilayer within a membrane-protein transporter. Proc. Natl. Acad. Sci. USA 2018, 115, 12985–12990. [Google Scholar] [CrossRef] [PubMed]
- King, M.S.; Boes, C.; Kunji, E.R. Membrane protein expression in Lactococcus lactis. Methods Enzymol. 2015, 556, 77–97. [Google Scholar] [CrossRef] [PubMed]
- Martens, C. Membrane Protein Production in Lactococcus lactis for Structural Studies. Methods Mol. Biol. 2020, 2127, 29–45. [Google Scholar] [CrossRef]
- Galluccio, M.; Mazza, T.; Scalise, M.; Sarubbi, M.C.; Indiveri, C. Bacterial over-expression of functionally active human CT2 (SLC22A16) carnitine transporter. Mol. Biol. Rep. 2022, 49, 8185–8193. [Google Scholar] [CrossRef]
- Schwarzbaum, P.J.; Schachter, J.; Bredeston, L.M. The broad range di- and trinucleotide exchanger SLC35B1 displays asymmetrical affinities for ATP transport across the ER membrane. J. Biol. Chem. 2022, 298, 101537. [Google Scholar] [CrossRef]
- Kantipudi, S.; Fotiadis, D. Yeast Cell-Based Transport Assay for the Functional Characterization of Human 4F2hc-LAT1 and -LAT2, and LAT1 and LAT2 Substrates and Inhibitors. Front. Mol. Biosci. 2021, 8, 676854. [Google Scholar] [CrossRef]
- Schmidt, T.G.; Skerra, A. The Strep-tag system for one-step purification and high-affinity detection or capturing of proteins. Nat. Protoc. 2007, 2, 1528–1535. [Google Scholar] [CrossRef]
- Kimple, M.E.; Brill, A.L.; Pasker, R.L. Overview of affinity tags for protein purification. Curr. Protoc. Protein Sci. 2013, 73. [Google Scholar] [CrossRef]
- Ishida, N.; Kuba, T.; Aoki, K.; Miyatake, S.; Kawakita, M.; Sanai, Y. Identification and characterization of human Golgi nucleotide sugar transporter SLC35D2, a novel member of the SLC35 nucleotide sugar transporter family. Genomics 2005, 85, 106–116. [Google Scholar] [CrossRef] [PubMed]
- Aller, S.G.; Unger, V.M. Projection structure of the human copper transporter CTR1 at 6-A resolution reveals a compact trimer with a novel channel-like architecture. Proc. Natl. Acad. Sci. USA 2006, 103, 3627–3632. [Google Scholar] [CrossRef] [PubMed]
- Scalise, M.; Mazza, T.; Pappacoda, G.; Pochini, L.; Cosco, J.; Rovella, F.; Indiveri, C. The Human SLC1A5 Neutral Amino Acid Transporter Catalyzes a pH-Dependent Glutamate/Glutamine Antiport, as Well. Front. Cell Dev. Biol. 2020, 8, 603. [Google Scholar] [CrossRef] [PubMed]
- Scalise, M.; Pochini, L.; Console, L.; Pappacoda, G.; Pingitore, P.; Hedfalk, K.; Indiveri, C. Cys Site-Directed Mutagenesis of the Human SLC1A5 (ASCT2) Transporter: Structure/Function Relationships and Crucial Role of Cys467 for Redox Sensing and Glutamine Transport. Int. J. Mol. Sci. 2018, 19, 648. [Google Scholar] [CrossRef] [PubMed]
- Scalise, M.; Pochini, L.; Pingitore, P.; Hedfalk, K.; Indiveri, C. Cysteine is not a substrate but a specific modulator of human ASCT2 (SLC1A5) transporter. FEBS Lett. 2015, 589, 3617–3623. [Google Scholar] [CrossRef]
- Scalise, M.; Pochini, L.; Panni, S.; Pingitore, P.; Hedfalk, K.; Indiveri, C. Transport mechanism and regulatory properties of the human amino acid transporter ASCT2 (SLC1A5). Amino Acids 2014, 46, 2463–2475. [Google Scholar] [CrossRef]
- Mazza, T.; Scalise, M.; Pappacoda, G.; Pochini, L.; Indiveri, C. The involvement of sodium in the function of the human amino acid transporter ASCT2. FEBS Lett. 2021, 595, 3030–3041. [Google Scholar] [CrossRef]
- Scalise, M.; Pochini, L.; Cosco, J.; Aloe, E.; Mazza, T.; Console, L.; Esposito, A.; Indiveri, C. Interaction of Cholesterol With the Human SLC1A5 (ASCT2): Insights Into Structure/Function Relationships. Front. Mol. Biosci. 2019, 6, 110. [Google Scholar] [CrossRef]
- Simons, C.H.; Weinglass, A.B.; Baldwin, S.A. Studies on the expression of the human erythrocyte glucose transporter (GLUT1) in the yeast Saccharomyces cerevisiae. Biochem. Soc. Trans. 1997, 25, 463S. [Google Scholar] [CrossRef]
- Schmidl, S.; Ursu, O.; Iancu, C.V.; Oreb, M.; Oprea, T.I.; Choe, J.Y. Identification of new GLUT2-selective inhibitors through in silico ligand screening and validation in eukaryotic expression systems. Sci. Rep. 2021, 11, 13751. [Google Scholar] [CrossRef]
- Kasahara, T.; Maeda, M.; Boles, E.; Kasahara, M. Identification of a key residue determining substrate affinity in the human glucose transporter GLUT1. Biochim. Biophys. Acta 2009, 1788, 1051–1055. [Google Scholar] [CrossRef] [PubMed]
- Iancu, C.V.; Bocci, G.; Ishtikhar, M.; Khamrai, M.; Oreb, M.; Oprea, T.I.; Choe, J.Y. GLUT3 inhibitor discovery through in silico ligand screening and in vivo validation in eukaryotic expression systems. Sci. Rep. 2022, 12, 1429. [Google Scholar] [CrossRef] [PubMed]
- Tripp, J.; Essl, C.; Iancu, C.V.; Boles, E.; Choe, J.Y.; Oreb, M. Establishing a yeast-based screening system for discovery of human GLUT5 inhibitors and activators. Sci. Rep. 2017, 7, 6197. [Google Scholar] [CrossRef]
- Costa, M.; Rosell, A.; Alvarez-Marimon, E.; Zorzano, A.; Fotiadis, D.; Palacin, M. Expression of human heteromeric amino acid transporters in the yeast Pichia pastoris. Protein Expr. Purif. 2013, 87, 35–40. [Google Scholar] [CrossRef] [PubMed]
- Kantipudi, S.; Jeckelmann, J.M.; Ucurum, Z.; Bosshart, P.D.; Fotiadis, D. The Heavy Chain 4F2hc Modulates the Substrate Affinity and Specificity of the Light Chains LAT1 and LAT2. Int. J. Mol. Sci. 2020, 21, 7573. [Google Scholar] [CrossRef]
- Kassem, N.; Kassem, M.M.; Pedersen, S.F.; Pedersen, P.A.; Kragelund, B.B. Yeast recombinant production of intact human membrane proteins with long intrinsically disordered intracellular regions for structural studies. Biochim. Biophys. Acta Biomembr. 2020, 1862, 183272. [Google Scholar] [CrossRef] [PubMed]
- Theis, S.; Doring, F.; Daniel, H. Expression of the myc/His-tagged human peptide transporter hPEPT1 in yeast for protein purification and functional analysis. Protein Expr. Purif. 2001, 22, 436–442. [Google Scholar] [CrossRef]
- Hu, Y.; Chen, X.; Smith, D.E. Species-dependent uptake of glycylsarcosine but not oseltamivir in Pichia pastoris expressing the rat, mouse, and human intestinal peptide transporter PEPT1. Drug Metab. Dispos. 2012, 40, 1328–1335. [Google Scholar] [CrossRef]
- Song, F.; Hu, Y.; Jiang, H.; Smith, D.E. Species Differences in Human and Rodent PEPT2-Mediated Transport of Glycylsarcosine and Cefadroxil in Pichia Pastoris Transformants. Drug Metab. Dispos. 2017, 45, 130–136. [Google Scholar] [CrossRef]
- Madeo, M.; Kovacs, A.D.; Pearce, D.A. The human synaptic vesicle protein, SV2A, functions as a galactose transporter in Saccharomyces cerevisiae. J. Biol. Chem. 2014, 289, 33066–33071. [Google Scholar] [CrossRef]
- Zhang, Y.; Tian, D.; Matsuyama, H.; Hamazaki, T.; Shiratsuchi, T.; Terada, N.; Hook, D.J.; Walters, M.A.; Georg, G.I.; Hawkinson, J.E. Human Adenine Nucleotide Translocase (ANT) Modulators Identified by High-Throughput Screening of Transgenic Yeast. J. Biomol. Screen. 2016, 21, 381–390. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rodriguez-Sanchez, L.; Rial, E. The distinct bioenergetic properties of the human UCP1. Biochimie 2017, 134, 51–55. [Google Scholar] [CrossRef] [PubMed]
- Zackova, M.; Jezek, P. Reconstitution of novel mitochondrial uncoupling proteins UCP2 and UCP3. Biosci. Rep. 2002, 22, 33–46. [Google Scholar] [CrossRef]
- Heidkaemper, D.; Winkler, E.; Muller, V.; Frischmuth, K.; Liu, Q.; Caskey, T.; Klingenberg, M. The bulk of UCP3 expressed in yeast cells is incompetent for a nucleotide regulated H+ transport. FEBS Lett. 2000, 480, 265–270. [Google Scholar] [CrossRef]
- Hamazaki, T.; Leung, W.Y.; Cain, B.D.; Ostrov, D.A.; Thorsness, P.E.; Terada, N. Functional expression of human adenine nucleotide translocase 4 in Saccharomyces cerevisiae. PLoS ONE 2011, 6, e19250. [Google Scholar] [CrossRef]
- Damaraju, V.L.; Mowles, D.; Yao, S.; Ng, A.; Young, J.D.; Cass, C.E.; Tong, Z. Role of human nucleoside transporters in the uptake and cytotoxicity of azacitidine and decitabine. Nucleosides Nucleotides Nucleic Acids 2012, 31, 236–255. [Google Scholar] [CrossRef] [PubMed]
- Damaraju, V.L.; Weber, D.; Kuzma, M.; Cass, C.E.; Sawyer, M.B. Selective Inhibition of Human Equilibrative and Concentrative Nucleoside Transporters by BCR-ABL Kinase Inhibitors: Identification of key hENT1 amino acid residues for interaction with BCR-ABL kinase inhibitors. J. Biol. Chem. 2016, 291, 18809–18817. [Google Scholar] [CrossRef]
- Damaraju, S.; Zhang, J.; Visser, F.; Tackaberry, T.; Dufour, J.; Smith, K.M.; Slugoski, M.; Ritzel, M.W.; Baldwin, S.A.; Young, J.D.; et al. Identification and functional characterization of variants in human concentrative nucleoside transporter 3, hCNT3 (SLC28A3), arising from single nucleotide polymorphisms in coding regions of the hCNT3 gene. Pharm. Genom. 2005, 15, 173–182. [Google Scholar] [CrossRef]
- SenGupta, D.J.; Unadkat, J.D. Glycine 154 of the equilibrative nucleoside transporter, hENT1, is important for nucleoside transport and for conferring sensitivity to the inhibitors nitrobenzylthioinosine, dipyridamole, and dilazep. Biochem. Pharmacol. 2004, 67, 453–458. [Google Scholar] [CrossRef]
- Vickers, M.F.; Kumar, R.; Visser, F.; Zhang, J.; Charania, J.; Raborn, R.T.; Baldwin, S.A.; Young, J.D.; Cass, C.E. Comparison of the interaction of uridine, cytidine, and other pyrimidine nucleoside analogues with recombinant human equilibrative nucleoside transporter 2 (hENT2) produced in Saccharomyces cerevisiae. Biochem. Cell Biol. 2002, 80, 639–644. [Google Scholar] [CrossRef]
- Vickers, M.F.; Mani, R.S.; Sundaram, M.; Hogue, D.L.; Young, J.D.; Baldwin, S.A.; Cass, C.E. Functional production and reconstitution of the human equilibrative nucleoside transporter (hENT1) in Saccharomyces cerevisiae. Interaction of inhibitors of nucleoside transport with recombinant hENT1 and a glycosylation-defective derivative (hENT1/N48Q). Biochem. J. 1999, 339, 21–32. [Google Scholar] [CrossRef] [PubMed]
- Visser, F.; Vickers, M.F.; Ng, A.M.; Baldwin, S.A.; Young, J.D.; Cass, C.E. Mutation of residue 33 of human equilibrative nucleoside transporters 1 and 2 alters sensitivity to inhibition of transport by dilazep and dipyridamole. J. Biol. Chem. 2002, 277, 395–401. [Google Scholar] [CrossRef] [PubMed]
- Aoki, K.; Ishida, N.; Kawakita, M. Substrate recognition by UDP-galactose and CMP-sialic acid transporters. Different sets of transmembrane helices are utilized for the specific recognition of UDP-galactose and CMP-sialic acid. J. Biol. Chem. 2001, 276, 21555–21561. [Google Scholar] [CrossRef]
- Newstead, S.; Kim, H.; von Heijne, G.; Iwata, S.; Drew, D. High-throughput fluorescent-based optimization of eukaryotic membrane protein overexpression and purification in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 2007, 104, 13936–13941. [Google Scholar] [CrossRef] [PubMed]
- Sun-Wada, G.H.; Yoshioka, S.; Ishida, N.; Kawakita, M. Functional expression of the human UDP-galactose transporters in the yeast Saccharomyces cerevisiae. J. Biochem. 1998, 123, 912–917. [Google Scholar] [CrossRef]
- Segawa, H.; Kawakita, M.; Ishida, N. Human and Drosophila UDP-galactose transporters transport UDP-N-acetylgalactosamine in addition to UDP-galactose. Eur. J. Biochem. 2002, 269, 128–138. [Google Scholar] [CrossRef]
- Ashikov, A.; Routier, F.; Fuhlrott, J.; Helmus, Y.; Wild, M.; Gerardy-Schahn, R.; Bakker, H. The human solute carrier gene SLC35B4 encodes a bifunctional nucleotide sugar transporter with specificity for UDP-xylose and UDP-N-acetylglucosamine. J. Biol. Chem. 2005, 280, 27230–27235. [Google Scholar] [CrossRef]
- Muraoka, M.; Kawakita, M.; Ishida, N. Molecular characterization of human UDP-glucuronic acid/UDP-N-acetylgalactosamine transporter, a novel nucleotide sugar transporter with dual substrate specificity. FEBS Lett. 2001, 495, 87–93. [Google Scholar] [CrossRef]
- Becares, E.R.; Pedersen, P.A.; Gourdon, P.; Gotfryd, K. Overproduction of Human Zip (SLC39) Zinc Transporters in Saccharomyces cerevisiae for Biophysical Characterization. Cells 2021, 10, 213. [Google Scholar] [CrossRef]
- Breen, C.J.; Martin, D.S.; Ma, H.; McQuaid, K.; O’Kennedy, R.; Findlay, J.B. Production of functional human vitamin A transporter/RBP receptor (STRA6) for structure determination. PLoS ONE 2015, 10, e0122293. [Google Scholar] [CrossRef]
- King, K.M.; Damaraju, V.L.; Vickers, M.F.; Yao, S.Y.; Lang, T.; Tackaberry, T.E.; Mowles, D.A.; Ng, A.M.; Young, J.D.; Cass, C.E. A comparison of the transportability, and its role in cytotoxicity, of clofarabine, cladribine, and fludarabine by recombinant human nucleoside transporters produced in three model expression systems. Mol. Pharmacol. 2006, 69, 346–353. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, L.; Batlle, A.; Di Venosa, G.; MacRobert, A.J.; Battah, S.; Daniel, H.; Casas, A. Study of the mechanisms of uptake of 5-aminolevulinic acid derivatives by PEPT1 and PEPT2 transporters as a tool to improve photodynamic therapy of tumours. Int. J. Biochem. Cell Biol. 2006, 38, 1530–1539. [Google Scholar] [CrossRef] [PubMed]
- Scalise, M.; Pochini, L.; Console, L.; Losso, M.A.; Indiveri, C. The Human SLC1A5 (ASCT2) Amino Acid Transporter: From Function to Structure and Role in Cell Biology. Front. Cell Dev. Biol. 2018, 6, 96. [Google Scholar] [CrossRef] [PubMed]
- Schmidl, S.; Iancu, C.V.; Choe, J.Y.; Oreb, M. Ligand Screening Systems for Human Glucose Transporters as Tools in Drug Discovery. Front. Chem. 2018, 6, 183. [Google Scholar] [CrossRef] [PubMed]
- Boles, E.; Oreb, M. A Growth-Based Screening System for Hexose Transporters in Yeast. Methods Mol. Biol. 2018, 1713, 123–135. [Google Scholar] [CrossRef] [PubMed]

| Host | Strain | Genotype | Feature | References |
|---|---|---|---|---|
| P. pastoris | KM71H | aox1::ARG4, arg4 | Strain with MutS phenotype | [58] |
| P. pastoris | SMD1168H | pep4 | Strain without protease A activity | [59] |
| S. cerevisiae | PAP1500 | MATα ura3-52 trp1::GAL10-GAL4 lys2-801 leu2Δ1 his3Δ200pep4::HIS3prb1Δ1.6Rcan1GAL | Overexpression of the Gal4 transcription factor | [60] |
| S. cerevisiae | AB11c | ena1-4Δnhx1Δnha1Δ | Deletion of endogenous cation/proton antiporters and pumps | [61] |
| S. cerevisiae | MSY6210 | MAT α leu2-3,112 ura3-52 his3200 trp1-901lys2-801suc2-9 smf1::HIS3,smf2::KANR | Deletion of Mg2+ transporters | [62] |
| S. cerevisiae | MSY6211 | MAT a leu2-3,112 ura3-52 his3200 trp1-901 ade2-101 suc2-9 smf3::LEU2 | Deletion of Mg2+ transporters | [62] |
| P. pastoris | GS115 | his4 | Deletion of histidinol dehydrogenase | [63] |
| S. cerevisiae | EBY.S7 | MATα hxt1-17Δgal2Δagt1Δstl1Δleu2-3,112 ura3-52 trp1-289 his3-Δ1 MAL2–8c SUC2 hxtΔfgy1 | Deletion of hexose transporters | [64] |
| S. cerevisiae | EBY.F4–1 | MATα hxt1-17Δgal2Δagt1Δstl1Δleu2-3,112 ura3-52 trp1-289 his3-Δ1 MAL2–8c SUC2 hxtΔfgy1 fgy41 | Deletion of hexose transporters | [64] |
| S. cerevisiae | EBY.VW4000 | MATa leu2-3,112 ura3-52 trp1-289 his3-1 MAL2-8c SUC2 Δhxt1-17 Δgal2 Δstl1::loxP Δagt1::loxP Δmph2::loxP Δmph3::loxP | Deletion of hexose transporters | [65] |
| S. cerevisiae | SDY.022 | MATa leu2-3,112 ura3-52 trp1-289 his3-∆1 MAL2-8C SUC2 ∆hxt1-17 ∆gal2 ∆agt1 ∆stl1 fgy1-1 erg4::kanMX | Deletion of hexose transporters | [66] |
| S. cerevisiae | BJ5457 | MATα ura3-52 trp1 lys2-801 leu2-Δ1 his3-Δ200 pep4:HIS3 prb1-delta1.6R can1 GAL | Protease deficient | [67] |
| S. cerevisiae | RE700A | MATa hxt1::HIS3::hxt4 hxt5::LEU2 hxt2::HIS3hxt3::LEU2::hxt6 hxt7::HIS3 | Deletion of hexose transporters | [68] |
| S. cerevisiae | BY4742 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 | Minimize homologous recombination | [59] |
| S. cerevisiae | BY4742 GEV | MATa, (PGAL10+gal1)Δ::loxP, leu2Δ0::PACT1-GEV-NatMX, gal4Δ::LEU2, HAP1+ | Minimize homologous recombination | [69] |
| P. pastoris | SMD1168H | pep4 | Protease A deficiency | [59] |
| S. cerevisiae | FAB158 | MATa his3- Δ200 leu2- Δ1 lys2-801 trp1- Δ1 ade2-101 ura3-52 tat2 Δ::HIS3 | Deletion of tryptophan transporter | [70] |
| S. cerevisiae | TMY203 | MATa his3- Δ200 leu2- Δ1 lys2-801 trp1- Δ1 ade2-101 ura3-52 tat1 Δ::kanMX4 tat2Δ::LEU2 | Deletion of tryptophan transporters | [70] |
| S. cerevisiae | FAY18A | MATa his3- Δ200 leu2- Δ1 lys2-801 trp1- Δ1 ade2-101 ura3-52 HPG1-1, Rsp5P514T | Deletion of Rsp5 ubiquitin ligase | [70] |
| S. cerevisiae | XPY1263a | MATa thi3Δ::natMX thi7D::kanMX | Deletion of thiamine transporter | [71] |
| S. cerevisiae | BY4741mp | MATa; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0; mir1Δ; pic2Δ | Deletion of phosphate and copper transporter | [72] |
| S. cerevisiae | BY4741 pic2Δ | MATa, leu2,met15, ura3, his3, PIC2::KANMX | Deletion of copper transporter | [73] |
| S. cerevisiae | WB-12 | MATα ade2-1 trp1-1 ura3-1 can1-100 aac1::LEU2 aac2::HIS3 | Deletion of adenine nucleotide carriers | [74] |
| S. cerevisiae | JL1-3Δ2 | Matα leu2-3,112 his3-11,15 ade2-1 trp1-1 ura3-1can1-100 anc1::LEU2 Δ anc2::HIS3 anc3::URA3 | Deletion of adenine nucleotide carriers | [75] |
| S. cerevisiae | W303-B1 | Mata; ade2-1; his3-11, -15; leu2-3, -112; ura3-1; canR; cyr+ | Poor leucine uptake | [76] |
| S. cerevisiae | W303-1A | MATa: ade2-2; trp1-1; can1-100; leu2-3, 112; his 3-11, 15; ura3-1 | Poor leucine uptake | [77] |
| S. cerevisiae | W303 | MATa/MATα (leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15) [phi+] | Poor leucine uptake | [78] |
| S. cerevisiae | W303 Wagc1Δ | Mat a/Mat a, ura3-1/ura3-1, trp1- Δ2/trp1- Δ2, leu2-3,112/leu2-3,112, his3-11/his3-11, ade2-1/ade2-1, can1-100/can1-100 | Poor leucine uptake; deletion of AGC1 carrier | [79] |
| S. cerevisiae | PW001 | BY4741 agc1∆::URA3 | Deletion of AGC1 carrier | [80] |
| S. cerevisiae | PW002 | BY4741 agc1∆::HIS3 | Deletion of AGC1 carrier | [80] |
| S. cerevisiae | ΔArg11 Y02386 | MATa; his3Δ1; leu2Δ0; met15Δ0; ura3D0; YOR130c::KanMX4 | Deletion of ornithine transporter 1 | [81] |
| S. cerevisiae | ΔAnt1 BJ1991 | MATa, leu2, trp1, ura3-251, prb1-1122, and pep4-3 | Deletion of ANT1 carrier | [82] |
| S. cerevisiae | TCY119 | MAT α ura3–52 leu2–3, 112 trp1-Δ1 ade2 his3-Δ1::hisG aac1-Δ1::hisG aac2-Δ1::kanMX6 aac3-Δ1::hisG [r+, TRP1] | Deletion of AAC1, AAC2 and AAC3 carriers | [83] |
| S. cerevisiae | W303 | his3-11,15; ade2-1; leu2-3,112; ura3-1; trp1-1; can1-100; RIM2/RIM2::kanMX | Poor leucine uptake; deletion of pyrimidine nucleotide carrier | [84] |
| S. cerevisiae | ST9352 | MATa, aro10Δ, pdc5Δ, pTEF1->ARO7, pPGK1->ARO4, pTEF1->FjTAL, pTEF1->HsSLC25A44 | Alteration of aromatic amino acid metabolism | [85] |
| S. cerevisiae | CEN.PK113-7D ndt1Δndt2Δ | MATa MAL2-8c SUC2 ndt1Δndt2Δ | Deletion of NAD transporter | [86] |
| S. cerevisiae | BY4742 ndt1Δndt2Δ | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0, ndt1Δndt2Δ | Deletion of NAD transporter | [87] |
| S. cerevisiae | FOH33 | MATα MAL2-8c SUC2 his3Δ ura3-52 hfd1Δ pox1Δ faa1Δ faa4Δ adh6Δ::kanMX gal80Δ gal1/10/7Δ::(GAL7p-MmCAR-ADH1t)+(GAL3p-npgA-FBA1t); pAOH9 | Deletion of alcohol dehydrogenase | [88] |
| S. cerevisiae | KY114 | MATα, gal, ura3-52, trp1, lys2, ade2, his d2000 | Deletion of thymidine transport | [89] |
| S. cerevisiae | BY4742-YBR021W | MATα, his3, leu2, lys2, ura3, ΔFUR4 | Deletion of uridine permease | [90] |
| S. cerevisiae | DY150 | Mata ade2-1 his3-11 leu2-3,112 trp1-1 ura3-52 can1-100(oc) | Deletion of Fe2+/Mn2+ transporter | [91] |
| S. cerevisiae | YPL1 | MATα fui1 Δ:: HIS3, ura3-52, lys2-801, HIS3 Δ | Deletion of uridine permease | [92] |
| S. cerevisiae | W303-Δpep4 | leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 hwas3-11,15 Δ pep4 MATα | Deletion of vacuolar endopeptidase Pep4 | [93] |
| S. cerevisiae | W303-1A | MATa ade2-1, can1-100, cyh2, his3-11,15, leu1-c, leu2-3,112, trp1-1, ura3-1 | No growth in absence of adenine | [94] |
| S. cerevisiae | FGY217 | MATα, ura3-52, lys2 Δ 201 and pep4 Δ | Deletion of vacuolar endopeptidase Pep4 | [95] |
| S. cerevisiae | BY4743 | Mata/α his3Δ1/ his3Δ1 leu2Δ0/ leu2Δ0 lys2Δ0/+ met15Δ0/+ ura3Δ0/ ura3Δ0 | Essential genes deletion | [96] |
| S. cerevisiae | BY4743 Δpmr1::KanMX | Mata/α his3Δ1/ his3Δ1 leu2Δ0/ leu2Δ0 lys2Δ0/+ met15Δ0/+ ura3Δ0/ ura3Δ0, Δpmr1::KanMX | Deletion of essential genes and Pmr1 Ca2+ and Mn2+ transporter | [97] |
| S. cerevisiae | ctr1 | MATa ura3 lys2 ade2 trp1 his3 leu2 Dctr1::LEU2 | Deletion of CTR1 transporter | [98] |
| S. cerevisiae | YPH499 | MATa ura3-52 lys2-801ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 | [99] | |
| S. cerevisiae | YPH500 | MAT α/ura3–52/lys2–801/ade2–101/trp1-Δ63/his3-Δ200/leu2-Δ1 | [100] | |
| S. cerevisiae | STY50 | MATa,his4-401,leu2-3,-112,trp1-1,ura3-52,HOL1-1,suc2::LEU2 | Deletion of invertase 2 | [101] |
| P. pastoris | CBS7435 | Wild type | [102] | |
| S. cerevisiae | YPH501 | MATa/MATα ura3-52/ura3-52 lys2-801/lys2-801 ade2-101/ade2-101 trp1-Δ63/ trp1-Δ63 his3- Δ200/his3- Δ200 Ieu2- Δ1/leu2- Δ1 | [100] | |
| S. cerevisiae | CB001L | MATα, leu2, trp1, ura3, prb−, pep4::LEU2 | Deletion of vacuolar proteinase A and B | [103] |
| S. cerevisiae | JRY472 | Δmpc1/2/3 | Deletion of pyruvate carriers | [104] |
| S. cerevisiae | BY4741 gdt1Δpmr1Δ | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 gdt1::KanMX4 pmr1::KanMX4 | Deletion of Ca2+ and Mg2+ transporters | [105] |
| Plasmid | Promoter | Type | Gene Product | Regulation | References |
|---|---|---|---|---|---|
| pPICZB | AOX | Inducible | Alcohol oxydase | Methanol (+) | [108] |
| pYES2 | GAL1 | Inducible | Galactokinase | Galactose (+)/Glucose (−) | [109] |
| YEp4H7 | HXT7 | Constitutive | hexose transporter HXT7 | Low glucose (+) | [110] |
| p426H7 | HXT7 | Constitutive | hexose transporter HXT7 | Low glucose (+) | [110] |
| pVT102 | ADH1 | Constitutive | Alcohol dehydrogenase I | Ethanol (+) | [111] |
| P426-GPD | GPD | Constitutive | Glyceraldehyde—3 -phosphate dehydrogenase | Glucose | [112,113] |
| pEMBLyex4 | CYC-GAL | Hybrid/inducible | Cytochrome C1, GAL1-GAL10 intergenic promoter | Galactose (+) | [114] |
| p426MET25 | MET25 | Inducible | methionine and cysteine synthase | methionine (−) | [115] |
| YEpM | Pma1 | Constitutive | Plasma membrane H+-ATPase | [67] | |
| pPGK | PGK1 | Constitutive | 3-phosphoglycerate kinase | Glucose | [61,116] |
| pYES2.1 | GAL1 | Inducible | Galactokinase | Galactose (+)/Glucose (−) | [109] |
| pYES | GAL1 | Inducible | Galactokinase | Galactose (+)/Glucose (−) | [109] |
| pPICHOLI | AOX | Inducible | Alcohol oxydase | Methanol (+) | [108] |
| pSM1052 | PGK1 | Constitutive | Phosphatidyl glycerol kinase—1 | Glucose | [61,116] |
| pPIC3L | AOX | Inducible | Alcohol oxydase | Methanol (+) | [108] |
| pPIC3.5K | AOX | Inducible | Alcohol oxydase | Methanol (+) | [108] |
| pRS315 | TDH3 | Constitutive | Glyceraldehyde—3-phosphate dehydrogenase | Glucose | [112,113] |
| pXP951 | THI7 | Thiamine transporter | [71] | ||
| pCM188 | CYC1 | Constitutive | Cytochrome c oxidase | glucose (−) | [117] |
| YEp352 | Lac | Inducible | Β-galactosidase | Lactose (+) | [72] |
| pRS314-YA2P | yAAC2 | Intrinsic | Yeast ADP/ATP translocase | Glucose (−) | [74] |
| pRS424 | yAAC2 | Intrinsic | Yeast ADP/ATP translocase | Glucose (−) | [74] |
| pYeDP-1/8-10 | GAL10-CYC1 | Hybrid/inducible | Cytochrome C1, GAL10 promoter | Galactose (+) | [114] |
| pCGS110 | GAL1 | Inducible | Galactokinase | Galactose (+)/Glucose (−) | [109] |
| pYeDP60 | GAL10-CYC1 | Hybrid/inducible | Cytochrome C1, GAL10 promoter | Galactose (+) | [114] |
| pGal110 | GAL1/GAL10 | Inducible | Galactokinase/epimerase | glucose (−)/galactose (+) | [109] |
| pCGS110 | GAL1 | Inducible | Galactokinase | Galactose (+)/Glucose (−) | [109] |
| pYX142 | TPI | Constitutive | Triose-phosphateisomerase | [118] | |
| pEL30 | CTA1 | Inducible | Catalase | Oleate (+) | [119] |
| pESC-Leu2d | GAL1 | Inducible | Galactokinase | Galactose (+)/Glucose (−) | [109] |
| pRS42H | RIM2 | Yeast mitochondrial pyrimidine nucleotide carrier | [84] | ||
| pcfB9056 | TEF | Constitutive | Translational elongation factor EF-1 α | [120] | |
| YCplac33-NAT_MCART1 | TEF | Constitutive | Translational elongation factor EF-1 α | [120] | |
| pRS415 | TEF | Constitutive | Translational elongation factor EF-1 α | [120] | |
| pIYH01 | HXT6, HXT7 | Constitutive | Hexose transporter 6 and 7 | Low glucose (+) | [88] |
| pYPGE15 | PGK1 | Constitutive | 3-phosphoglycerate kinase | Glucose | [61,116] |
| pDDGFP-2 | GAL1 | Inducible | Galactokinase | Galactose (+)/Glucose (−) | [109] |
| pPICZ | AOX | Inducible | Alcohol oxydase | Methanol (+) | [108] |
| pYES-DEST52 | GAL1 | Inducible | Galactokinase | Galactose (+)/Glucose (−) | [109] |
| pDB20 | ADH1 | Constitutive | Alcohol dehydrogenase I | Ethanol (+) | [121] |
| pYEX-BX | CUP1 | Inducible | metallothionein | Cu(II) (+) | [122] |
| pPIC9K | AOX | Inducible | Alcohol oxydase | Methanol (+) | [108] |
| pKTΔATG | GPD | Constitutive | Glyceraldehyde—3-phosphate dehydrogenase | Glucose | [113] |
| p426GFP | GPD | Constitutive | Glyceraldehyde—3-phosphate dehydrogenase | Glucose | [112,113] |
| pYEScupFLAGK or pYEScupFLAGE | CUP1 | Inducible | Metallothionein | Cu(II) (+) | [122] |
| pYEX-BESN | CUP1 | Inducible | Metallothionein | Cu(II) (+) | [122] |
| pBEVY-GU | GAL1 | Inducible | Galactokinase | Galactose (+)/Glucose (−) | [109] |
| pRS416 | TPI1 | Constitutive | Triose-phosphate isomerase, | [105] | |
| pPICZA | AOX | Inducible | Alcohol oxydase | Methanol (+) | [108] |
| Protein/Alias | Strain Feature/Deleted Gene | Tag | System of Study | Detection Method | Findings | References |
|---|---|---|---|---|---|---|
| SLC1A5/ASCT2 | None | C-Ter 6His | PL | R, FM | F, K, I | [30,33,195,196,197,198,199,200] |
| SLC2A1/GLUT1 | Tolerates copper induction/Pep4 | 6His | - | WB | E | [201] |
| SLC2A1/GLUT1 | hxt0 | - | IC | C, R | L, K | [64] |
| SLC2A1/GLUT1 | hxt0 | - | IC | R | I | [202] |
| SLC2A1/GLUT1 | - | C-Ter 8His | PL | R | P, TA | [180] |
| SLC2A1/GLUT1 | hxt0 | N-Ter 6His | IC | R | K, I | [203] |
| SLC2A1/GLUT1 | hxt0 | C-Ter HA-6His | IC | C, R | L, K, I | [68] |
| SLC2A1/GLUT2 | hxt0 | - | IC | R | I | [204] |
| SLC2A2/GLUT2 | - | C-Ter-GFP-8His | - | FM, FSEC | L, P | [60] |
| SLC2A2/GLUT2 | hxt0 | - | IC | FM, R | L, I | [66,202] |
| SLC2A2/GLUT2 | hxt0 | - | IC | R | I | [204] |
| SLC2A3/GLUT3 | hxt0 | - | IC | R | I | [66,202] |
| SLC2A3/GLUT3 | hxt0 | - | IC | R | I | [204] |
| SLC2A4/GLUT4 | hxt0 | - | IC | C, R | L, K | [64,202] |
| SLC2A4/GLUT4 | hxt0 | - | IC | R | I | [204] |
| SLC2A5/GLUT5 | hxt0 | GFP | IC | FM, R | L, K, I | [202,205] |
| SLC2A5/GLUT5 | hxt0 | - | IC | R | I | [204] |
| SLC4A1/AE1 | Pep4 | - | PL | C, FM, R | L, P, F | [67] |
| SLC4A1/AE1 | Pep4 | N-Ter HA; Internal HA; N-ter yeGFP | IC | FM, AEC | L, F | [59] |
| SLC4A1/AE1 | end3 | N-Ter HA; Internal HA; N-ter yeGFP | IC | FM, EM, WB | L | [69] |
| SLC5A1/SGLT1 | - | eGFP | PLM | FM, CM | L, P, F | [168] |
| SLC5A6/SMVT | - | C-Ter FLAG 6His | PL | WB, R | P, TA, K | [58] |
| SLC7A5/LAT1 | - | N-Ter StrepTagII | - | WB | E | [206] |
| SLC7A5/LAT1 | - | N-Ter 10His | IC | R | K, I, TA | [190,207] |
| SLC7A6/y+LAT2 | - | N-Ter StrepTagII | ND | WB | E | [206] |
| SLC7A8/LAT2 | - | N-Ter StrepTagII | IC | WB, R | P, TA, I, K | [190,207,208] |
| SLC9A1/NHE1 | - | C-Ter yeGFP | IC, N | FM, IMAC, CD | L, P | [208] |
| SLC9B2/NHA2 | ena1-4, nhx1, nha1 | N-Ter 9His, or N-Ter GFP | IC | SM | TA, I | [61] |
| SLC11A1/Nramp1 | smf1, smf2, smf3, bsd2, rer1 | C-Ter HA or GFP | IC | FM, PG | L, TI | [62] |
| SLC15A1/PEPT1 | his4 | - | IC | R | K | [63] |
| SLC15A1/PEPT1 | his4 | C-Ter Myc, 6His | IC | RFT | F, K, I | [209] |
| SLC15A1/PEPT1 | - | - | IC | R | F, K, I | [210] |
| SLC15A2/PEPT2 | - | - | IC | RFT | F, K, I | [211] |
| SLC16A10/MCT10 | Tat2 | N-ter 3HA, C-Ter GFP | IC | FM, R | L, TA | [99] |
| SLC19A3/Thiamine transporter 2 | thi3, thi7 | - | IC | PG, R | F, I | [71] |
| SLC22B1/SV2A | hxt0 | - | IC | PG, R | F, K, I | [212] |
| SLC25A3/phosphate carrier | mir1, pic2 | - | - | PG | CG | [72] |
| SLC25A3/phosphate carrier | pic2 | - | M | SM | TA | [73] |
| SLC25A4/ANT1 | aacs | - | M | R | TA, I | [74,213] |
| SLC25A4/ANT1 | aacs | - | M | SM, R | K, I | [75,213] |
| SLC25A5/ANT2 | aacs | - | M | SM, R | K,I | [75,213] |
| SLC25A6/ANT3 | aacs | - | M | SM, R | K, I | [75,213] |
| SLC25A7/UCP1 | - | - | M | RA | MR | [214] |
| SLC25A8/UCP2 | - | - | PL | FM | I, A | [215] |
| SLC25A9/UCP3L | - | - | IC | FM, O | MP, MR | [76] |
| SLC25A9/UCP3L | - | - | IC | FC, XTT | MP, MR | [77] |
| SLC25A9/UCP3(L,S) | - | - | M | FM, EA | MP, OP | [216] |
| SLC25A9/UCP3 | - | - | PL | FM | I, A | [215] |
| SLC25A12/Aralar1 | agc1 | - | IC | SM | TI | [79] |
| SLC25A13/Citrin | agc1 | - | IC | SM | TI | [79] |
| SLC25A13/Citrin | agc1 | C-Ter GFP | IC | F | L | [80] |
| SLC25A15/ORNT1 | Arg11 | C-Ter V5, His | IC | PG | CG | [81] |
| SLC25A16/ORNT2 | Arg11 | C-Ter V5, His | IC | PG | CG | [81] |
| SLC25A17/PMP34 | Ant1 | C-Ter 6His | PL | IA, R | L, TA | [82] |
| SLC25A31/ANT4 | Aacs | C-Ter V5 | IC, M | CG, R | TA, I | [213,217] |
| SLC25A33/PNC1 | Rim2 | C-Ter 6His | IC | O | MR | [84] |
| SLC25A36 | Rim2 | C-Ter 6His | IC | O | MR | [84] |
| SLC25A44 | - | - | IC | H | TA | [85] |
| SLC25A51/MCART1 | ndt1, ndt2 | - | IC | LC-MS | TI | [86] |
| SLC25A51/MCART1 | ndt1, ndt2 | - | M | R | TI, TA | [87] |
| SLC27A1/FATP1 | High fatty alcohol production | - | - | GC, SA | YS | [88] |
| SLC28A1/CNT1 | Thymidine transporter defective | - | IC | PG | CG | [89] |
| SLC28A1/CNT1 | fui1 | - | IC | R | I | [218,219] |
| SLC28A2/CNT2 | fur4, fui1 | - | IC | IA, R | TA, K, I | [90,219,220] |
| SLC28A3/CNT3 | fui1 | - | IC | FM, R | L, F, K | [220] |
| SLC28A3/CNT3 | fui1 | - | IC | CM, R | L, I, K | [90] |
| SLC28A3/CNT3 | fui1 | - | IC | R | I | [219,220,221] |
| SLC29A1/ENT1 | Pep4 | N-Ter 10His-MBP | - | IMAC | P | [93] |
| SLC29A1/ENT1 | fui1 | - | IC | R | TA, K, I, | [219,220,221,222] |
| SLC29A1/ENT1 | Expression of thyimidine kinase | - | IC, PL | R | TA, I, EB | [223] |
| SLC29A1/ENT1 | Fui1 | N-Ter GFP | IC | R, FM | TA, K, L | [92] |
| SLC29A1/ENT1 | - | - | IC | R | K, I | [221] |
| SLC29A1/ENT1 | Fui1 | - | IC | R | TA, I | [222] |
| SLC29A1/ENT1 | Fui1 | - | IC | R | K, I | [94] |
| SLC29A2/ENT2 | Pep4 | N-Ter 10His-MBP | - | IMAC | P | [93] |
| SLC29A2/ENT2 | Fui1 | - | IC | R | I | [219,220,221] |
| SLC29A2/ENT2 | Fui1 | - | IC | R | TA, I, K | [222,224] |
| SLC30A1/ZnT1 | Pep4 | C-Ter His, C-Ter Strep | PL | IMAC, FM | P, TA | [95] |
| SLC30A8/ZnT8 | - | G3-tev-G3-H10-G3-FLAG | IC, PL | R | TA, F | [181] |
| SLC30A10/ZnT10 | Pmr1 | C-ter V5,6His | - | PG | CG | [97] |
| SLC31A1/CTR1 | Ctr1 | - | - | PG | CG | [98] |
| SLC35A1/CST1 | - | - | V | R | TA, I | [225] |
| SLC35A1/CST1 | Pep4 | GFP, 8His | IC | FM, FSEC | P, L | [226] |
| SLC35A1/CST1 | - | GFP, 8His | - | FM, FSEC | L | [178] |
| SLC35A2/UGT1 | - | V | R | TA | [225] | |
| SLC35A2/UGT1 | - | - | V | R | TA, F, K, I | [227,228] |
| SLC35B1/UGTrel1 | - | GFP | PL | R | F, K, I | [189] |
| SLC35B4 | UDP-Glc NAc transporter | N-Ter GFP | V | FM, R | L, TA | [229] |
| SLC35D1/ UGTrel7 | Pep4 | C-Ter 8His, C-Ter HA | V, PL | R | TA, K | [103,230] |
| SLC35D2/UGTrel8 | - | C-Ter HA | V | FM, R | L, TA | [193] |
| SLC39A1/ZIP1 | Pep4 | C-Ter TEV-GFP-His | - | IMAC, FSEC | P | [231] |
| SLC39A2/ZIP2 | Pep4 | C-Ter TEV-GFP-His | - | FSEC | P | [231] |
| SLC39A11/ZIP11 | Pep4 | C-Ter TEV-GFP-His | - | - | E | [231] |
| SLC39A13/ZIP13 | Pep4 | C-Ter TEV-GFP-His | - | FSEC | P | [231] |
| SLC54A1/MPC1 | mpc1, mpc2, mpc3 | C-Ter 8His | M, PL | FM, IMAC, R, EA | L, P, TA | [104] |
| SLC54A2/MPC2 | mpc1, mpc2, mpc3 | C-Ter GFP | M, PL | IMAC, FM, R, EA, O | P, L, TA, MR | [104] |
| SLC64/TMEM165 | Gdt1p, Pmr1 | - | - | PG | CG | [105] |
| STRA6 | None | C-Ter GFP | IC | FM, FSEC | L, P | [232] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pochini, L.; Galluccio, M. Heterologous (Over) Expression of Human SoLute Carrier (SLC) in Yeast: A Well-Recognized Tool for Human Transporter Function/Structure Studies. Life 2022, 12, 1206. https://doi.org/10.3390/life12081206
Pochini L, Galluccio M. Heterologous (Over) Expression of Human SoLute Carrier (SLC) in Yeast: A Well-Recognized Tool for Human Transporter Function/Structure Studies. Life. 2022; 12(8):1206. https://doi.org/10.3390/life12081206
Chicago/Turabian StylePochini, Lorena, and Michele Galluccio. 2022. "Heterologous (Over) Expression of Human SoLute Carrier (SLC) in Yeast: A Well-Recognized Tool for Human Transporter Function/Structure Studies" Life 12, no. 8: 1206. https://doi.org/10.3390/life12081206
APA StylePochini, L., & Galluccio, M. (2022). Heterologous (Over) Expression of Human SoLute Carrier (SLC) in Yeast: A Well-Recognized Tool for Human Transporter Function/Structure Studies. Life, 12(8), 1206. https://doi.org/10.3390/life12081206







