Abstract
Background: Retrotransposons with long terminal repeats (LTR retrotransposons) are widespread in all groups of eukaryotes and are often both the cause of new mutations and the source of new sequences. Apart from their high activity in generative and differentiation-stage tissues, LTR retrotransposons also become more active in response to different stressors. The precise causes of LTR retrotransposons’ activation in response to stress, however, have not yet been thoroughly investigated. Methods: We used RT-PCR to investigate the transcriptional profile of LTR retrotransposons and piRNA clusters in response to oxidative and chronic heat stresses. We used Oxford Nanopore sequencing to investigate the genomic environment of new insertions of the retrotransposons. We used bioinformatics methods to find the stress-induced transcription factor binding sites in LTR retrotransposons. Results: We studied the transposition activity and transcription level of LTR retrotransposons in response to oxidative and chronic heat stress and assessed the contribution of various factors that can affect the increase in their expression under stress conditions: the state of the piRNA-interference system, the influence of the genomic environment on individual copies, and the presence of the stress-induced transcription factor binding sites in retrotransposon sequences. Conclusions: The main reason for the activation of LTR retrotransposons under stress conditions is the presence of transcription factor binding sites in their regulatory sequences, which are triggered in response to stress and are necessary for tissue regeneration processes. Stress-induced transposable element activation can function as a trigger mechanism, triggering multiple signal pathways and resulting in a polyvariant cell response.
1. Introduction
Transposition significantly affects the structure of the host genome and demonstrates a fundamental importance in the formation of genetic variability [1]. One of the most interesting types of transposable elements (TEs) for research is long-terminal repeat retrotransposons (LTR retrotransposons) since they have a common origin with retroviruses. Therefore, understanding the mechanisms of interaction between the host genome and LTR retrotransposons significantly contributes to understanding the mechanisms of interaction between the host genome and retroviruses. LTR retrotransposons insert into a new site through a replicative mechanism in which revertase synthesizes a new copy of TE on the matrix of its transcript and the integrase then inserts this copy into a new site [2].
The eukaryotic cell has a series of mechanisms to prevent transposition [3,4,5]. All of them are aimed at suppressing transcription and translation by RNA interference (mostly si- and piRNA). The highest activity of LTR retrotransposons in eukaryotes is observed in germline cells; therefore, the piRNA interference system, the main mechanism for suppressing TEs in germline cells, works intensively in these tissues [6]. Drosophila melanogaster is a model object on which the piRNA interference system has been investigated in detail. In Drosophila, this process is different in the somatic and generative ovary tissues.
In somatic tissues of D. melanogaster, piRNA precursors are transcripts of uni-strand clusters (aggregations of old copies of TEs transcribed in the antisense direction), such as flamenco. These long polyadenylated transcripts undergo alternative splicing and are sent to the protein complex on the outer mitochondrial membrane, Yb-body, where they are cut into smaller fragments and, after methylation, are transported to the nucleus in a complex with the PIWI protein, which finds the transcribed retrotransposon with the piRNA sequence and attracts proteins that provide a silencing of this region [6,7,8,9,10].
piRNA clusters, transcribed in two directions, undergo transcription initiation from non-canonical sites within the clusters by the RDC complex (Rhino, Deadlock, Cutoff) [11]. This complex not only drives RNA polymerase to start transcription from a non-canonic site but also prevents polyadenylation and capping of the transcript, due to which transcripts are delivered with no changes to the Nuage perinuclear area, where the Aub and AGO3 proteins start the ping-pong cycle. During this process, long transcripts of piRNA clusters interfere with TE transcripts, and their duplexes are cleaved into small fragments. This results in the formation of primary and secondary piRNAs, some of which serve as a template for TE RNA cutting and others for the cleavage of piRNA precursors [12,13,14]. In addition, several piRNAs from the ping-pong cycle are also used by PIWI to suppress TE activity at the transcription level [15,16,17]. Thus, the transposition activity of TEs and the subsequent mutation process directly depend on the stability of the piRNA system.
In addition to endogenous genomic factors, TE activity is often regulated by external stress conditions, such as heat and oxidative stress. It has been shown for different groups of organisms that certain TEs are able to increase their expression in response to biotic and abiotic stresses. For Drosophila, it has been shown that the 412 element copy number correlates with the temperature increase in wild populations of Drosophila [18]. It is also shown that in D. melanogaster, some TEs are able to increase expression, while others, on the contrary, decrease it under various types of stress. For example, four TEs increased their expression, while the other two decreased in response to dioxin, and only three of the studied TEs responded with an increase in expression under formaldehyde [19]. Thus, different TEs respond to different stresses individually. The hypothesis that TEs are adapted to stressful influences was proposed in the work of Barbara McClintock [20]. It is obvious that such adaptation occurs due to the accumulation of mutational changes, some of which may contribute to the conservation and spread of TEs. Another aspect of the problem is the possible role of TEs in the formation of the adaptive response of the organism to the influence of various stressors. While TE activation under strong stress conditions can lead to transposition, cause multiple damage to the genome, and lead to cell death, under weak stress conditions, TE activation can play the role of a trigger mechanism that starts several processes at once and provides a polyvariant reaction of a living cell in response to the stressor.
There are several molecular cascades involved in the response to various abiotic stresses, in particular, to oxidative stress: Jak/STAT, ERK, JNK, FOXO, MAPK, NF-kB, keap1/CncC, and PI(3)K/Akt, as well as proteins Hsp22, p53, and p38 [21,22,23]. Therefore, the presence of binding sites for certain transcription factors of various signaling cascades could explain the reason for individual TE responses to various oxidizing agents and other abiotic factors. Such binding sites were found with Chip-seq in many TEs of Drosophila and humans [24]. Under the influence of evolutionary processes, cis-regulatory elements of TE may become promoters, insulators, and enhancers for the genes of the host organism, in particular, Drosophila. And among these cis-regulatory elements, the vast majority belong to LTR retrotransposons [25]. Thus, the reasons for the activation of retrotransposons in response to stress may be associated with piRNA interference system functionality, the position of copies in the host genome, and the presence of their own binding sites for transcription factors.
In this work, we investigated the causes of changes in the expression of four well-studied LTR retrotransposons, copia, gypsy, Tirant, and springer, in response to oxidative and chronic heat stress. We analyzed the expression dynamics of selected TEs, piRNA clusters, and three markers of oxidative stress (upd3, sid, and hsp22). After that, we searched for conservative and unique insertions of these retrotransposons in the genomes of two strains, SS7K (the strain with impaired transposition control) and two wild-type strains, and analyzed the genomic environment of these insertions and their potential effect on the expression of these retrotransposons. We compared the change in the expression of two copies of Tirant and their genomic environment, searched for potential transcription factors binding sites, and then validated their conservatism among different copies.
2. Materials and Methods
2.1. Drosophila melanogaster Strains and Cultivation Conditions
The SS7K strain has a mutation in the flamenco locus that controls the transposition of the TE gypsy [26]; Canton-S is a wild-type strain; D32 is a wild-type laboratory strain from the collection of the Department of Genetics of Moscow State University. All strains were cultivated at 25 °C on the standard nutrient agar medium.
2.2. Induction of Oxidative and Chronic Heat Stress
To induce oxidative stress, adult seven-day-old females were incubated on a fresh nutrient agar medium containing 0.1 M ammonium persulfate (APS) for 24 h at 25 °C. After that, total RNA was isolated. To rest after stress, the flies were incubated on a standard medium for 24 and 48 h. To induce chronic temperature stress, adults were incubated on a fresh nutrient agar medium and left overnight at 25 °C. The next day, the parents were removed and the embryos were incubated at 29 °C until metamorphosis to imago. After hatching, the imago was incubated at 29 °C for 7 days, after which RNA was isolated from the females.
2.3. RNA Isolation, Reverse Transcription, and Real-Time PCR
RNA isolation was performed from the tissues of the ovaries, heads, and bodies using the ExtractRNA reagent (Evrogen, Moscow, Russia) in PBS buffer. RNA samples were treated with DNase I (Thermo Fisher Scientific, Waltham, MA, USA). The MMLV-RT Kit (Evrogen, Moscow, Russia) was used for reverse transcription. Reverse transcription was performed with a random primer for all samples (because the expression of most clusters in our experiment did not exceed the expression of low-copy TEs, and primers were designed for unique genome regions). PCR was performed in the presence of SYBR Green I (Evrogen, Moscow, Russia) on a MiniOpticon Real-Time PCR System (Bio-Rad, Hercules, CA, USA). In the experiment, we analyzed the relative expression of gypsy, Tirant, copia, and springer retrotransposons, 38C, 20A, 42AB, and flamenco clusters, normalized to the expression of the αTub84D, Rpl40, and EloB genes (Table 1). Statistical processing of PCR results was performed using the Mann–Whitney test. Primers for the 42AB cluster were designed earlier in [27]; primers for the 38C and 20A clusters were designed in [16]; primers for the flamenco cluster were designed for the unspliced form of the transcript according to the sequence from the NCBI database (Gene ID: 26067356). For primer design, we used the flamenco locus sequences that did not contain TEs. The forward primer was placed in the first exon and the reverse in the first intron. The selected primers cover the transcription region; however, they do not match the TEs in this cluster and therefore do not anneal to the cDNA of TEs.

Table 1.
Primers used to evaluate TEs, genes, and piRNA cluster expression, as well as to count the copy number of the studied transposable elements.
2.4. DNA Isolation and Nanopore Sequencing
DNA isolation was carried out according to the standard method [28]. The DNA concentration in the sample was measured using a NanoDrop spectrophotometer (Peqlab, Erlangen, Germany). After that, the library was prepared according to the protocol of the Oxford Nanopore. The library was estimated using a NanoDrop spectrophotometer and loaded into the Nanopore MiniION Oxford sequencer. The sequencing data were mapped to the D. melanogaster reference genome (dm 6) in a virtual kernel (WSL) of Ubuntu for Windows 11 using miniconda2 (https://docs.conda.io/projects/miniconda/en (accessed on 3 December 2021)) with settings for reads obtained using Nanopore sequencing. The results were visualized using the IGV program [29]. For unique insertions of the studied TEs, we used the TLDR program [30]. The library of searched TEs was based on sequences from BDGP (https://www.fruitfly.org/p_disrupt/TE.html (accessed on 22 December 2021)). The results of the TLDR program were sorted by the length of the regions of new insertions matching the TE sequences, and long matches were checked using pairwise alignment in BlastN with the reference sequence [31]. The LASAGNA database [32], JASPAR, and TRANSFAC models of TF binding sites were used to search for binding sites with TF binding sites in the regulatory sequences of TEs. For analysis, only those binding sites were used that were predicted with a p < 0.001. The functions and expression patterns of TFs were checked in the FlyBase database [33]. The conservatism of the found binding sites was checked using multiple alignments in the Ugene [34]. We also searched for conservative TE insertions utilizing the Blast function in FlyBase and validated these positions in SS7K and Canton-S in IGV.
2.5. Evaluation of the Number of Copies of Transposable Elements by PCR
The reaction was run on a MiniOpticon Real-Time PCR System from Bio-Rad Laboratories. In the experiment, we analyzed the relative number of copies of the TE copia, gypsy, springer, and Tirant normalized on the number of single-copy genes αTub84D and BoYb. DNA from 20 females was used to determine the average copy number of LTR retrotransposons; 80, 40, and 20 ng DNA were taken into the PCR reaction, with two repeats for each concentration. After normalization on single-copy genes, the average number of copies of each TE between three dilutions for each strain was calculated.
3. Results
3.1. Transcription Analysis of LTR Retrotransposons under Oxidative and Chronic Heat Stress Conditions
We measured the expression of four LTR retrotransposons, copia, gypsy, Tirant, and springer, in response to oxidative and chronic heat stress. These TEs represent different groups of LTR retrotransposons with different copy numbers. Ammonium persulfate (APS) was used as an oxidizing agent, which activates oxidative stress-responding genes [35]. We measured the transcription level of three genes as a control for the induction of oxidative stress: upd3, sid, and hsp22 (Figure 1). The expression of these genes increased significantly after a 24 h incubation of 7-day-old Drosophila females on 0.1 M APS in all three strains.

Figure 1.
Relative expression of genes involved in the oxidative stress response during 24 h incubation on media with 0.1 M APS in females of SS7K, Canton-S, and D32 strains. (* p < 0.5, ** p < 0.01, according to the Mann–Whitney test; ns, not significant—statistically insignificant change).
After validating the induction of oxidative stress, we measured the transcription level of studied TEs in the same samples (Figure 2). TEs reacted to oxidative stress in most cases by increasing their expression; however, this was not a consistent pattern for all strains. Thus, most of the TEs in the D32 strain did not demonstrate a statistically significant change in expression in response to oxidative stress. At the same time, both without stress and during stress, we found a large scatter in the values of the expression level in different samples. This may indicate the genetic heterogeneity of this strain, which causes different levels of TE expression individually.
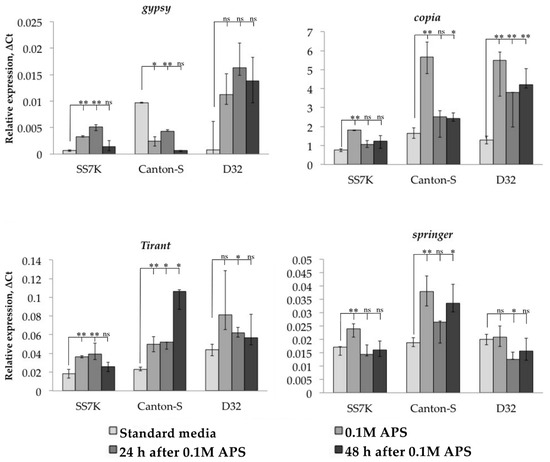
Figure 2.
Dynamics of changes in TE expression in 7-day-old imago females exposed to APS and during rest after the stress (* p < 0.5, ** p < 0.01, according to the Mann–Whitney test; ns, not significant—statistically insignificant change).
Additionally, we tracked the dynamics of the expression of particular retrotransposons 24 and 48 h after the stress was removed. We observed that LTR retrotransposon expression did not immediately return to the initial level in some cases. The differences between flies incubated on a standard media and flies exposed to an oxidizing agent were significant in the following 24 or 48 h of the rest after stress: gypsy and Tirant in SS7K did not decrease expression within 24 h; Tirant in Canton-S and copia in D32 did not decrease expression within 48 h.
Since TE transcription may depend on the piRNA interference system, the expression of various transcription factors, and the position in the genome of a particular strain, we analyzed each of these factors. First of all, we studied the dynamics of oxidative stress marker gene expression in the Canton-S strain before, under, and after stress exposure (Figure 3). The expression of these genes was significantly increased under oxidative stress but returned to a normal level 48 h after stress. At the same time, as was shown above, the expression of LTR retrotransposons at this time point in some cases was significantly higher than the baseline level. Thus, it can be argued that the regulation mechanisms of the studied genes and TEs are different since the expression of TEs after the onset of stress persists longer. Thus, TEs may have a mechanism that maintains their expression even during the recovery period.
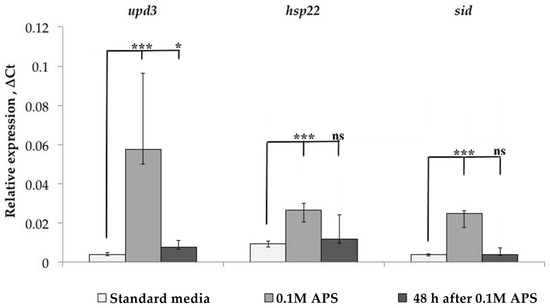
Figure 3.
Dynamics of changes in the expression of genes-markers of oxidative stress in the Canton-S strain under treatment with APS and during rest (* p < 0.5, *** p < 0.001, according to the Mann–Whitney test; ns, not significant—statistically insignificant change).
After that, we evaluated the functionality of the piRNA interference system, the main mechanism of TE control in Drosophila ovaries, under oxidative stress. At least 20 proteins are involved in piRNA interference system functioning and their number is constantly being refined upwards [6]. Therefore, we decided not to study the expression of individual genes related to the piRNA interference system, but to evaluate its work by the level of piRNA cluster expression, i.e., the ratio of spliced and unspliced forms of transcripts of these clusters. We measured the dynamics of flamenco, 42AB, 38C, and 20A cluster expression and evaluated the spliced and unspliced transcript form expression for cluster 42AB, but such an analysis turned out to be impossible due to the absence of flamenco transcript spliced forms in the Canton-S strain, as well as in the D32 strain. For clusters 20A and 38C, we analyzed only unspliced transcript forms (Figure 4).
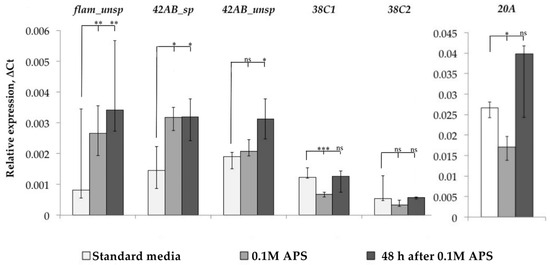
Figure 4.
Dynamics of piRNA cluster expression. (* p < 0.5, ** p < 0.01, *** p < 0.001, according to the Mann–Whitney test; ns, not significant—statistically insignificant change).
As a result, we found that flamenco and 42AB cluster expression was increased in response to oxidative stress, and we detected an increase in the amount of spliced forms in the case of the 42AB cluster, which can affect the functionality of the piRNA interference system since the mechanisms that prevent the splicing of this cluster transcripts are active under normal conditions because their introns contain sequences that are the source of piRNA. Analyzing the other two clusters, we did not detect an increase in 20A cluster expression, while 38C, on the contrary, reacted with a decrease in expression. Thus, the overall change in piRNA interference clusters in some cases (flamenco and 42AB) remained unchanged even 48 h after stress, and TE expression in response to stress may be associated with a misregulation of individual piRNA interference system components.
To induce chronic heat stress, we incubated flies at 29 °C. Since this stress is poorly studied, and marker genes for chronic heat stress are not known (in this situation, we do not consider heat shock), we estimated the heat stress presence by male sterility. Measuring LTR retrotransposon expression under chronic heat stress, we found that only Tirant significantly increased expression in the SS7K and Canton-S strains, while the expression of other TEs does not change under these conditions, and the expression of Tirant is maintained at a high level even after stress (Figure 5).
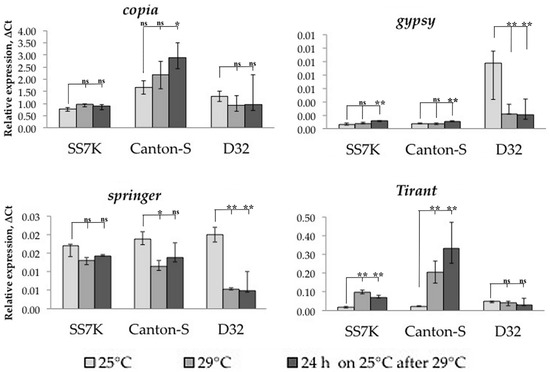
Figure 5.
Dynamics of LTR retrotransposon expression in seven-day-old adult females under chronic heat stress and rest (* p < 0.5, ** p < 0.01, according to the Mann–Whitney test; ns, not significant—statistically insignificant change).
Therefore, changes in TE expression under stress conditions are not always regular. Some TEs respond to only one stressor, while others are capable of responding to a range of extreme conditions with different effects.
3.2. Analysis of the Relationship between Transposable Element Copy Number and Their Position in the Genome with Their Transcription Activation
Obviously, it is unreasonable to compare the transcription of different TEs between strains because there are many factors that reduce the effectiveness of this approach. These factors will be discussed below. However, we can assume that the expression level of each individual TE may be related to the copy number in the genome: the more copies the higher the expression level. The expediency of TE expression normalization on its copy number is questioned. Since it is impossible to choose a universal primer that would anneal on all studied TE copies, we can only estimate the total expression of TEs that this primer is complementary to. In the same way, by isolating DNA from 20 flies of each strain, we estimated the number of copies whose expression was measured with RT-PCR (Table 2).

Table 2.
Number of TE copies in the SS7K, Canton-S, and D32 strains.
Comparing this data with the TEs’ expression histograms, we observe that their expression may depend on the number of copies, but this correlation is not linear. Therefore, normalization of TE expression on the number of its copies in the genome is not an effective approach.
Expression of TEs under normal and stress conditions can also be determined by the position of their copies in the genome of studied strains. Therefore, we analyzed the position of TE copies in the studied genomes, where they demonstrated a significant expression change in response to environmental conditions: Canton-S and SS7K. For position analysis, we first chose the TE copies that were absent in the reference genome, which are, therefore, new and, probably, functionally active. The next requirement to select copies was that they contain the sequences of the used primers. All insertions were in intergenic space, introns, or 3′-UTR of genes (Table 3).

Table 3.
Unique insertions of LTR retrotransposons gypsy, copia, Tirant, and springer in the SS7K and Canton-S.
Most of the unique TE insertions localized in genes were not involved in the stress response. Not one of the insertions in the genes induced under stress was collinear to the gene transcription direction. Therefore, in this case, the effect of the position should be minimal.
We also identified the position of the TE copies that were present in the reference genome and analyzed the presence of these copies in the Canton-S and SS7K genomes (Table 4). However, most of these regions in the sequenced genomes did not contain uniquely mapped reads or had poor coverage. But we also frequently observed the absence of TE sequences in reads, probably due to the fact that these insertions are unique in the reference genome and may not be present in our strains. Almost all of the TE copies presented in the reference genome were heterozygous in our strains.

Table 4.
Insertions of the LTR retrotransposons gypsy, copia, Tirant, and springer common to reference genome and SS7K and Canton-S strains.
After analyzing the insertions of studied LTR retrotransposons common to the reference genome and these two strains, we revealed the same trend as for unique insertions. TE copies were located in the intergenic space or in the introns of the genes, as a rule, and most of them were located in a chain complementary to the direction of gene transcription. Most of the genes that contained copies of the TEs, according to FlyBase, did not increase their expression in response to oxidative stress or heat shock; almost all of the insertions were heterozygous. We did not find any copy of the springer of gypsy in stress-activated genes. Therefore, in this case, we can assume that LTR retrotransposon expression is not determined by its insertion site. Moreover, normalizing expression to the number of copies is impractical, because each RNA sample contains the number of transcripts that correspond to a different number of original copies of TEs due to strain heterogeneity.
To get an idea of how the position of the TE might affect its expression, we took two different variants of the Tirant (according to the sequence of our laboratory strain SS7K), designed allele-specific primers for these copies, and compared the change in their expression in response to stress impact with changes in the expression of the genes in which these TE variants were inserted: Nuak and cactus. Since the change in the total expression of Tirant is higher under chronic heat stress, the chance to catch the change in the expression of individual copies under these conditions is much more probable than under oxidative stress, so we analyzed its expression only in the case of chronic heat stress (Figure 6).
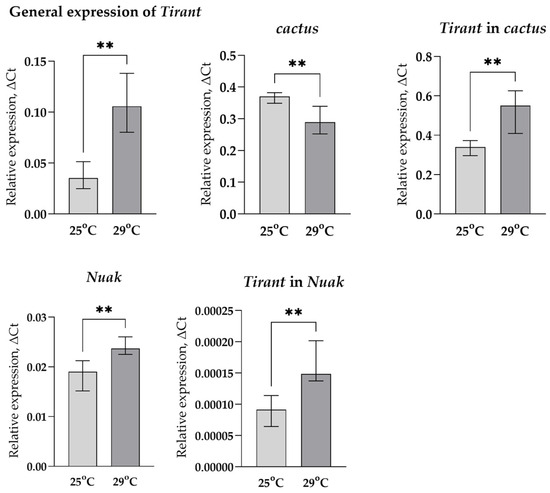
Figure 6.
Changes in the expression of LTR retrotransposon Tirant, cactus, and Nuak genes in response to chronic heat stress (** p < 0.01, according to the Mann–Whitney test).
We observed an increase in Nuak expression and a decrease in cactus expression under chronic heat stress, while the total expression of Tirant and both of its copies increased. This suggests that the Tirant probably has its own binding sites of transcription factors (TFs), which activate its transcription in response to stress.
3.3. Search for Transcription Factor Binding Sites in the Regulatory Regions of LTR Retrotransposons
To verify the presence of TF binding sites in the studied retrotransposons, we took their LTR and 5′-UTR sequences and analyzed them for the presence of potential binding sites using the LASAGNA resource. As a result, we obtained a list of potential TF binding sites, among which we were interested in stress-induced TFs (the same transcription factors were responsible for early embryonic development of Drosophila). After that, we performed multiple alignments of recent TE insertion sequences using Ugene and checked the conservatism of these binding sites (Table 5).

Table 5.
Transcription factor binding sites in new LTR retrotransposon copies.
Thus, all four TEs demonstrated conserved binding sites of TFs activated in response to oxidative and chronic heat stress. Although, for low-copy LTR retrotransposons gypsy and Tirant, this analysis is not clear. The presence of such sites, as well as the activation of individual alleles of Tirant in response to chronic heat stress, indicates that the regulation of retrotransposons under extreme conditions depends on the features of the regulatory sequences of their individual copies and does not always depend on the position in the genome. Moreover, impaired splicing of transcription of double-stranded clusters, such as 42AB, also may be a consequence of stress and be one of the reasons for LTR retrotransposon repression weakening.
4. Discussion
We observed the activation of individual LTR retrotransposons under oxidative stress caused by ammonium persulfate, as well as under chronic heat stress. Moreover, different TEs were activated to different degrees; however, the activation itself mostly did not depend on the initial state of the piRNA interference system. For example, in the wild-type strains, D32 and Canton-S, the expression of copia increased no less than in the strain with a mutation in the flamenco cluster, SS7K. We also registered an increase in Tirant expression regardless of the initial state of the piRNA interference system under chronic heat stress.
We could assume that TE transcription activation depended on changes that occur in the piRNA interference system during stress exposure. However, we have shown that individual clusters (uni-strand flamenco and dual-strand 42AB) also increased their expression, which might be a response to elevated TE activity under stress conditions. According to the modern concept, piRNAs can be cleaved from transcripts of repressed TEs; therefore, we considered clusters as piRNA sources in the genome (since it is impossible to validate the transcription of piRNA sources that are represented as satellite copies of retrotransposons) [36]. Also, using the 42AB cluster, we have shown that stress leads to a misbalance in the splicing mechanism and disruption of piRNA biogenesis. We cannot definitively conclude whether the imbalance between spliced and unspliced forms is associated with the disruption of the functioning of the RDC complex genes under conditions of oxidative stress. There are other factors that influence piRNA splicing. The heat stress protein, Hsp70, has previously been shown to play a role in piRNA biogenesis; during heat shock, this protein localizes to Nuage, where it binds to AGO3 and Aub. The release of this protein from the complex occurs in three days after stress is removed [37]. However, we observed that not all TEs that were activated in response to oxidative stress demonstrated the same activity under chronic heat stress. It is known that mutations of the main genes involved in piRNA interference (aub and ago3) do not lead to an increase of absolutely all TEs’ expression but only to several retrotransposons (gypsy, roo, copia, Tirant, and blood) [38]. Thus, the regulation of TEs controlled by the same piRNA interference system differs under a range of conditions. This may indicate the existence of alternative regulatory subsystems for different TEs or the repression of individual components of the piRNA interference system under stress conditions. Other researchers have shown a similar behavior of TEs under different types of stress: different chemical and physical agents cause an increase in different TE expressions, and the number of activated retrotransposons rises with stress intensification [19].
The second problem investigated in this work was the applicability of the normalization of TE expression on TE copy number. We have seen that the expression of LTR retrotransposons did not linearly correlate with the amount of TE copies. Moreover, laboratory strains that are not subjected to selection or gene drift for a long time become a polymorphic cause of the insertion of new TE copies; therefore, the amount of RNA was transcribed from an initially different number of TE copies in each sample, which also prevents the normalization of expression to the average number copies in the genome. Also, using the expression level of individual insertions of Tirant, we observed that the change in the total expression of this TE was more significant than the change in the expression of individual copies. Thus, taking into account the potential individual features of individual copies regulation, we can conclude that the normalization of any TE expression on the number of its insertions can result in misrepresentation.
We considered two hypotheses for the possible regulation of TE expression: the influence of the genomic environment and the presence of transcription factor binding sites in TE regulatory regions. After analyzing the positions of TE insertions in the SS7K and Canton-S genomes, we did not find insertions that could be affected by the genomic environment: almost all insertions were in intergenic space, and most genes that contained insertions in introns, according to the FlyBase, were not responsible for stress response. Next, we selected allele-specific primers for two copies of Tirant in the SS7K strain, since we implemented sequencing of this stain in our laboratory, and, therefore, we could detect these copies using PCR. After the induction of chronic heat stress (Tirant increased its expression higher in these stress conditions), we observed that the genes in which the insertions were located did not respond to stress. But the general expression of Tirant, as well as the expression of its individual copies, changed significantly. Thus, most TE insertions are capable of self-regulation rather than being co-expressed with the genes in which they are located.
To test the conservatism of TFs’ binding sites in LTRs and 5′UTRs, we performed a multiple alignment of all new insertions revealed in analyzed genomes and found that some sites predicted by the LASAGNA program were indeed conserved among the new TE insertions. Despite the fact that all these TFs are activated in response to stress, they have another role in explaining the maintenance of LTR retrotransposon expression. The role of these TFs is in the development of the organism, regulation of differentiation processes, and maintenance of stem cells. For example, HSF is necessary for early larval development and oogenesis, as well as the development of Drosophila [39]; cad is important for the determination of the anterior–posterior axis of the embryo, as well as maintaining stem cells in the gut [40]; slbo and Trlare are involved in cell migration during oogenesis [41]; and Lag-1 regulates differentiation of stem cells [42]. Thus, TE activation under stress does not occur directly in response to stress but under the influence of tissue regeneration processes that are triggered by this stress. This conclusion is also based on the fact that Hsp70, which, like TE, was expressed in the cell for three days after stress was removed, is triggered by HSF, which is the activator of heat shock proteins [37,43]. Cad demonstrates a pleiotropic effect and is important for stress response in adults: it prevents the production of antimicrobial peptides in the hindgut, which are induced under stress conditions by Relish. Relish is one of the participants in the IMD pathway and is bound by negative feedback with cad [44]. The activation of antimicrobial peptides themselves is associated with the IMD and Jak-STAT pathways [45]. Both pathways respond to stress under ammonium persulfate treatment [35].
The presence of TF binding sites explains why LTR retrotransposons are active in generative tissues during the development of the organism, respond to stress, and do not decrease their expression immediately after the removal of the stress inducer. The reason for the non-total increase in LTR retrotransposon expression in response to various poisons and oxidizing agents precisely underlies the specific action of different oxidants, which leads to the involvement of different molecular cascades in response and, consequently, several LTR retrotransposons.
We assume that the primary reason for the activation of TEs in response to stress is the presence of TF-binding sites in their regulatory regions. Due to the presence of these sites, the cell moderates TE activation during the regeneration of damaged tissues. But to prove it, it is necessary to conduct ChIP-seq experiments. The position of individual copies in the genome is a minor reason for TE activation. However, the TE activation process should also be beneficial for the host organism. This suggests that only those TEs that contain binding sites for specific TFs in their regulatory regions were selected during evolution. Apparently, the co-activation of TEs with stress response genes contributes to the overall mobilization of the stress response, which was previously shown by other researchers [24,25,46]. However, in this case, we observed that TE expression continues for the next 48 h following stress removal. In addition, all TFs, for which the binding sites are present in the studied TEs, respond not only to stress but also take an active part in early embryonic development; thus, it is possible that the process leading to an increase in TE expression is not so much the stress itself but the recovery process.
The TEs underwent extensive selection, during which their relationship with the host genome evolved from parasitic to mutually beneficial. Therefore, the L1 element in mammals is necessary for embryonic development and neurogenesis, as it regulates the expression of many genes [47]; other TEs participate in tissue-specific regulation of transcription [48]. In addition, TEs can play the role of ncRNA sources, which suppress the activity of certain genes, such as miR-431 [49,50]. This ncRNA is required for axon regeneration after injury. The effect of active axon growth after damage was also observed in the nematode with a mutation of the piwi gene, a core participant of the piRNA interference system, which suggests that this process is conservative [51]. Also, mosaicism caused by TE transpositions during the development of the nervous tissue is also a necessary condition for the normal functioning of the nervous system. Therefore, TEs studied in our case and in a number of other similar studies are TEs that have already passed through several stages of domestication and are controlled by the host genome.
5. Conclusions
Under stress, TE activation can lead to transposition, causing extensive genomic damage and resulting in cell death. On the other hand, TE activation can serve as a trigger mechanism, driving multiple processes simultaneously and providing a multivariate response of a living cell to negative factors. Thus, the activation of TEs during stress is not associated with the resistance of the cell to an oxidizing agent or any other stressor, but with regeneration processes that start at this moment and continue after stress. The regulation of TEs in response to stress and during development seems to be the same. This is a matter for further research.
Author Contributions
P.A.M.: investigation, writing—original draft preparation; I.V.K.: investigation; A.I.K.: conceptualization; L.N.N.: conceptualization, writing—review and editing, funding acquisition. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Russian Science Foundation, grant number 22-24-00305.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Additional information regarding the manuscript will be welcomed by the authors.
Acknowledgments
We thank Makhnovskii P.A. for the assistance in bioinformatics. We also thank Lavrenov A.R. for the design of primers for flamenco and Kuzmin I.V. for statistical consultations.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Casacuberta, E.; González, J. The Impact of Transposable Elements in Environmental Adaptation. Mol. Ecol. 2013, 22, 1503–1517. [Google Scholar] [CrossRef] [PubMed]
- Kidwell, M.G. Transposable Elements and the Evolution of Genome Size in Eukaryotes. Genetica 2002, 115, 49–63. [Google Scholar] [CrossRef] [PubMed]
- Park, E.G.; Ha, H.; Lee, D.H.; Kim, W.R.; Lee, Y.J.; Bae, W.H.; Kim, H.-S. Genomic Analyses of Non-Coding RNAs Overlapping Transposable Elements and Its Implication to Human Diseases. Int. J. Mol. Sci. 2022, 23, 8950. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, D.S.; Hutvágner, G.; Haley, B.; Zamore, P.D. Evidence That SiRNAs Function as Guides, Not Primers, in the Drosophila and Human RNAi Pathways. Mol. Cell 2002, 10, 537–548. [Google Scholar] [CrossRef] [PubMed]
- Guerreiro, M.P.G. What Makes Transposable Elements Move in the Drosophila Genome. Heredity 2012, 108, 461–468. [Google Scholar] [CrossRef]
- Théron, E.; Dennis, C.; Brasset, E.; Vaury, C. Distinct Features of the PiRNA Pathway in Somatic and Germ Cells: From PiRNA Cluster Transcription to PiRNA. Process. Amplif. 2014, 5, 28. [Google Scholar] [CrossRef] [PubMed]
- Yashiro, R.; Murota, Y.; Nishida, K.M.; Yamashiro, H.; Fujii, K.; Ogai, A.; Yamanaka, S.; Negishi, L.; Siomi, H.; Siomi, M.C. Piwi Nuclear Localization and Its Regulatory Mechanism in Drosophila Ovarian Somatic Cells. Cell Rep. 2018, 23, 3647–3657. [Google Scholar] [CrossRef]
- Yamanaka, S.; Siomi, M.C.; Siomi, H. PiRNA Clusters and Open Chromatin Structure. Mob. DNA 2014, 5, 22. [Google Scholar] [CrossRef]
- Schnabl, J.; Wang, J.; Hohmann, U.; Gehre, M.; Batki, J.; Andreev, V.I.; Purkhauser, K.; Fasching, N.; Duchek, P.; Novatchkova, M.; et al. Molecular Principles of Piwi-Mediated Cotranscriptional Silencing through the Dimeric SFiNX Complex. Genes Dev. 2021, 35, 392–409. [Google Scholar] [CrossRef]
- Onishi, R.; Sato, K.; Murano, K.; Negishi, L.; Siomi, H.; Siomi, M.C. Piwi Suppresses Transcription of Brahma-Dependent Transposons via Maelstrom in Ovarian Somatic Cells. Sci. Adv. 2020, 6, eaaz7420. [Google Scholar] [CrossRef]
- Mohn, F.; Sienski, G.; Handler, D.; Brennecke, J. The rhino-deadlock-cutoff complex licenses noncanonical transcription of dual-strand piRNA clusters in Drosophila. Cell 2014, 157, 1364–1379. [Google Scholar] [CrossRef] [PubMed]
- Aravin, A.A.; Hannon, G.J.; Brennecke, J. The Piwi-PiRNA Pathway Provides an Adaptive Defense in the Transposon Arms Race. Science 2007, 318, 761–764. [Google Scholar] [CrossRef] [PubMed]
- Brennecke, J.; Aravin, A.A.; Stark, A.; Dus, M.; Kellis, M.; Sachidanandam, R.; Hannon, G.J. Discrete Small RNA-Generating Loci as Master Regulators of Transposon Activity in Drosophila. Cell 2007, 128, 1089–1103. [Google Scholar] [CrossRef] [PubMed]
- Siomi, M.C.; Sato, K.; Pezic, D.; Aravin, A.A. PIWI-Interacting Small RNAs: The Vanguard of Genome Defence. Nat. Rev. Mol. Cell Biol. 2011, 12, 246–258. [Google Scholar] [CrossRef] [PubMed]
- Sato, K.; Siomi, M.C. Two Distinct Transcriptional Controls Triggered by Nuclear Piwi-PiRISCs in the Drosophila PiRNA Pathway. Curr. Opin. Struct. Biol. 2018, 53, 69–76. [Google Scholar] [CrossRef]
- Hur, J.K.; Luo, Y.; Moon, S.; Ninova, M.; Marinov, G.K.; Chung, Y.D.; Aravin, A.A. Splicing-Independent Loading of TREX on Nascent RNA Is Required for Efficient Expression of Dual-Strand PiRNA Clusters in Drosophila. Genes Dev. 2016, 30, 840–855. [Google Scholar] [CrossRef]
- Kneuss, E.; Munafò, M.; Eastwood, E.L.; Deumer, U.-S.; Preall, J.B.; Hannon, G.J.; Czech, B. Specialization of the Drosophila Nuclear Export Family Protein Nxf3 for PiRNA Precursor Export. Genes Dev. 2019, 33, 1208–1220. [Google Scholar] [CrossRef]
- Vieira, C.; Aubry, P.; Lepetit, D.; Bié Mont, C. A Temperature Cline in Copy Number for 412 but Not Roo/B104 Retrotransposons in Populations of Drosophila simulans. Proc. R. Soc. Lond. B 1998, 265, 1161–1165. [Google Scholar] [CrossRef]
- Oliveira, D.S.; Rosa, M.T.; Vieira, C.; Loreto, E.L.S. Oxidative and Radiation Stress Induces Transposable Element Transcription in Drosophila melanogaster. J. Evol. Biol. 2021, 34, 628–638. [Google Scholar] [CrossRef]
- McClintock, B. The Significance of Responses of the Genome to Challenge. Science 1984, 226, 792–801. [Google Scholar] [CrossRef]
- Finkel, T.; Holbrook, N.J. Oxidants, Oxidative Stress and the Biology of Ageing. Nature 2000, 408, 239–247. [Google Scholar] [CrossRef] [PubMed]
- Zou, S.; Meadows, S.; Sharp, L.; Jan, L.Y.; Jan, Y.N. Genome-Wide Study of Aging and Oxidative Stress Response in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 2000, 97, 13726–13731. [Google Scholar] [CrossRef] [PubMed]
- Tettweiler, G.; Miron, M.; Jenkins, M.; Sonenberg, N.; Lasko, P.F. Starvation and Oxidative Stress Resistance in Drosophila Are Mediated through the EIF4E-Binding Protein, D4E-BP. Genes Dev. 2005, 19, 1840–1843. [Google Scholar] [CrossRef] [PubMed]
- Villanueva-Cañas, J.L.; Horvath, V.; Aguilera, L.; González, J. Diverse Families of Transposable Elements Affect the Transcriptional Regulation of Stress-Response Genes in Drosophila melanogaster. Nucleic Acids Res. 2019, 47, 6842–6857. [Google Scholar] [CrossRef] [PubMed]
- Moschetti, R.; Palazzo, A.; Lorusso, P.; Viggiano, L.; Massimiliano Marsano, R. “What You Need, Baby, I Got It”: Transposable Elements as Suppliers of Cis-Operating Sequences in Drosophila. Biology 2020, 9, 25. [Google Scholar] [CrossRef] [PubMed]
- Kim, A.I.; Belyaeva, E.S.; Larkina, Z.G.; Aslanyan, M.M. Genetic Instability and Transposition of the Mobile Element MDG4 in the Drosophila melanogaster Mutator Line. Sov. Genet. 1989, 25, 1747–1756. [Google Scholar]
- Zhang, Z.; Wang, J.; Schultz, N.; Zhang, F.; Parhad, S.S.; Tu, S.; Vreven, T.; Zamore, P.D.; Weng, Z.; Theurkauf, W.E. The HP1 Homolog Rhino Anchors a Nuclear Complex That Suppresses PiRNA Precursor Splicing. Cell 2014, 157, 1353–1363. [Google Scholar] [CrossRef]
- Nefedova, L.N.; Urusov, F.A.; Romanova, N.I.; Shmel’kova, A.O.; Kim, A.I. Study of the Transcriptional and Transpositional Activities of the Tirant Retrotransposon in Drosophila melanogaster Strains Mutant for the flamenco Locus. Russ. J. Genet. 2012, 48, 1089–1096. [Google Scholar] [CrossRef]
- Robinson, J.T.; Thorvaldsdóttir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative Genomics Viewer. Nat. Biotechnol. 2011, 29, 24–26. [Google Scholar] [CrossRef]
- Ewing, A.D.; Smits, N.; Sanchez-Luque, F.J.; Faivre, J.; Brennan, P.M.; Richardson, S.R.; Cheetham, S.W.; Faulkner, G.J. Nanopore Sequencing Enables Comprehensive Transposable Element Epigenomic Profiling. Mol. Cell 2020, 80, 915–928.e5. [Google Scholar] [CrossRef]
- Sayers, E.W.; Bolton, E.E.; Brister, J.R.; Canese, K.; Chan, J.; Comeau, D.C.; Connor, R.; Funk, K.; Kelly, C.; Kim, S.; et al. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2022, 50, D20–D26. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.; Huang, C.-H. LASAGNA-Search: An integrated web tool for transcription factor binding site search and visualization. BioTechniques 2013, 54, 141–153. [Google Scholar] [CrossRef] [PubMed]
- Gramates, L.S.; Agapite, J.; Attrill, H.; Calvi, B.R.; Crosby, M.A.; dos Santos, G.; Goodman, J.L.; Goutte-Gattat, D.; Jenkins, V.K.; Kaufman, T.; et al. FlyBase: A guided tour of highlighted features. Genetics 2022, 220, iyac035. [Google Scholar] [CrossRef] [PubMed]
- Okonechnikov, K.; Golosova, O.; Fursov, M. Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef] [PubMed]
- Makhnovskii, P.; Balakireva, Y.; Nefedova, L.; Lavrenov, A.; Kuzmin, I.; Kim, A. Domesticated Gag Gene of Drosophila LTR Retrotransposons Is Involved in Response to Oxidative Stress. Genes 2020, 11, 396. [Google Scholar] [CrossRef] [PubMed]
- Cui, M.; Bai, Y.; Li, K.; Rong, Y.S. Taming Active Transposons at Drosophila Telomeres: The Interconnection between HipHop’s Roles in Capping and Transcriptional Silencing. PLoS Genet. 2021, 17, e1009925. [Google Scholar] [CrossRef] [PubMed]
- Cappucci, U.; Noro, F.; Casale, A.M.; Fanti, L.; Berloco, M.; Alagia, A.A.; Grassi, L.; Le Pera, L.; Piacentini, L.; Pimpinelli, S. The Hsp70 Chaperone Is a Major Player in Stress-Induced Transposable Element Activation. Proc. Natl. Acad. Sci. USA 2019, 116, 17943–17950. [Google Scholar] [CrossRef] [PubMed]
- Perrat, P.N.; DasGupta, S.; Wang, J.; Theurkauf, W.; Weng, Z.; Rosbash, M.; Waddell, S. Transposition-Driven Genomic Heterogeneity in the Drosophila. Brain Sci. 2013, 340, 91–95. [Google Scholar] [CrossRef]
- Jedlicka, P.; Mortin, M.A.; Wu, C. Multiple Functions of Drosophila Heat Shock Transcription Factor In Vivo. EMBO J. 1997, 16, 2452–2462. [Google Scholar] [CrossRef]
- Wu, K.; Tang, Y.; Zhang, Q.; Zhuo, Z.; Sheng, X.; Huang, J.; Ye, J.; Li, X.; Liu, Z.; Chen, H. Aging-Related Upregulation of the Homeobox Gene Caudal Represses Intestinal Stem Cell Differentiation in Drosophila. PLoS Genet. 2021, 17, e1009649. [Google Scholar] [CrossRef]
- Ogienko, A.A.; Yarinich, L.A.; Fedorova, E.V.; Dorogova, N.V.; Bayborodin, S.I.; Baricheva, E.M.; Pindyurin, A.V. GAGA Regulates Border Cell Migration in Drosophila. Int. J. Mol. Sci. 2020, 21, 7468. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Mohammad, A.; Pazdernik, N.; Huang, H.; Bowman, B.; Tycksen, E.; Schedl, T. GLP-1 Notch—LAG-1 CSL Control of the Germline Stem Cell Fate Is Mediated by Transcriptional Targets Lst-1 and Sygl-1. PLoS Genet. 2020, 16, e1008650. [Google Scholar] [CrossRef] [PubMed]
- Van Oosten-Hawle, P. Organismal Roles of Hsp90. Biomolecules 2023, 13, 251. [Google Scholar] [CrossRef] [PubMed]
- Stączek, S.; Cytryńska, M.; Zdybicka-Barabas, A. Unraveling the Role of Antimicrobial Peptides in Insects. Int. J. Mol. Sci. 2023, 24, 5753. [Google Scholar] [CrossRef] [PubMed]
- Loch, G.; Zinke, I.; Mori, T.; Carrera, P.; Schroer, J.; Takeyama, H.; Hoch, M. Antimicrobial Peptides Extend Lifespan in Drosophila. PLoS ONE 2017, 12, e0176689. [Google Scholar] [CrossRef] [PubMed]
- Salces-Ortiz, J.; Vargas-Chavez, C.; Guio, L.; Rech, G.E.; González, J. Transposable Elements Contribute to the Genomic Response to Insecticides in Drosophila melanogaster. Philos. Trans. R. Soc. B 2020, 375, 20190341. [Google Scholar] [CrossRef] [PubMed]
- Jachowicz, J.W.; Bing, X.; Pontabry, J.; Bošković, A.; Rando, O.J.; Torres-Padilla, M.-E. LINE-1 Activation after Fertilization Regulates Global Chromatin Accessibility in the Early Mouse Embryo. Nat. Genet. 2017, 49, 1502–1510. [Google Scholar] [CrossRef]
- Trizzino, M.; Kapusta, A.; Brown, C.D. Transposable Elements Generate Regulatory Novelty in a Tissue-Specific Fashion. BMC Genom. 2018, 19, 468. [Google Scholar] [CrossRef]
- Nampoothiri, S.S.; Rajanikant, G.K. Decoding the Ubiquitous Role of MicroRNAs in Neurogenesis. Mol. Neurobiol. 2017, 54, 2003–2011. [Google Scholar] [CrossRef]
- Mustafin, R.N.; Khusnutdinova, E.K. Involvement of Transposable Elements in Neurogenesis. Vestn. VOGiS 2020, 24, 209–218. [Google Scholar] [CrossRef]
- Kim, K.W. PIWI Proteins and PiRNAs in the Nervous System. Mol. Cells 2019, 42, 828–835. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).