Epigenetics Role in Spermatozoa Function: Implications in Health and Evolution—An Overview
Abstract
1. Introduction
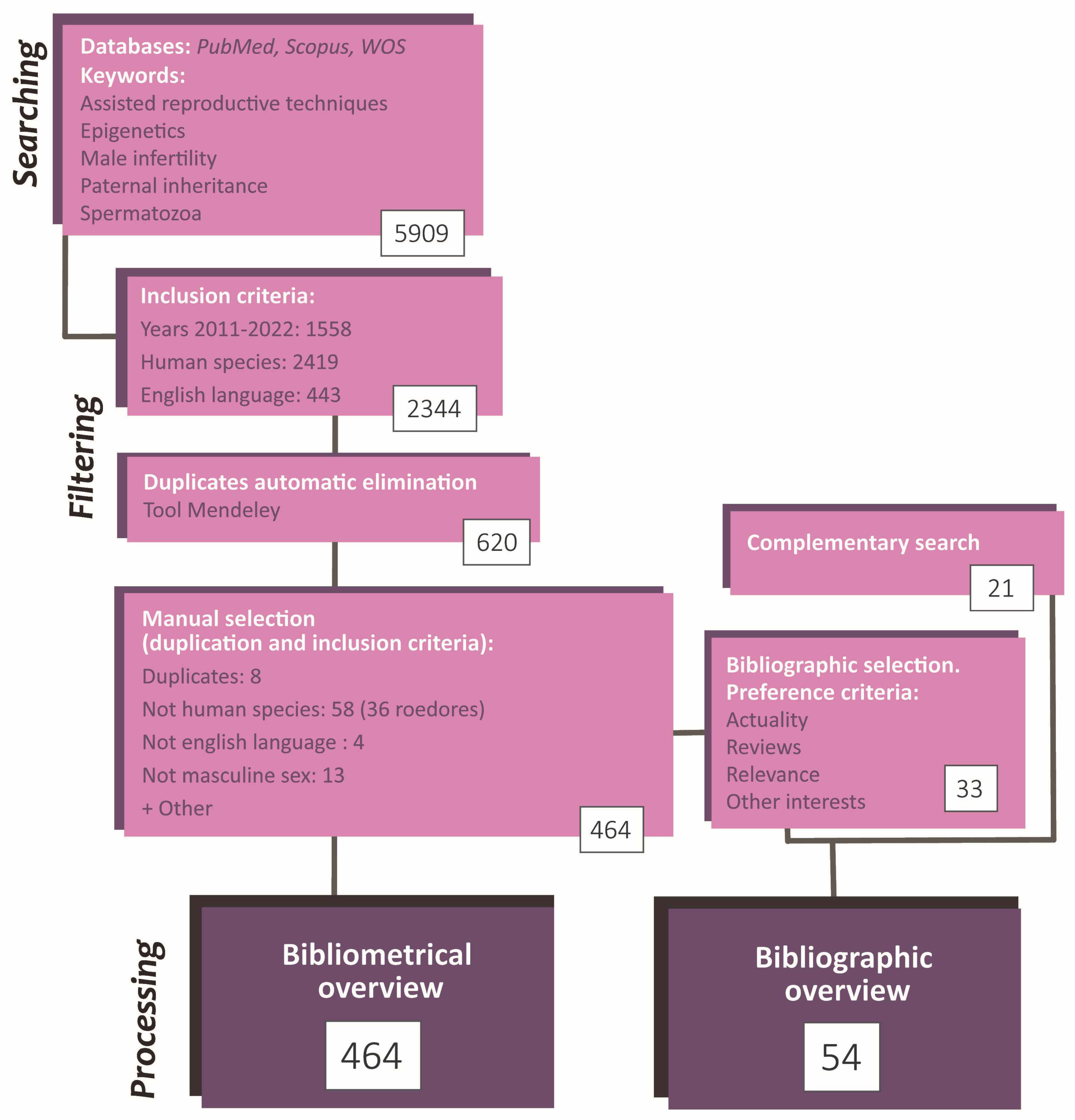
2. Materials and Methods
3. Results
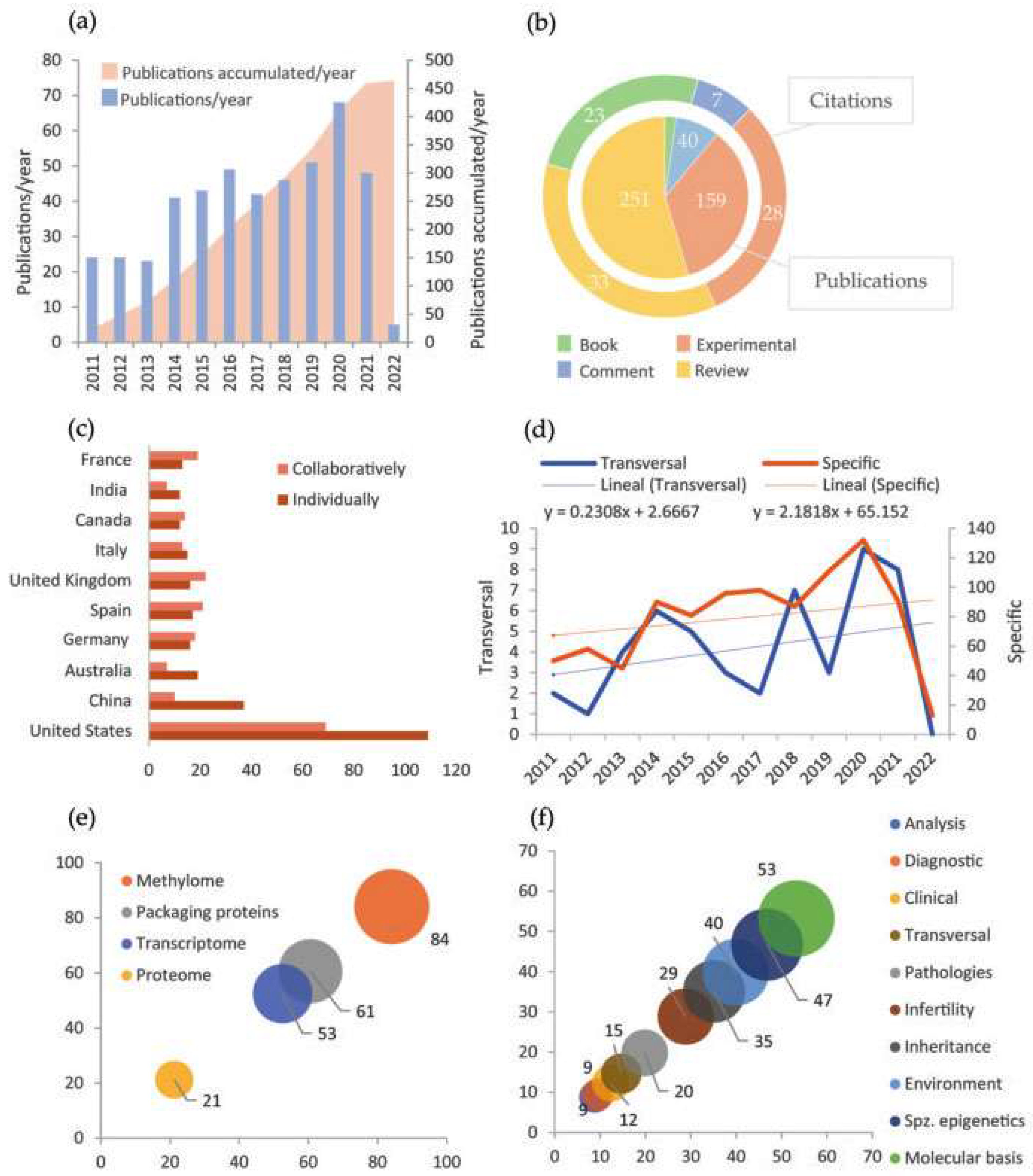
3.1. Bibliometric Results
3.2. Review Results
3.2.1. Spermatozoa Epigenetics
DNA Methylation
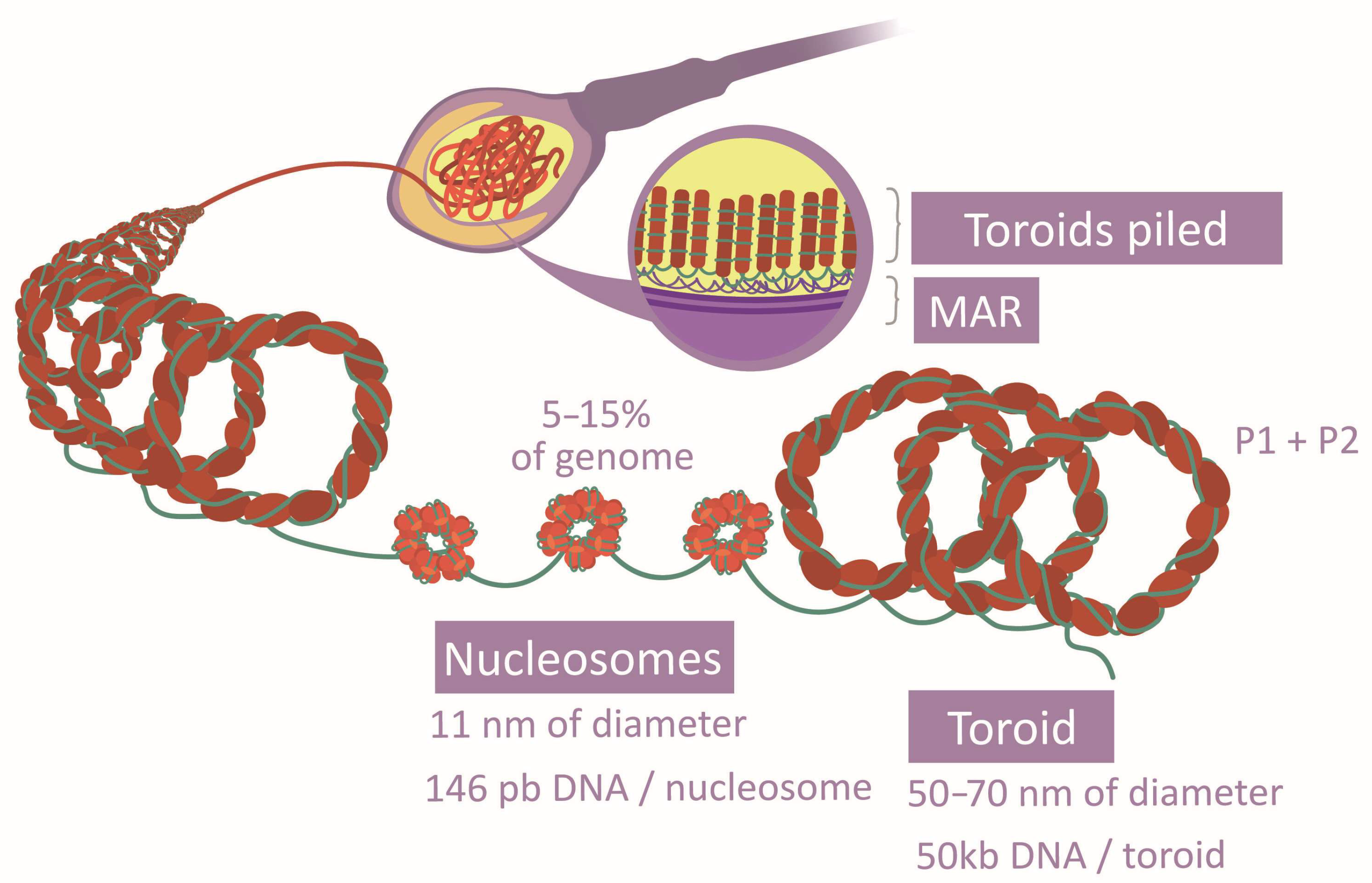
Packaging Proteins
Transcriptome
Proteome
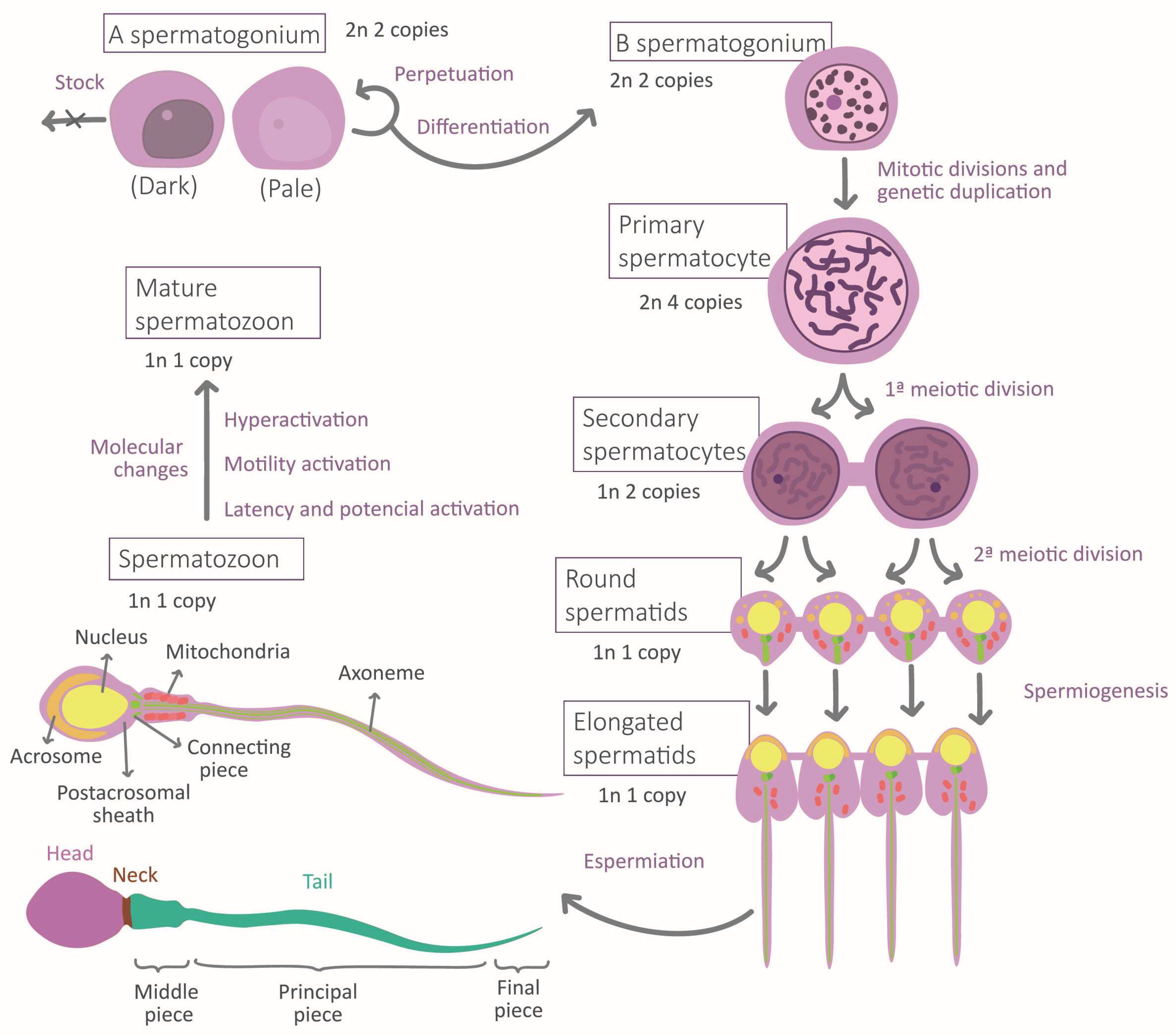
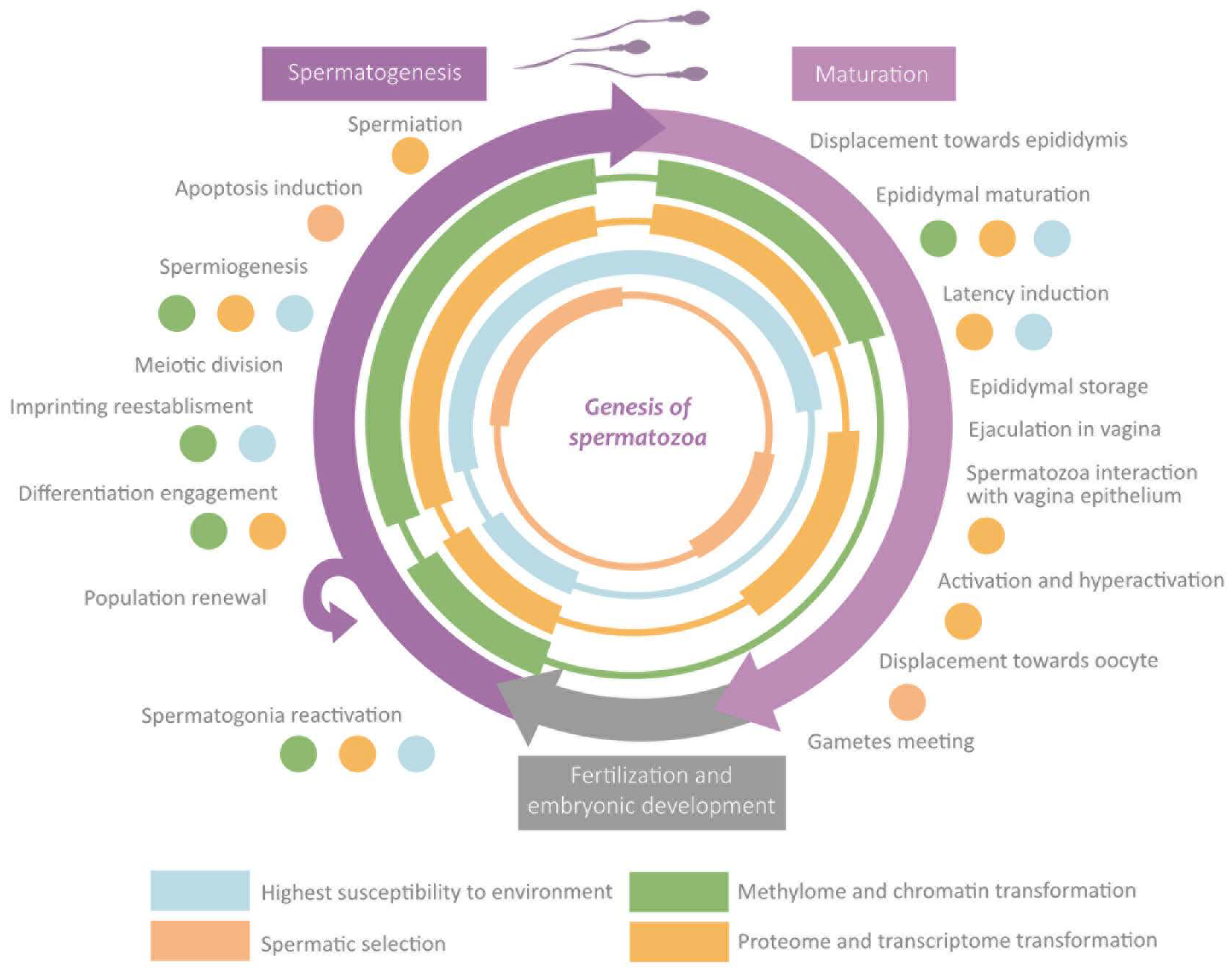
3.2.2. Epigenetics during Spermatozoa Genesis
3.2.3. Spermatogenesis
Germ Cells during Embryonic Development
Initiation of Spermatogenesis
Chromatin Reorganization
Methylome Reestablishment
Proteome and Transcriptome Transformation
3.2.4. Maturation
Epidydimal Transit
Transit through the Female Reproductive System
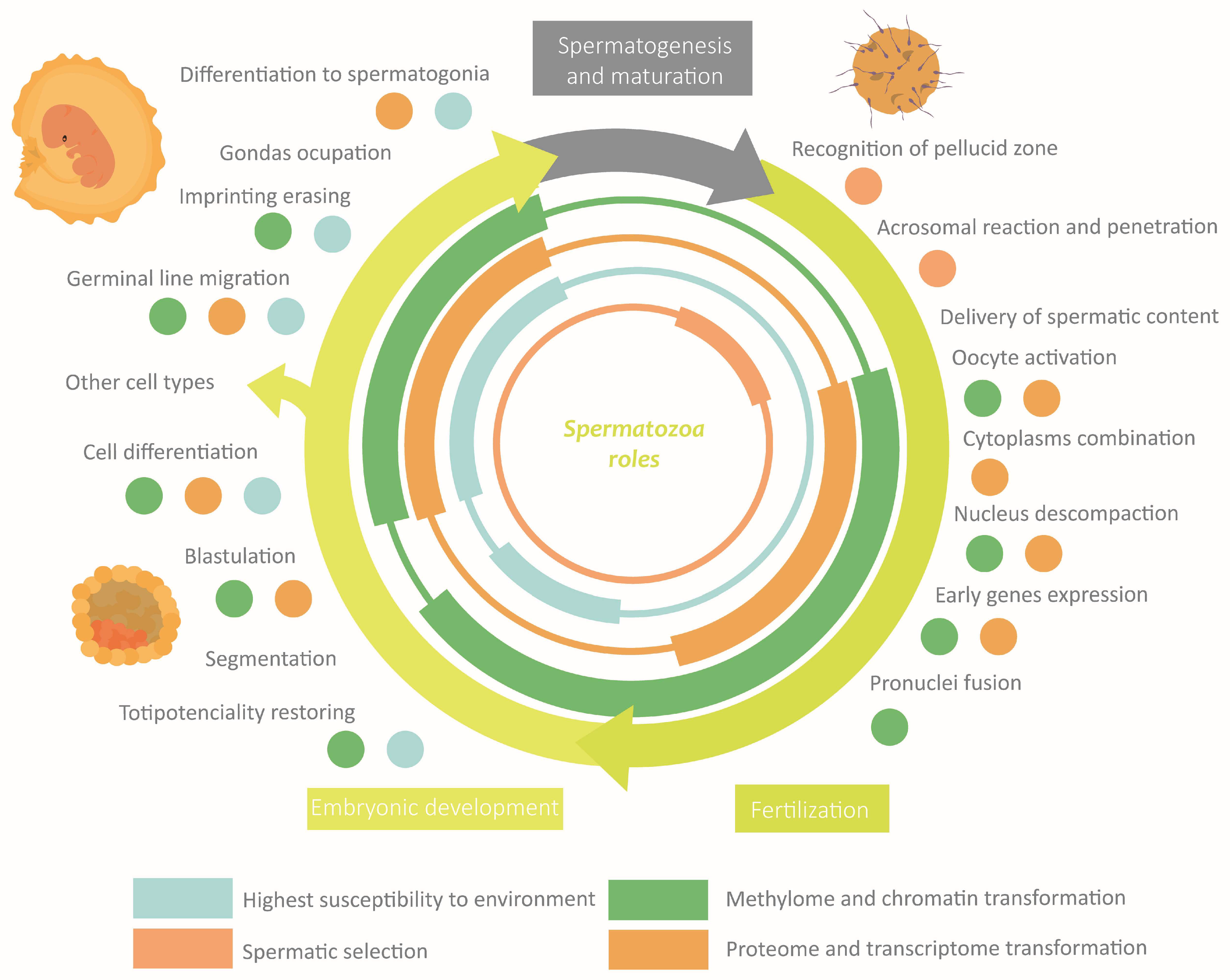
3.2.5. Epigenetics in Spermatozoa Functions
Fertilization
Embryonic Development
3.2.6. Spermatozoa Epigenetics in Evolution
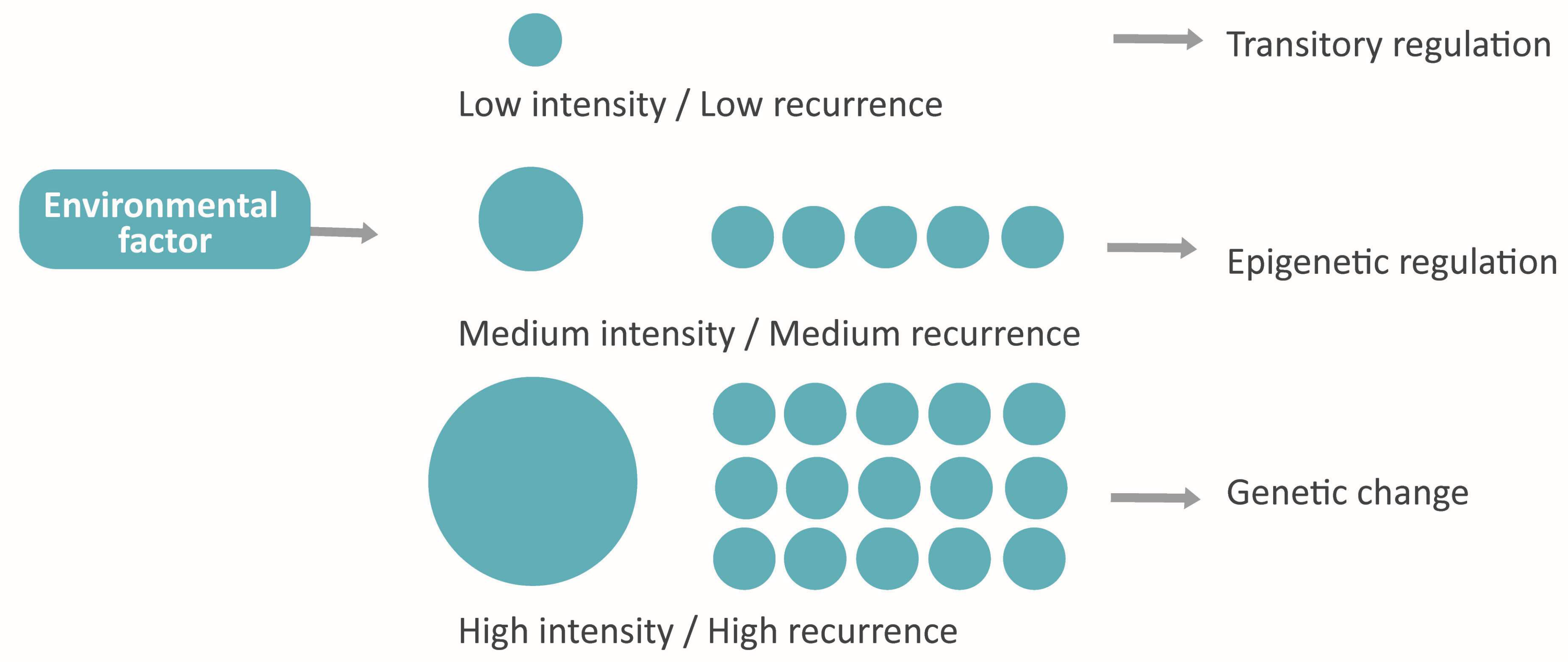
Environmental Factors
Epigenetic Inheritance
Accelerated Evolution
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Allis, C.D.; Caparros, M.-L.; Jenuwein, T.; Reinberg, D. Epigenetics, 6th ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2007. [Google Scholar]
- Gonzalès, J. Histoire Du Spermatozoïde et Mobilité Des Idées. Gynécologie Obs. Fertil. 2006, 34, 819–826. [Google Scholar] [CrossRef]
- Peixoto, P.; Cartron, P.-F.; Serandour, A.A.; Hervouet, E. From 1957 to Nowadays: A Brief History of Epigenetics. Int. J. Mol. Sci. 2020, 21, 7571. [Google Scholar] [CrossRef] [PubMed]
- Cordonnier, M.-N. La epigenética, moduladora clave de la evolución. Investig. Cienc. 2020, 520, 70–75. [Google Scholar]
- Neto, F.T.L.; Bach, P.V.; Najari, B.B.; Li, P.S.; Goldstein, M. Spermatogenesis in Humans and Its Affecting Factors. Semin. Cell Dev. Biol. 2016, 59, 10–26. [Google Scholar] [CrossRef] [PubMed]
- Gui, Y.; Yuan, S. Epigenetic Regulations in Mammalian Spermatogenesis: RNA-m6A Modification and Beyond. Cell. Mol. Life Sci. 2021, 78, 4893–4905. [Google Scholar] [CrossRef] [PubMed]
- Ross, M.H.; Pawlina, W. Histología: Texto y Atlas. Correlación con Biología Celular y Molecular, 7th ed.; Wolters Kluwer: Barcelona, Spain, 2016; pp. 849–895. [Google Scholar]
- Bashiri, Z.; Amidi, F.; Amiri, I.; Zandieh, Z.; Maki, C.B.; Mohammadi, F.; Amiri, S.; Koruji, M. Male Factors: The Role of Sperm in Preimplantation Embryo Quality. Reprod. Sci. 2021, 28, 1788–1811. [Google Scholar] [CrossRef] [PubMed]
- Ozkocer, S.E.; Konac, E. The Current Perspective on Genetic and Epigenetic Factors in Sperm Maturation in the Epididymis. Andrologia 2021, 53, e13989. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Yan, W.; Duan, E. Epigenetic Inheritance of Acquired Traits through Sperm RNAs and Sperm RNA Modifications. Nat. Rev. Genet. 2016, 17, 733–743. [Google Scholar] [CrossRef]
- Hamdi, S.M.; Vieitez, G.; Jaspard, B.; Barbaras, R.; Perret, B.; Mieusset, R.; Parinaud, J.; Collet, X. Effects of Human Follicular Fluid and High-Density Lipoproteins on Early Spermatozoa Hyperactivation and Cholesterol Efflux. J. Lipid Res. 2010, 51, 1363–1369. [Google Scholar] [CrossRef]
- Hernández Alvarado, S.R.; Guzmán-Grenfell, A.M.; Hicks Gómez, J.J. Communication of Gametes at Distance. Chemotaxis and Chemokinesis in Spermatozoa in Mammals. Ginecol. Obstet. Mex. 1995, 63, 323–327. [Google Scholar]
- Tourmente, M.; Varea-Sánchez, M.; Roldan, E.R.S. Faster and More Efficient Swimming: Energy Consumption of Murine Spermatozoa under Sperm Competition. Biol. Reprod. 2019, 100, 420–428. [Google Scholar] [CrossRef] [PubMed]
- Rickard, J.P.; Pool, K.R.; Druart, X.; de Graaf, S.P. The Fate of Spermatozoa in the Female Reproductive Tract: A Comparative Review. Theriogenology 2019, 137, 104–112. [Google Scholar] [CrossRef] [PubMed]
- Bastiaan, H.; Franken, D. The Influence of Homogenous Zona Pellucida on Human Spermatozoa Hyperactivation, Acrosome Reaction and Zona Binding. Andrologia 2007, 39, 7–11. [Google Scholar] [CrossRef] [PubMed]
- Schagdarsurengin, U.; Steger, K. Epigenetics in Male Reproduction: Effect of Paternal Diet on Sperm Quality and Offspring Health. Nat. Rev. Urol. 2016, 13, 584–595. [Google Scholar] [CrossRef]
- Cescon, M.; Chianese, R.; Tavares, R.S. Environmental Impact on Male (in)Fertility via Epigenetic Route. J. Clin. Med. 2020, 9, 2520. [Google Scholar] [CrossRef]
- Chioccarelli, T.; Pierantoni, R.; Manfrevola, F.; Porreca, V.; Fasano, S.; Chianese, R.; Cobellis, G. Histone Post-Translational Modifications and Circrnas in Mouse and Human Spermatozoa: Potential Epigenetic Marks to Assess Human Sperm Quality. J. Clin. Med. 2020, 9, 640. [Google Scholar] [CrossRef]
- Protopapas, N.; Hamilton, L.E.; Warkentin, R.; Xu, W.; Sutovsky, P.; Oko, R. The Perforatorium and Postacrosomal Sheath of Rat Spermatozoa Share Common Developmental Origins and Protein Constituents. Biol. Reprod. 2019, 100, 1461–1472. [Google Scholar] [CrossRef]
- Nicoglou, A.; Merlin, F. Epigenetics: A Way to Bridge the Gap between Biological Fields. Stud. Hist. Philos. Sci. Part C Stud. Hist. Philos. Biol. Biomed. Sci. 2017, 66, 73–82. [Google Scholar] [CrossRef]
- Patel, D.P.; Jenkins, T.G.; Aston, K.I.; Guo, J.; Pastuszak, A.W.; Hanson, H.A.; Hotaling, J.M. Harnessing the Full Potential of Reproductive Genetics and Epigenetics for Male Infertility in the Era of “Big Data”. Fertil. Steril. 2020, 113, 478–488. [Google Scholar] [CrossRef]
- Gunes, S.; Esteves, S.C. Role of Genetics and Epigenetics in Male Infertility. Andrologia 2021, 53, e13586. [Google Scholar] [CrossRef]
- Escher, J. How Family Histories Can Inform Research about Germ Cell Exposures: The Example of Autism. Biol. Reprod. 2021, 105, 767–773. [Google Scholar] [CrossRef] [PubMed]
- Finelli, R.; Mottola, F.; Agarwal, A. Impact of Alcohol Consumption on Male Fertility Potential: A Narrative Review. Int. J. Environ. Res. Public Health 2022, 19, 328. [Google Scholar] [CrossRef] [PubMed]
- Milliken-Smith, S.; Potter, C.M. Paternal Origins of Obesity: Emerging Evidence for Incorporating Epigenetic Pathways into the Social Determinants of Health Framework. Soc. Sci. Med. 2021, 271, 112066. [Google Scholar] [CrossRef] [PubMed]
- Laugesen, A.; Højfeldt, J.W.; Helin, K. Molecular Mechanisms Directing PRC2 Recruitment and H3K27 Methylation. Mol. Cell. 2019, 74, 8–18. [Google Scholar] [CrossRef] [PubMed]
- Soubry, A.; Hoyo, C.; Jirtle, R.L.; Murphy, S.K. A Paternal Environmental Legacy: Evidence for Epigenetic Inheritance through the Male Germ Line. BioEssays 2014, 36, 359–371. [Google Scholar] [CrossRef]
- Jodar, M.; Selvaraju, S.; Sendler, E.; Diamond, M.P.; Krawetz, S.A. The Presence, Role and Clinical Use of Spermatozoal RNAs. Hum. Reprod. Update 2013, 19, 604–624. [Google Scholar] [CrossRef]
- Marzouni, E.T.; Ilkhani, H.; Harchegani, A.B.; Shafaghatian, H.; Layali, I.; Shahriary, A. Epigenetic Modifications, A New Approach to Male Infertility Etiology: A Review. Int. J. Fertil. Steril. 2022, 16, 1–9. [Google Scholar] [CrossRef]
- Bodden, C.; Hannan, A.J.; Reichelt, A.C. Diet-Induced Modification of the Sperm Epigenome Programs Metabolism and Behavior. Trends Endocrinol. Metab. 2020, 31, 131–149. [Google Scholar] [CrossRef]
- Cecere, G. Small RNAs in Epigenetic Inheritance: From Mechanisms to Trait Transmission. FEBS Lett. 2021, 95, 2953–2977. [Google Scholar] [CrossRef]
- Craig, J.R.; Jenkins, T.G.; Carrell, D.T.; Hotaling, J.M. Obesity, Male Infertility, and the Sperm Epigenome. Fertil. Steril. 2017, 107, 848–859. [Google Scholar] [CrossRef]
- Donkin, I.; Barrès, R.; Barres, R.; Barrès, R. Sperm Epigenetics and Influence of Environmental Factors. Mol. Metab. 2018, 14, 1–11. [Google Scholar] [CrossRef]
- Agarwal, A.; Mulgund, A.; Hamada, A.; Chyatte, M.R. A unique view on male infertility around the globe. Reprod. Biol. Endocrinol. 2015, 13, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Levine, H.; Jørgensen, N.; Martino-Andrade, A.; Mendiola, J.; Weksler-Derri, D.; Jolles, M.; Pinotti, R.; Swan, S.H. Temporal trends in sperm count: A systematic review and meta-regression analysis of samples collected globally in the 20th and 21st centuries. Hum. Reprod. Update 2022, dmac035. [Google Scholar] [CrossRef] [PubMed]
- Velez, D.; Hwang, K. Personalized Medicine for the Infertile Male. Urol. Clin. N. Am. 2020, 47, 523–536. [Google Scholar] [CrossRef] [PubMed]
- Brotons, B.; Fernández-Peinado, A.A.; Moya, A.; Ibañez, P.; Vidal-Iglesias, F.J.; Solla-Gullón, J.; Iniesta, J. Epigenética en técnicas de reproducción asistida: Razones y evidencias para una reflexión. Revista Asebir. 2014, 19, 38–46. [Google Scholar]
- Salas-Huetos, A.; Aston, K.I. Defining New Genetic Etiologies of Male Infertility: Progress and Future Prospects. Transl. Androl. Urol. 2021, 10, 1486–1498. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Feng, S.; Qin, W.; Wang, X.; Tang, Y.; Yuan, S. Epigenetic Regulation of Spermatogonial Stem Cell Homeostasis: From DNA Methylation to Histone Modification. Stem Cell Rev. Rep. 2021, 17, 562–580. [Google Scholar] [CrossRef]
- Asenius, F.; Danson, A.F.; Marzi, S.J.; Åsenius, F.; Danson, A.F.A.F.; Marzi, S.J.S.J. DNA Methylation in Human Sperm: A Systematic Review. Hum. Reprod. Update 2020, 26, 841–873. [Google Scholar] [CrossRef]
- Balhorn, R. The Protamine Family of Sperm Nuclear Proteins. Genome Biol. 2007, 8, 227. [Google Scholar] [CrossRef]
- Carrell, D.T. Epigenetics of the Male Gamete. Fertil. Steril. 2012, 97, 267–274. [Google Scholar] [CrossRef]
- Cannarella, R.; Crafa, A.; Condorelli, R.A.; Mongioì, L.M.; La Vignera, S.; Calogero, A.E. Relevance of Sperm Imprinted Gene Methylation on Assisted Reproductive Technique Outcomes and Pregnancy Loss: A Systematic Review. Syst. Biol. Reprod. Med. 2021, 67, 251–259. [Google Scholar] [CrossRef] [PubMed]
- Thanassoulas, A.; Swann, K.; Lai, F.A.; Nomikos, M. SPERM FACTORS AND EGG ACTIVATION: The Structure and Function Relationship of Sperm PLCZ1. Reproduction 2022, 164, F1–F8. [Google Scholar] [CrossRef]
- Eshghifar, N.; Dehghan, B.K.; Do, A.A.; Koukhaloo, S.Z.; Habibi, M.; Pouresmaeili, F. Infertility Cell Therapy and Epigenetic Insights. Hum. Antibodies 2021, 29, 17–26. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Pavón, E.; Mendoza, H.; García-Ferreyra, J. Trisomy 21 and Assisted Reproductive Technologies: A Review. JBRA Assist. Reprod. 2022, 26, 129–141. [Google Scholar] [CrossRef]
- Iwao, Y. Mechanisms of Egg Activation and Polyspermy Block in Amphibians and Comparative Aspects with Fertilization in Other Vertebrates. Zoolog. Sci. 2000, 17, 699–709. [Google Scholar] [CrossRef]
- Jablonski, K.P.; Carron, L.; Mozziconacci, J.; Forné, T.; Hütt, M.-T.; Lesne, A. Contribution of 3D Genome Topological Domains to Genetic Risk of Cancers: A Genome-Wide Computational Study. Hum. Genom. 2020, 16, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Khambata, K.; Raut, S.; Deshpande, S.; Mohan, S.; Sonawane, S.; Gaonkar, R.; Ansari, Z.; Datar, M.; Bansal, V.; Patil, A.; et al. DNA Methylation Defects in Spermatozoa of Male Partners from Couples Experiencing Recurrent Pregnancy Loss. Hum. Reprod. 2021, 36, 48–60. [Google Scholar] [CrossRef]
- Silver, M.J. Intergenerational Influences on Child Development: An Epigenetic Perspective. Nestle Nutr. Inst. Workshop Ser. 2020, 93, 145–152. [Google Scholar] [CrossRef]
- Selvaraju, V.; Baskaran, S.; Agarwal, A.; Henkel, R. Environmental Contaminants and Male Infertility: Effects and Mechanisms. Andrologia 2021, 53, e13646. [Google Scholar] [CrossRef]
- Itoh, M. Testicular Autoimmunity: A Cause of Male Infertility, 1st ed.; Springer: Tokyo, Japan, 2017; pp. 1–232. [Google Scholar] [CrossRef]
- Moshrefi, M.; Ghasemi-Esmailabad, S.; Ali, J.; Findikli, N.; Mangoli, E.; Khalili, M.A. The Probable Destructive Mechanisms behind COVID-19 on Male Reproduction System and Fertility. J. Assist. Reprod. Genet. 2021, 38, 1691–1708. [Google Scholar] [CrossRef]
- Portera, M.; Mandrioli, M.; Portera, M.; Mandrioli, M. Who’s Afraid of Epigenetics? Habits, Instincts, and Charles Darwin’s Evolutionary Theory. Hist. Philos. Life Sci. 2021, 43, 20. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Andreu-Noguera, J.; López-Botella, A.; Sáez-Espinosa, P.; Gómez-Torres, M.J. Epigenetics Role in Spermatozoa Function: Implications in Health and Evolution—An Overview. Life 2023, 13, 364. https://doi.org/10.3390/life13020364
Andreu-Noguera J, López-Botella A, Sáez-Espinosa P, Gómez-Torres MJ. Epigenetics Role in Spermatozoa Function: Implications in Health and Evolution—An Overview. Life. 2023; 13(2):364. https://doi.org/10.3390/life13020364
Chicago/Turabian StyleAndreu-Noguera, Julia, Andrea López-Botella, Paula Sáez-Espinosa, and María José Gómez-Torres. 2023. "Epigenetics Role in Spermatozoa Function: Implications in Health and Evolution—An Overview" Life 13, no. 2: 364. https://doi.org/10.3390/life13020364
APA StyleAndreu-Noguera, J., López-Botella, A., Sáez-Espinosa, P., & Gómez-Torres, M. J. (2023). Epigenetics Role in Spermatozoa Function: Implications in Health and Evolution—An Overview. Life, 13(2), 364. https://doi.org/10.3390/life13020364




