Diversity of Antimicrobial Peptides in Silkworm
Abstract
:1. Introduction
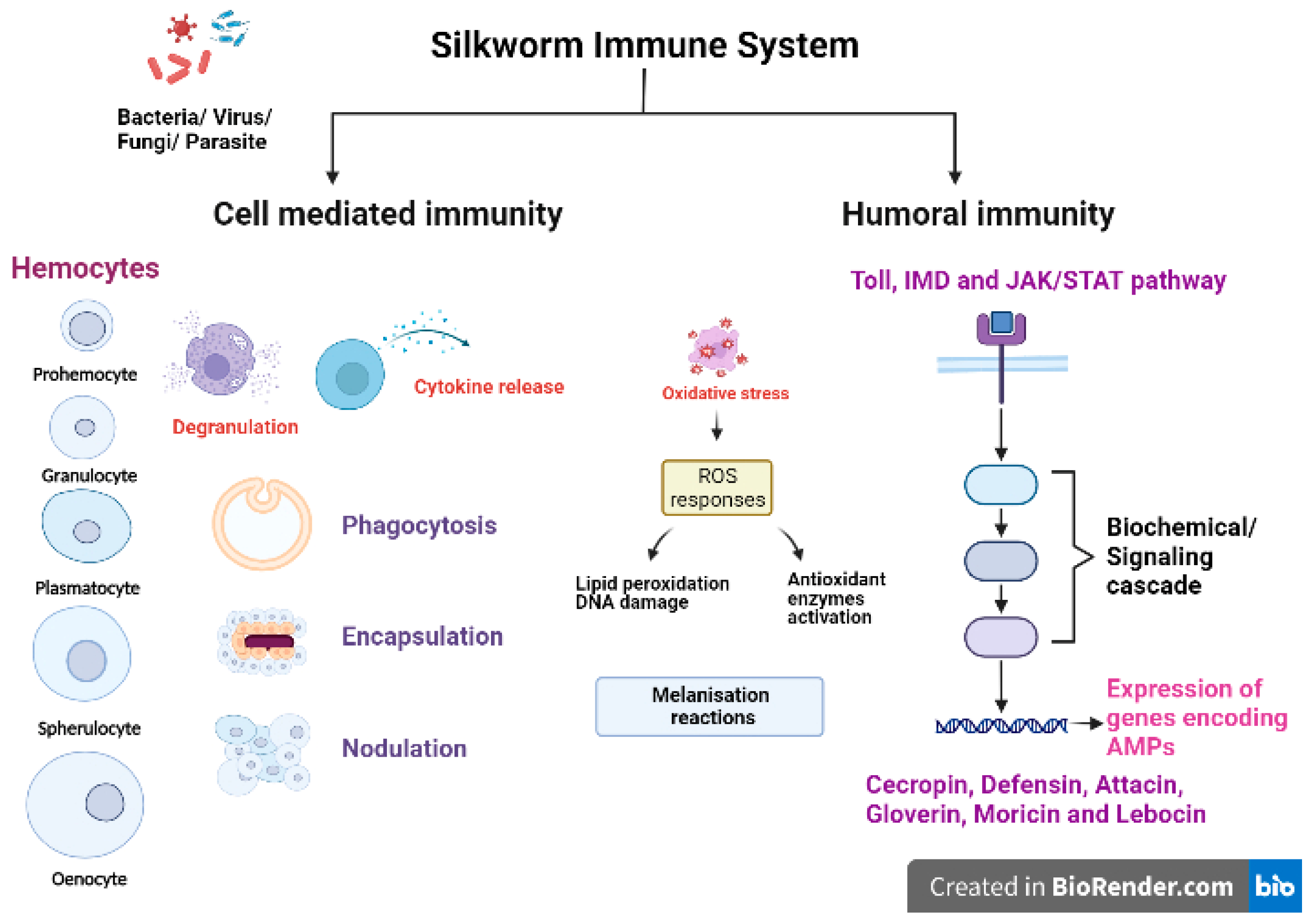
2. Silkworm and Immune Responses
3. Isolation of AMPs from Silkworms
4. Different AMPs in Silkworms
4.1. AMPs Reported from Mulberry Silkworm B. mori
4.1.1. Cecropins
4.1.2. Defensins
4.1.3. Moricins
4.1.4. Gloverins
4.1.5. Attacins
4.1.6. Lebocins
| AMP | Characteristics | Structure | Mode of action | Microorganisms | MIC | LC | Ref. |
|---|---|---|---|---|---|---|---|
| Cecropins | Cationic | α-helix | Pore forming; Forms a carpet structure on lipid bilayer surface and disintegrates the bacterial membrane | ||||
| BmcecA1 | B. subtilis | 2.5 µM | [117] | ||||
| B. thuringiensis | 2.5 µM | ||||||
| E. coli | 2.5 µM | ||||||
| P. aeruginosa | 2.5 µM | ||||||
| Ralstonia dolaanacearum | 2.5 µM | ||||||
| Cecropin B1 | P. fluorescens | 1.6 µM | [118] | ||||
| Xanthomonas campestris | 1.2 µM | ||||||
| Chromobacterium iodinum | 0.85 µM | ||||||
| Agrobacterium tumefaciens | 0.41 µM | ||||||
| Alcaligenes faecalis | 0.49 µM | ||||||
| Achromobacter polymorph | 1.8 µM | ||||||
| E. coli K12 | 0.38 µM | ||||||
| S. marcescens | 0.67 µM | ||||||
| M. luteus | 0.59 µM | ||||||
| S. aureus | >10 µM | ||||||
| Brevibacterium ammoniagenes | 0.49 µM | ||||||
| Lactobacillus plantarum | 0.62 µM | ||||||
| Arthrobacter simplex | 0.46 µM | ||||||
| B. subtilis | 3.6 µM | ||||||
| B. sphaericus | 4.4 µM | ||||||
| Cecropin B | B. megaterium | 1.7 µM | [89] | ||||
| E. coli | 0.35 µM | ||||||
| M. luteus | >207 µM | ||||||
| P. aeruginosa | 10 µM | ||||||
| S. marcescens | 17.22 µM | ||||||
| BmcecB6 | B. bombysepticus | 2.5 µM | [117] | ||||
| B. subtilis | 2.5 µM | ||||||
| B. thuringiensis | 1.25 µM | ||||||
| B. thuringiensis subsp. galleriae | 1.25 µM | ||||||
| E. coli, S. marcescens, P. aeruginosa | 0.625 µM | ||||||
| R. dolaanacearum | 1.25 µM | ||||||
| BmcecD | B. bombysepticus | 2.5 µM | [117] | ||||
| B. subtilis | 2.5 µM | ||||||
| B. thuringiensis | 1.25 µM | ||||||
| B. thuringiensis subsp. galleriae | 2.5 µM | ||||||
| E. coli, P. aeruginosa, R. dolaanacearum | 1.25 µM | ||||||
| S. marcescens | 2.5 µM | ||||||
| BmcecE | B. thuringiensis | 1.25 µM | [117] | ||||
| E. coli, S. marcescens, P. aeruginosa, R. dolaanacearum | 2.5 µM | ||||||
| Modified Cecropins CecXJ-37C | E. coli ATCC 25922 | 3.9 µM | [91] | ||||
| P. aeruginosa ATCC 27853 | 3.9 µM | ||||||
| K. pneumoniae ATCC 700603 | 15.7 µM | ||||||
| S. aureus ATCC 25923 | 0.25 µM | ||||||
| S. aureus ATCC 29213 | 1 µg/mL | ||||||
| S. aureus ATCC 43300 | 1 µg/mL | ||||||
| B. subtilis ATCC 6633 | 1 µg/mL | ||||||
| S. epidermidis ATCC 12228 | 1 µg/mL | ||||||
| CecXJ-37N | E. coli ATCC 25922 | 1 µM | [91] | ||||
| P. aeruginosa ATCC 27853 | 1 µM | ||||||
| K. pneumoniae ATCC 700603 | 7.8 µM | ||||||
| S. aureus ATCC 25923 | 0.25 µM | ||||||
| S. aureus ATCC 29213 | 1 µg/mL | ||||||
| S. aureus ATCC 43300 | 1 µg/mL | ||||||
| B. subtilis ATCC 6633 | 1 µg/mL | ||||||
| S. epidermidis ATCC 12228 | 1 µg/mL | ||||||
| Defensins | Cationic Cysteine rich Hydrophilic peptide | Cysteine-stabilized αβ motif | Disrupts bacterial cell membrane; Formation of voltage dependent anion-selective channels in cell membranes | S. aureus | NP | [101,102,103] | |
| Moricins | Basic Amphipathic | α-helix | Alters the membrane permeability; Formation of voltage dependent pores | E. coli JM 109 | 0.31 µM | [77,105] | |
| Acinetobacter sp. NISL B-4653 | 0.27 µM | ||||||
| P. fluorescens IAM 1179 | 0.53 µM | ||||||
| P. aeruginosa IAM 15140 | 0.81 µM | ||||||
| B. subtilis IAM 1107 | 0.19 µM | ||||||
| B. megaterium IAM 1030 | 0.09 µM | ||||||
| B. cereus IFO 3457 | 0.38 µM | ||||||
| S. aureus ATCC 6538P | 0.21 µM | ||||||
| S. aureus ATCC 6538Pa | 0.22 µM | ||||||
| S. aureus IFO 3083 | 0.46 µM | ||||||
| S. xylosus IAM 1312 | 0.27 µM | ||||||
| S. epidermidis IFO 12993 | 0.18 µM | ||||||
| S. pyogenes ATCC 21547 | 0.25 µM | ||||||
| Bmmor | S. aureus, B. subtilis | 0.625 µM | [117] | ||||
| B. bombysepticus, B. thuringiensis, B. thuringiensis subsp. galleriae, E. coli, P. aeruginosa, R. dolaanacearum | 1.25 µM | ||||||
| S. marcescens | 0.625 µM | ||||||
| Gloverins | Glycine rich Acidic to neutral (pI: 5.5–7.2) | Random coil | Pore forming | [117] | |||
| Bmglv1 | B. thuringiensis | 1.4 µM | |||||
| B. thuringiensis subsp. galleriae | 1.6 µM | ||||||
| E. coli, P. aeruginosa, R. dolaanacearum | 1.4 µM | ||||||
| S. marcescens | 1.2 µM | ||||||
| X. campestris | 1.6 µM | ||||||
| Bmglv2 | B. thuringiensis, B. thuringiensis subsp. galleriae, E. coli, S. marcescens, R. dolaanacearum | 1.6 µM | |||||
| X. campestris, P. aeruginosa | 1.8 µM | ||||||
| Bmglv3 | B. thuringiensis, B. thuringiensis subsp. galleriae, S. marcescens, R. dolaanacearum | 1.6 µM | |||||
| E. coli | 1.4 µM | ||||||
| P. aeruginosa, X. campestris | 1.8 µM | ||||||
| Bmglv4 | B. thuringiensis, E. coli, S. marcescens, R. dolaanacearum | 1.4 µM | |||||
| B. thuringiensis subsp. galleriae, P. Aeruginosa, X. campestris | 1.6 µM | ||||||
| Attacins | Glycine-rich | Random coil structure in aqueous solution | Altering cell membrane permeability; Hampers synthesis of plasma membrane proteins of bacterial cell | Gram negative and Gram positive bacteria | NP | [119] | |
4.2. AMPs Reported from Non-Mulberry Silkworms
5. Factors Affecting the Activity of AMPs
6. Current Status: AMPs under Clinical Investigation
7. Conclusions
8. Future Perspectives/Challenges
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ramzah, N.H.H.L.; Yenn, T.W.; Lee, W.H.; Loo, C.Y.; Tan, W.N.; Ring, L.C. Antimicrobial peptides, an alternative antimicrobial agent against multi-drug-resistant microbes: Source, application, and potential. In Advancements in Materials Science and Technology Led by Women. Advanced Structured Materials; Ismail, A., Nur Zulkipli, F., Husin, H.S., Öchsner, A., Eds.; Springer: Cham, Switzerland, 2023; Volume 165, pp. 235–259. [Google Scholar]
- Manniello, M.D.; Moretta, A.; Salvia, R.; Scieuzo, C.; Lucchetti, D.; Vogel, H.; Sgambato, A.; Falabella, P. Insect antimicrobial peptides: Potential weapons to counteract the antibiotic resistance. Cell Mol. Life Sci. 2021, 78, 4259–4282. [Google Scholar] [CrossRef] [PubMed]
- Huan, Y.; Kong, Q.; Mou, H.; Yi, H. Antimicrobial peptides: Classification, design, application and research progress in multiple fields. Front. Microbiol. 2020, 11, 582779. [Google Scholar] [CrossRef] [PubMed]
- Nwobodo, D.C.; Ugwu, M.C.; Anie, C.O.; Al-Ouqaili, M.T.S.; Ikem, J.C.; Chigozie, U.V.; Saki, M. Antibiotic resistance: The challenges and some emerging strategies for tackling a global menace. J. Clin. Lab Anal. 2022, 36, e24655. [Google Scholar] [CrossRef]
- Strathdee, S.A.; Davies, S.C.; Marcelin, J.R. Confronting antimicrobial resistance beyond the COVID-19 pandemic and the 2020 US election. Lancet 2020, 396, 1050–1053. [Google Scholar] [CrossRef]
- Armin, S.; Fallah, F.; Karimi, A.; Karbasiyan, F.; Alebouyeh, M.; Rafiei Tabatabaei, S.; Rajabnejad, M.; Ghanaie, R.M.; Fahimzad, S.A.; Abdollahi, N.; et al. Antibiotic susceptibility patterns for carbapenem-resistant Enterobacteriaceae. Int. J. Microbiol. 2023, 2023, 8920977. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.H.; Lu, T.K. Development and challenges of antimicrobial peptides for therapeutic applications. Antibiotics 2020, 9, 24. [Google Scholar] [CrossRef] [PubMed]
- Hancock, R.E.; Falla, T.J. Antimicrobial peptides: Broad-spectrum antibiotics from nature. Clin. Microbiol. Infect. 1996, 1, 226–229. [Google Scholar] [CrossRef]
- Uddin, S.J.; Shilpi, J.A.; Nahar, L.; Sarker, S.D.; Göransson, U. Editorial: Natural antimicrobial peptides: Hope for new antibiotic lead molecules. Front. Pharmacol. 2021, 12, 640938. [Google Scholar] [CrossRef]
- Dini, I.; De Biasi, M.-G.; Mancusi, A. An Overview of the potentialities of antimicrobial peptides derived from natural sources. Antibiotics 2022, 11, 1483. [Google Scholar] [CrossRef]
- Moretta, A.; Scieuzo, C.; Petrone, A.M.; Salvia, R.; Manniello, M.D.; Franco, A.; Lucchetti, D.; Vassallo, A.; Vogel, H.; Sgambato, A.; et al. Antimicrobial peptides: A new hope in biomedical and pharmaceutical fields. Front. Cell Infect. Microbiol. 2021, 11, 668632. [Google Scholar] [CrossRef]
- Mahlapuu, M.; Björn, C.; Ekblom, J. Antimicrobial peptides as therapeutic agents: Opportunities and challenges. Crit. Rev. Biotechnol. 2020, 40, 978–992. [Google Scholar] [CrossRef] [PubMed]
- Ganz, T. The role of antimicrobial peptides in innate immunity. Integr. Comp. Biol. 2003, 43, 300–304. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zuo, S.; Wang, B.; Zhang, K.; Wang, Y. Antimicrobial mechanisms and clinical application prospects of antimicrobial peptides. Molecules 2022, 27, 2675. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.J.; Kim, D.H.; Mishig-Ochir, T.; Lee, B.J. Antimicrobial peptides: Their physicochemical properties and therapeutic application. Arch. Pharm. Res. 2012, 35, 409–413. [Google Scholar] [CrossRef]
- Huang, Y.; Huang, J.; Chen, Y. Alpha-helical cationic antimicrobial peptides: Relationships of structure and function. Protein Cell 2010, 1, 143–152. [Google Scholar] [CrossRef]
- Vanzolini, T.; Bruschi, M.; Rinaldi, A.C.; Magnani, M.; Fraternale, A. Multitalented synthetic antimicrobial peptides and their antibacterial, antifungal and antiviral mechanisms. Int. J. Mol. Sci. 2022, 23, 545. [Google Scholar] [CrossRef]
- Azmiera, N.; Krasilnikova, A.; Sahudin, S.; Al-Talib, H.; Heo, C.C. Antimicrobial peptides isolated from insects and their potential applications. J. Asia Pac. Entomol. 2022, 25, 101892. [Google Scholar] [CrossRef]
- Talapko, J.; Meštrović, T.; Juzbašić, M.; Tomas, M.; Erić, S.; Horvat Aleksijević, L.; Bekić, S.; Schwarz, D.; Matić, S.; Neuberg, M.; et al. Antimicrobial peptides—Mechanisms of action, antimicrobial effects and clinical applications. Antibiotics 2022, 11, 1417. [Google Scholar] [CrossRef]
- Pereira, H.A. Novel therapies based on cationic antimicrobial peptides. Curr. Pharm. Biotechnol. 2006, 7, 229–234. [Google Scholar] [CrossRef]
- Stork, N.E. How many species of insects and other terrestrial arthropods are there on earth? Annu. Rev. Entomol. 2018, 63, 31–45. [Google Scholar] [CrossRef]
- Ejiofor, A.O. Insect Biotechnology. In Short Views on Insect Genomics and Proteomics. Entomology in Focus; Raman, C., Goldsmith, M.R., Agunbiade, T.A., Eds.; Springer International Publishing: Cham, Switzerland, 2016; Volume 4, pp. 185–210. [Google Scholar]
- Yi, H.Y.; Chowdhury, M.; Huang, Y.D.; Yu, X.Q. Insect antimicrobial peptides and their applications. Appl. Microbiol. Biotechnol. 2014, 98, 5807–5822. [Google Scholar] [CrossRef] [PubMed]
- Mylonakis, E.; Podsiadlowski, L.; Muhammed, M.; Vilcinskas, A. Diversity, evolution and medical applications of insect antimicrobial peptides. Phil. Trans. R. Soc. B 2016, 371, 20150290. [Google Scholar] [CrossRef] [PubMed]
- Dijksteel, G.S.; Ulrich, M.M.W.; Middelkoop, E.; Boekema, B.K.H.L. Review: Lessons Learned from Clinical Trials Using Antimicrobial Peptides (AMPs). Front. Microbiol. 2021, 12, 616979. [Google Scholar] [CrossRef] [PubMed]
- Assoni, L.; Milani, B.; Carvalho, M.R.; Nepomuceno, L.N.; Waz, N.T.; Guerra, M.E.S.; Converso, T.R.; Darrieux, M. Resistance mechanisms to antimicrobial peptides in Gram-positive bacteria. Front. Microbiol. 2020, 11, 593215. [Google Scholar] [CrossRef]
- Moravej, H.; Moravej, Z.; Yazdanparast, M.; Heiat, M.; Mirhosseini, A.; Moosazadeh, M.M.; Mirnejad, R. Antimicrobial Peptides: Features, Action, and Their Resistance Mechanisms in Bacteria. Microb. Drug Resist. 2018, 24, 747–767. [Google Scholar] [CrossRef]
- Erdem Büyükkiraz, M.; Kesmen, Z. Antimicrobial peptides (AMPs): A promising class of antimicrobial compounds. J. Appl. Microbiol. 2022, 132, 1573–1596. [Google Scholar] [CrossRef]
- Shi, J.; Chen, C.; Wang, D.; Wang, Z.; Liu, Y. The antimicrobial peptide LI14 combats multidrug-resistant bacterial infections. Commun. Biol. 2022, 5, 926. [Google Scholar] [CrossRef]
- Lee, H.; Yang, S. Dimerization of cell-penetrating buforin II enhances antimicrobial properties. J. Anal. Sci. Technol. 2021, 12, 9. [Google Scholar] [CrossRef]
- Liu, P.; Zeng, X.; Wen, X. Design and Synthesis of New Cationic Antimicrobial Peptides with Low Cytotoxicity. Int. J. Pept. Res. Ther. 2021, 27, 831–840. [Google Scholar] [CrossRef]
- Ko, S.J.; Park, E.; Asandei, A.; Choi, J.Y.; Lee, S.C.; Seo, C.H.; Luchian, T.; Park, Y. Bee venom-derived antimicrobial peptide melectin has broad-spectrum potency, cell selectivity, and salt-resistant properties. Sci. Rep. 2020, 10, 10145. [Google Scholar] [CrossRef]
- Zhu, H.; Ding, X.; Li, W.; Lu, T.; Ma, C.; Xi, X.; Wang, L.; Zhou, M.; Burden, R.; Chen, T. Discovery of two skin-derived dermaseptins and design of a TAT-fusion analogue with broad-spectrum antimicrobial activity and low cytotoxicity on healthy cells. PeerJ 2018, 6, e5635. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.; Liu, Z.; Cao, S.; Wang, H.; Jiang, C.; Hussain, M.A.; Pang, S. Broad-Spectrum Antimicrobial Activity and Low Cytotoxicity against Human Cells of a Peptide Derived from Bovine αS1-Casein. Molecules 2018, 23, 1220. [Google Scholar] [CrossRef]
- Lee, J.K.; Park, S.C.; Hahm, K.S.; Park, Y. A helix-PXXP-helix peptide with antibacterial activity without cytotoxicity against MDRPA-infected mice. Biomaterials 2014, 35, 1025–1039. [Google Scholar] [CrossRef]
- Barańska-Rybak, W.; Pikula, M.; Dawgul, M.; Kamysz, W.; Trzonkowski, P.; Roszkiewicz, J. Safety profile of antimicrobial peptides: Camel, citropin, protegrin, temporin a and lipopeptide on HaCaT keratinocytes. Acta Pol. Pharm. 2013, 70, 795–801. [Google Scholar] [PubMed]
- Kausar, S.; Abbas, M.N.; Zhao, Y.; Cui, H. Immune strategies of silkworm, Bombyx mori against microbial infections. Invertebr. Surviv. J. 2019, 16, 130–140. [Google Scholar]
- Zha, X.-L.; Wang, H.; Sun, W.; Zhang, H.-Y.; Wen, J.; Huang, X.-Z.; Lu, C.; Shen, Y.-H. Characteristics of the peritrophic matrix of the silkworm, Bombyx mori and factors influencing its formation. Insects 2021, 12, 516. [Google Scholar] [CrossRef]
- Chen, K.; Lu, Z. Immune responses to bacterial and fungal infections in the silkworm, Bombyx mori. Dev. Comp. Immunol. 2018, 83, 3–11. [Google Scholar] [CrossRef]
- Takahasi, K.; Ochiai, M.; Horiuchi, M.; Kumeta, H.; Ogura, K.; Ashida, M.; Inagaki, F. Solution structure of the silkworm betaGRP/GNBP3 N-terminal domain reveals the mechanism for beta-1,3-glucan-specific recognition. Proc. Natl. Acad. Sci. USA 2009, 106, 11679–11684. [Google Scholar] [CrossRef]
- Strand, M.R. The insect cellular immune response. Insect Sci. 2008, 15, 1–14. [Google Scholar] [CrossRef]
- Akira, S.; Uematsu, S.; Takeuchi, O. Pathogen recognition and innate immunity. Cell 2006, 124, 783–801. [Google Scholar] [CrossRef]
- Jiang, L.; Liu, W.; Guo, H.; Dang, Y.; Cheng, T.; Yang, W.; Sun, Q.; Wang, B.; Wang, Y.; Xie, E.; et al. Distinct functions of Bombyx mori peptidoglycan recognition protein 2 in immune responses to bacteria and viruses. Front. Immunol. 2019, 10, 776. [Google Scholar] [CrossRef] [PubMed]
- Patnaik, B.B.; Saha, A.K.; Bindroo, B.B.; Han, Y.S. Lectins as possible candidates towards anti-microbial defense in silkworm, Bombyx mori L. Afr. J. Biotechnol. 2012, 11, 11045–11052. [Google Scholar]
- Tan, J.; Xu, M.; Zhang, K.; Wang, X.; Chen, S.; Li, T.; Xiang, Z.; Cui, H. Characterization of hemocytes proliferation in larval silkworm, Bombyx mori. J. Insect Physiol. 2013, 59, 595–603. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, C.; Brehelin, M. Insect haemocytes: What type of cell is that? J. Insect. Physiol. 2006, 52, 417–429. [Google Scholar] [CrossRef]
- Lavine, M.D.; Strand, M.R. Insect hemocytes and their role in immunity. Insect Biochem. Mol. Biol. 2002, 32, 1295–1309. [Google Scholar] [CrossRef] [PubMed]
- Minakhina, S.; Steward, R. Hematopoietic stem cells in Drosophila. Development 2010, 137, 27–31. [Google Scholar] [CrossRef] [PubMed]
- Prabhuling, S.H.; Makwana, P.; Pradeep, A.N.R.; Vijayan, K.; Mishra, R.K. Release of mediator enzyme β-hexosaminidase and modulated gene expression accompany hemocyte degranulation in response to parasitism in the silkworm Bombyx mori. Biochem. Genet. 2021, 59, 997–1017. [Google Scholar] [CrossRef]
- Suzuki, A.; Yoshizawa, Y.; Tanaka, S.; Kitami, M.; Sato, R. Extra-and intracellular signaling pathways regulating nodule formation by hemocytes of the silkworm, Bombyx mori (Lepidoptera: Bombicidae). J. Insect Biotechnol. Sericol. 2011, 80, 49–56. [Google Scholar]
- Ling, E.; Shirai, K.; Kanehatsu, R.; Kiguchi, K. Reexamination of phenoloxidase in larval circulating hemocytes of the silkworm, Bombyx mori. Tissue Cell 2005, 37, 101–107. [Google Scholar] [CrossRef]
- Dubovskiy, I.M.; Kryukova, N.A.; Glupov, V.V.; Ratcliffe, N.A. Encapsulation and nodulation in insects. Invertebr. Surviv. J. 2016, 13, 229–246. [Google Scholar]
- Wu, G.; Li, M.; Liu, Y.; Ding, Y.; Yi, Y. The specificity of immune priming in silkworm, Bombyx mori, is mediated by the phagocytic ability of granular cells. J. Insect. Physiol. 2015, 81, 60–68. [Google Scholar] [CrossRef] [PubMed]
- Kawakami, K. Phagocytosis in muscardine-diseased larvae of the silkworm, Bombyx mori (Linnaeus). J. Invertebr. Pathol. 1965, 7, 203–208. [Google Scholar] [CrossRef]
- Shambhavi, H.P.; Makwana, P.; Surendranath, B.; Ponnuvel, K.M.; Mishra, R.K.; Pradeep, A.N.R. Phagocytic events, associated lipid peroxidation and peroxidase activity in hemocytes of silkworm Bombyx mori induced by microsporidian infection. Caryologia 2020, 73, 93–106. [Google Scholar]
- Satyavathi, V.V.; Minz, A.; Nagaraju, J. Nodulation: An unexplored cellular defense mechanism in insects. Cell. Signal. 2014, 26, 1753–1763. [Google Scholar] [CrossRef] [PubMed]
- Gandhe, A.S.; John, S.H.; Nagaraju, J. Noduler, a novel immune up-regulated protein mediates nodulation response in insects. J. Immunol. 2007, 179, 6943–6951. [Google Scholar] [CrossRef]
- Tsakas, S.; Marmaras, V.J. Insect immunity and its signalling: An overview. Invertebr. Surviv. J. 2010, 7, 228–238. [Google Scholar]
- Yang, Y.; Tang, H.; Zhang, Y.; Zhu, F.; Lü, P.; Yao, Q.; Chen, K. Research progress on the immune mechanism of the silkworm Bombyx mori. Physiol. Entomol. 2018, 43, 159–168. [Google Scholar] [CrossRef]
- Li, B.; Ye, C.J.; Meng, Y.; Fan, T.; Chen, F. Prokaryotic expression of Bombyx mori serine protease inhibitor Bmserpin6 and its regulation on prophenoloxidase activity and antimicrobial peptide expression. Sci. Seric. 2016, 2, 237–242. [Google Scholar]
- Li, J.; Ma, L.; Lin, Z.; Zou, Z.; Lu, Z. Serpin-5 regulates prophenoloxidase activation and antimicrobial peptide pathways in the silkworm, Bombyx mori. Insect Biochem. Mol. Biol. 2016, 73, 27–37. [Google Scholar] [CrossRef]
- Li, B.; Yu, H.-Z.; Ye, C.-J.; Ma, Y.; Li, X.; Fan, T.; Chen, F.-S.; Xu, J.-P. Bombyx mori Serpin6 regulates prophenoloxidase activity and the expression of antimicrobial proteins. Gene 2017, 610, 64–70. [Google Scholar] [CrossRef] [PubMed]
- Pradeep, A.N.R.; Anitha, J.; Awasthi, A.K.; Babu, M.A.; Geetha, M.N.; Arun, H.K.; Chandrashekhar, S.; Rao, G.C.; Vijayaprakash, N.B. Activation of autophagic programmed cell death and innate immune gene expression reveals immuno-competence of integumental epithelium in Bombyx mori infected by a dipteran parasitoid. Cell Tissue Res. 2013, 352, 371–385. [Google Scholar] [CrossRef] [PubMed]
- Jayaram, A.; Pradeep, A.N.R.; Awasthi, A.K.; Murthy, G.N.; Ponnuvel, K.M.; Sasibhushan, S.; Rao, G.C. Coregulation of host–response genes in integument: Switchover of gene expression correlation pattern and impaired immune responses induced by dipteran parasite infection in the silkworm, Bombyx mori. J. Appl. Genet. 2014, 55, 209–221. [Google Scholar] [CrossRef] [PubMed]
- Lekha, G.; Vijayagowri, E.; Sirigineedi, S.; Sivaprasad, V.; Ponnuvel, K.M. Differential level of host gene expression associated with nucleopolyhedrovirus infection in silkworm races of Bombyx mori. Int. J. Indust. Entomol. 2014, 29, 145–152. [Google Scholar] [CrossRef]
- Lekha, G.; Gupta, T.; Awasthi, A.K.; Murthy, G.N.; Trivedy, K.; Ponnuvel, K.M. Genome wide microarray based expression profiles associated with BmNPV resistance and susceptibility in Indian silkworm races of Bombyx mori. Genomics 2015, 106, 393–403. [Google Scholar] [CrossRef]
- Makwana, P.; Dubey, H.; Pradeep, A.N.R.; Sivaprasad, V.; Ponnuvel, K.M.; Mishra, R.K. Dipteran endoparasitoid infestation actively suppressed host defense components in hemocytes of silkworm Bombyx mori for successful parasitism. Anim. Gene 2021, 22, 200118. [Google Scholar] [CrossRef]
- Hoffmann, J.A. The immune response of Drosophila. Nature 2003, 426, 33–38. [Google Scholar] [CrossRef]
- De Gregorio, E.; Spellman, P.T.; Tzou, P.; Rubin, G.M.; Lemaitre, B. The Toll and Imd pathways are the major regulators of the immune response in Drosophila. EMBO J. 2002, 21, 2568–2579. [Google Scholar] [CrossRef]
- Williams, M.J. Drosophila hemopoiesis and cellular immunity. J. Immunol. 2007, 178, 4711–4716. [Google Scholar] [CrossRef]
- Kaneko, Y.; Furukawa, S.; Tanaka, H.; Yamakawa, M. Expression of antimicrobial peptide genes encoding Enbocin and Gloverin isoforms in the silkworm, Bombyx mori. Biosci. Biotechnol. Biochem. 2007, 71, 2233–2241. [Google Scholar] [CrossRef]
- Tanaka, H.; Ishibashi, J.; Fujita, K.; Nakajima, Y.; Sagisaka, A.; Tomimoto, K.; Suzuki, N.; Yoshiyama, M.; Kaneko, Y.; Iwasaki, T.; et al. A genome-wide analysis of genes and gene families involved in innate immunity of Bombyx mori. Insect Biochem. Mol. Biol. 2008, 38, 1087–1110. [Google Scholar] [CrossRef]
- Arbouzova, N.I.; Zeidler, M.P. JAK/STAT signalling in Drosophila: Insights into conserved regulatory and cellular functions. Development 2006, 133, 2605–2616. [Google Scholar] [CrossRef] [PubMed]
- Bang, I.S. JAK/STAT signaling in insect innate immunity. Entomol. Res. 2019, 49, 339–353. [Google Scholar] [CrossRef]
- Rahul, K.; Moamongba, K.S.; Rabha, M.; Sivaprasad, V. Identification and characterization of bacteria causing flacherie in mulberry silkworm, Bombyx mori L. J. Crop Weed 2019, 15, 178–181. [Google Scholar] [CrossRef]
- Kajiwara, H.; Itou, Y.; Imamaki, A.; Nakamura, M.; Mita, K.; Ishizaka, M. Proteomic analysis of silkworm fat body. J. Insect Biotechnol. Sericology 2006, 75, 47–56. [Google Scholar]
- Hara, S.; Yamakawa, M. Moricin, a novel type of antibacterial peptide isolated from the silkworm, Bombyx mori. J. Biol. Chem. 1995, 270, 29923–29927. [Google Scholar] [CrossRef]
- Perkins, D.N.; Pappin, J.C.; Creasy, D.M.; Cottrell, J.S. Probability-based protein identification by searching database using mass spectrometry data. Electrophoresis 1999, 20, 3551–3567. [Google Scholar] [CrossRef]
- Orsburn, B.C. Proteome Discoverer—A community enhanced data processing suite for protein informatics. Proteomes 2021, 9, 15. [Google Scholar] [CrossRef]
- Ma, B.; Zhang, K.; Hendrie, C.; Liang, C.; Li, M.; Doherty-Kirby, A.; Lajoie, G. PEAKS: Powerful software for peptide de novo sequencing by tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2003, 17, 2337–2342. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Carlson, A.; Sinitcyn, P.; Mann, M.; Cox, J. Visualization of LC-MS/MS proteomics data in MaxQuant. Proteomics 2015, 15, 1453–1456. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Cox, J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 2016, 11, 2301–2319. [Google Scholar] [CrossRef]
- Balouiri, M.; Sadiki, M.; Ibnsouda, S.K. Methods for in vitro evaluating antimicrobial activity: A review. J. Pharm. Anal. 2016, 6, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Steiner, H.; Hultmark, D.; Engström, A.; Bennich, H.; Boman, H.G. Sequence and specificity of two antibacterial proteins involved in insect immunity. Nature 1981, 292, 246–248. [Google Scholar] [CrossRef] [PubMed]
- Cheng, T.; Zhao, P.; Liu, C.; Xu, P.; Gao, Z.; Xia, Q.; Xiang, Z. Structures, regulatory regions, and inductive expression patterns of antimicrobial peptide genes in the silkworm Bombyx mori. Genomics 2006, 87, 356–365. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Park, B.S.; Yun, E.Y.; Je, Y.H.; Woo, S.D.; Kang, S.W.; Kim, K.Y.; Kang, S.K. Cloning and expression of a novel gene encoding a new antibacterial peptide from silkworm, Bombyx mori. Biochem. Biophys. Res. Commun. 1998, 246, 388–392. [Google Scholar] [CrossRef] [PubMed]
- Romoli, O.; Mukherjee, S.; Mohid, S.A.; Dutta, A.; Montali, A.; Franzolin, E.; Brady, D.; Zito, F.; Bergantino, E.; Rampazzo, C.; et al. Enhanced Silkworm Cecropin B Antimicrobial Activity against Pseudomonas aeruginosa from Single Amino Acid Variation. ACS Infect. Dis. 2019, 5, 1200–1213. [Google Scholar] [CrossRef] [PubMed]
- Sato, H.; Feix, J.B. Peptide–membrane interactions and mechanisms of membrane destruction by amphipathic-helical antimicrobial peptides. Biochim. Biophys. Acta. 2006, 1758, 1245–1256. [Google Scholar] [CrossRef]
- Gazit, E.; Lee, W.J.; Brey, P.T.; Shai, Y. Mode of action of the antibacterial cecropin B2: A spectrofluorometric study. Biochemistry 1994, 33, 10681–10692. [Google Scholar] [CrossRef]
- Efimova, S.S.; Schagina, L.V.; Ostroumova, O.S. Channel-forming activity of cecropins in lipid bilayers: Effect of agents modifying the membrane dipole potential. Langmuir 2014, 30, 7884–7892. [Google Scholar] [CrossRef]
- Liu, D.; Liu, J.; Li, J.; Xia, L.; Yang, J.; Sun, S.; Ma, J.; Zhang, F. A potential food biopreservative, CecXJ-37N, non-covalently intercalates into the nucleotides of bacterial genomic DNA beyond membrane attack. Food Chem. 2017, 217, 576–584. [Google Scholar] [CrossRef]
- Bulet, P.; Charlet, M.; Hetru, C. Antimicrobial peptides in insect immunity. In Innate Immunity. Infectious Disease; Ezekowitz, R.A.B., Hoffmann, J.A., Eds.; Humana Press: Totowa, NJ, USA, 2003; pp. 89–107. [Google Scholar]
- Brady, D.; Grapputo, A.; Romoli, O.; Sandrelli, F. Insect Cecropins, Antimicrobial Peptides with Potential Therapeutic Applications. Int. J. Mol. Sci. 2019, 20, 5862. [Google Scholar] [CrossRef]
- Xia, L.; Wu, Y.; Ma, J.I.; Yang, J.; Zhang, F. The antibacterial peptide from Bombyx mori cecropinXJ induced growth arrest and apoptosis in human hepatocellular carcinoma cells. Oncol. Lett. 2016, 12, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Cerón, J.M.; Contreras-Moreno, J.; Puertollano, E.; de Cienfuegos, G.Á.; Puertollano, M.A.; de Pablo, M.A. The antimicrobial peptide cecropin A induces caspase-independent cell death in human promyelocytic leukemia cells. Peptides 2010, 31, 1494–1503. [Google Scholar] [CrossRef] [PubMed]
- Tornesello, A.L.; Borrelli, A.; Buonaguro, L.; Buonaguro, F.M.; Tornesello, M.L. Antimicrobial Peptides as Anticancer Agents: Functional Properties and Biological Activities. Molecules 2020, 25, 2850. [Google Scholar] [CrossRef]
- Matsuyama, K.; Natori, S. Purification of three antibacterial proteins from the culture medium of NIH-Sape-4, an embryonic cell line of Sarcophaga peregrina. J. Biol. Chem. 1988, 263, 17112–17116. [Google Scholar] [CrossRef] [PubMed]
- Koehbach, J. Structure-activity relationships of insect defensins. Front Chem. 2017, 5, 45. [Google Scholar] [CrossRef]
- Kaneko, Y.; Tanaka, H.; Ishibashi, J.; Iwasaki, T.; Yamakawa, M. Gene expression of a novel defensin antimicrobial peptide in the silkworm, Bombyx mori. Biosci. Biotechnol. Biochem. 2008, 72, 2353–2361. [Google Scholar] [CrossRef]
- Song, K.J.; Park, B.R.; Kim, S.Y.; Park, K.S. Molecular characterization of anionic defensin-like peptide in immune response of silkworm, Bombyx mori L. (Lepidoptera). Genes Genom. 2010, 32, 447–453. [Google Scholar] [CrossRef]
- Lehrer, R.I.; Lu, W. Alpha-Defensins in human innate immunity. Immunol. Rev. 2012, 245, 84–112. [Google Scholar] [CrossRef]
- Kagan, B.L.; Selsted, M.E.; Ganz, T.; Lehrer, R.I. Antimicrobial defensin peptides form voltage-dependent ion-permeable channels in planar lipid bilayer membranes. Proc. Natl. Acad. Sci. USA 1990, 87, 210–214. [Google Scholar] [CrossRef]
- Lehrer, R.I.; Barton, A.; Daher, K.A.; Harwig, S.S.; Ganz, T.; Selsted, M.E. Interaction of human defensins with Escherichia coli. Mechanism of bactericidal activity. J. Clin. Investig. 1989, 84, 553–561. [Google Scholar] [CrossRef]
- Baxter, A.A.; Poon, I.K.H.; Hulett, M.D. The lure of the lipid: How defensins exploit membrane phospholipids to induce cytolysis in target cells. Cell Death Dis. 2017, 8, 22712. [Google Scholar] [CrossRef] [PubMed]
- Hemmi, H.; Ishibashi, J.; Hara, S.; Yamakawa, M. Solution structure of moricin, an antibacterial peptide, isolated from the silkworm Bombyx mori. FEBS Lett. 2002, 518, 33–38. [Google Scholar] [CrossRef]
- Axen, A.; Carlsson, A.; Engstrǒm, A.; Bennich, H. Gloverin, an antibacterial protein from the immune hemolymph of Hyalophora pupae. Eur. J. Biochem. 1997, 247, 614–619. [Google Scholar] [CrossRef] [PubMed]
- Yi, H.Y.; Deng, X.J.; Yang, W.Y.; Zhou, C.Z.; Cao, Y.; Yu, X.Q. Gloverins of the silkworm Bombyx mori: Structural and binding properties and activities. Insect Biochem. Mol. Biol. 2013, 43, 612–625. [Google Scholar] [CrossRef]
- Lü, D.; Geng, T.; Hou, C.; Qin, G.; Gao, K.; Guo, X. Expression profiling of Bombyx mori gloverin2 gene and its synergistic antifungal effect with cecropin A against Beauveria bassiana. Gene 2017, 600, 55–63. [Google Scholar] [CrossRef]
- Wang, Q.; Guo, P.; Wang, Z.; Liu, H.; Zhang, Y.; Jiang, S.; Han, W.; Xia, Q.; Zhao, P. Antibacterial mechanism of gloverin2 from silkworm, Bombyx mori. Int. J. Mol. Sci. 2018, 19, 2275. [Google Scholar] [CrossRef] [PubMed]
- Hedengren, M.; Borge, K.; Hultmark, D. Expression and evolution of the Drosophila attacin/diptericin gene family. Biochem. Biophys. Res. Commun. 2000, 279, 574–581. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Aksoy, S. An antimicrobial peptide with trypanocidal activity characterized from Glossina morsitans morsitans. Insect Biochem. Mol. Biol. 2005, 35, 105–115. [Google Scholar] [CrossRef] [PubMed]
- Kwon, Y.M.; Kim, H.J.; Kim, Y.I.; Kang, Y.J.; Lee, I.H.; Jin, B.R.; Han, Y.S.; Cheon, H.M.; Ha, N.G.; Seo, S.J. Comparative analysis of two attacin genes from Hyphantria cunea. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2008, 151, 213–220. [Google Scholar] [CrossRef]
- Carlsson, A.; Engstrom, P.; Palva, E.T.; Bennich, H. Attacin, an antibacterial protein from Hyalophora cecropia, inhibits synthesis of outer membrane proteins in Escherichia coli by interfering with OMP gene transcription. Infect. Immun. 1991, 59, 3040–3045. [Google Scholar] [CrossRef]
- Engstrom, P.; Carlsson, A.; Engstrom, A.; Tao, Z.J.; Bennich, H. The antibacterial effect of attacins from the silk moth Hyalophora cecropia is directed against the outer membrane of Escherichia coli. EMBO J. 1984, 3, 3347–3351. [Google Scholar] [CrossRef]
- Furukawa, S.; Taniai, K.; Ishibashi, J.; Hara, S.; Shono, T.; Yamakawa, M. A novel member of lebocin gene family from the silkworm, Bombyx mori. Biochem. Biophys. Res. Commun. 1997, 238, 769–774. [Google Scholar] [CrossRef] [PubMed]
- Rao, X.J.; Xu, X.X.; Yu, X.Q. Functional analysis of two lebocin-related proteins from Manduca sexta. Insect Biochem. Mol. Biol. 2012, 42, 231–239. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Cheng, T.; Ye, M.; Deng, X.; Yi, H.; Huang, Y.; Tan, X.; Han, D.; Wang, B.; Xiang, Z.; et al. Functional divergence among silkworm antimicrobial peptide paralogs by the activities of recombinant proteins and the induced expression profiles. PLoS ONE 2011, 6, e18109. [Google Scholar] [CrossRef]
- Morishima, I.; Suginaka, S.; Ueno, T.; Hirano, H. Isolation and structure of cecropins, inducible antibacterial peptides, from the silkworm. Bombyx Mori. Comp. Biochem. Physiol. B 1990, 95, 551–554. [Google Scholar] [CrossRef]
- Buonocore, F.; Fausto, A.M.; Della Pelle, G.; Roncevic, T.; Gerdol, M.; Picchietti, S. Attacins: A promising class of insect antimicrobial peptides. Antibiotics 2021, 10, 212. [Google Scholar] [CrossRef]
- Hashimoto, K.; Yamano, Y.; Morishima, I. Cloning and expression of a gene encoding gallerimycin, a cysteine-rich antifungal peptide, from eri-silkworm, Samia cynthia ricini. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2008, 150, 229–232. [Google Scholar] [CrossRef] [PubMed]
- Bao, Y.; Yamano, Y.; Morishima, I. A novel lebocin-like gene from eri-silkworm, Samia cynthia ricini, that does not encode the antibacterial peptide lebocin. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2005, 140, 127–131. [Google Scholar] [CrossRef]
- Kishimoto, K.; Fujimoto, S.; Matsumoto, K.; Yamano, Y.; Morishima, I. Protein purification, cDNA cloning and gene expression of attacin, an antibacterial protein, from eri-silkworm, Samia cynthia ricini. Insect Biochem. Mol. Biol. 2002, 32, 881–887. [Google Scholar] [CrossRef]
- Li, Q.; Zhang, J.; Sun, Y.; Wang, L.; Qian, C.; Wei, G.; Zhu, B.; Liu, C. Immunological function of the antibacterial peptide Attacin-like in the Chinese oak silkworm, Antheraea pernyi. Protein Pept. Lett. 2020, 27, 953–961. [Google Scholar] [CrossRef]
- Nayak, T.; Mandal, S.M.; Neog, K.; Ghosh, A.K. Characterization of a gloverin-like antimicrobial peptide isolated from muga silkworm, Antheraea assamensis. Int. J. Pept. Res. Ther. 2018, 24, 337–346. [Google Scholar] [CrossRef]
- Dutta, S.R.; Gauri, S.S.; Ghosh, T.; Halder, S.K.; DasMohapatra, P.K.; Mondal, K.C.; Ghosh, A.K. Elucidation of structural and functional integration of a novel antimicrobial peptide from Antheraea mylitta. Bioorganic Med. Chem. Lett. 2017, 27, 1686–1692. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, T.; Mandal, S.M.; Kumari, R.; Ghosh, A.K. Purification and characterization of a novel antimicrobial peptide (QAK) from the hemolymph of Antheraea mylitta. Biochem. Biophys. Res. Commun. 2020, 527, 411–417. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.R.; Goo, T.W.; Choi, K.H.; Park, S.W.; Kim, S.W.; Hwang, J.S.; Kang, S.W. Isolation and purification of a cecropin-like antimicrobial peptide from the Japanese oak silkworm, Antheraea yamamai. J. Sericultural Entomol. Sci. 2012, 50, 145–149. [Google Scholar] [CrossRef]
- Dutta, S.R.; Gauri, S.S.; Mondal, B.; Vemula, A.; Haider, S.K.; Mondal, K.C.; Ghosh, A.K. Screening of antimicrobial peptides from hemolymph extract of tasar silkworm Antheraea mylitta against urinary tract and wound infecting multidrug-resistant bacteria. Acta Biol. Szeged. 2016, 60, 49–55. [Google Scholar]
- Ebbensgaard, A.; Mordhorst, H.; Overgaard, M.T.; Nielsen, C.G.; Aarestrup, F.M.; Hansen, E.B. Comparative evaluation of the antimicrobial activity of different antimicrobial peptides against a range of pathogenic bacteria. PLoS ONE 2015, 10, e0144611. [Google Scholar] [CrossRef]
- Heymich, M.L.; Srirangan, S.; Pischetsrieder, M. Stability and activity of the antimicrobial peptide Leg1 in solution and on meat and its optimized generation from chickpea storage protein. Foods 2021, 10, 1192. [Google Scholar] [CrossRef]
- Vishnepolsky, B.; Zaalishvili, G.; Karapetian, M.; Nasrashvili, T.; Kuljanishvili, N.; Gabrielian, A.; Rosenthal, A.; Hurt, D.E.; Tartakovsky, M.; Grigolava, M.; et al. De novo design and in vitro testing of antimicrobial peptides against Gram-negative bacteria. Pharmaceuticals 2019, 12, 82. [Google Scholar] [CrossRef]
- Wei, D.; Zhang, X. Biosynthesis, bioactivity, biotoxicity and applications of antimicrobial peptides for human health. Biosaf. Health 2022, 4, 118–134. [Google Scholar] [CrossRef]
- Gan, B.H.; Gaynord, J.; Rowe, S.M.; Deingruber, T.; Spring, D.R. The multifaceted nature of antimicrobial peptides: Current synthetic chemistry approaches and future directions. Chem. Soc. Rev. 2021, 50, 7820–7880. [Google Scholar] [CrossRef]
- Li, Y.; Wang, Y.; Wei, Q.; Zheng, X.; Tang, L.; Kong, D.; Gong, M. Variant fatty acid-like molecules Conjugation, novel approaches for extending the stability of therapeutic peptides. Sci. Rep. 2015, 5, 18039. [Google Scholar] [CrossRef] [PubMed]
- Shi, G.; Kang, X.; Dong, F.; Liu, Y.; Zhu, N.; Hu, Y.; Xu, H.; Lao, X.; Zheng, H. DRAMP 3.0: An enhanced comprehensive data repository of antimicrobial peptides. Nucleic Acids Res. 2022, 50, D488–D496. [Google Scholar] [CrossRef] [PubMed]
- Divyashree, M.; Mani, M.K.; Reddy, D.; Kumavath, R.; Ghosh, P.; Azevedo, V.; Barh, D. Clinical Applications of Antimicrobial Peptides (AMPs): Where do we Stand Now? Protein Pept. Lett. 2020, 27, 120–134. [Google Scholar] [CrossRef] [PubMed]
- Heidary, M.; Khosravi, A.D.; Khoshnood, S.; Nasiri, M.J.; Soleimani, S.; Goudarzi, M. Daptomycin. J. Antimicrob. Chemother. 2018, 73, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Rosenthal, S.; Decano, A.G.; Bandali, A.; Lai, D.; Malat, G.E.; Bias, T.E. Oritavancin (Orbactiv): A new-generation lipoglycopeptide for the treatment of acute bacterial skin and skin structure infections. PT 2018, 43, 143–179. [Google Scholar]

| Silkworm Disease | Causative Microorganism/s | Symptoms |
|---|---|---|
| Bacterial flacherie |
|
|
| Muscardine |
|
|
| Grasserie |
|
|
| Pebrine |
|
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Makwana, P.; Rahul, K.; Ito, K.; Subhadra, B. Diversity of Antimicrobial Peptides in Silkworm. Life 2023, 13, 1161. https://doi.org/10.3390/life13051161
Makwana P, Rahul K, Ito K, Subhadra B. Diversity of Antimicrobial Peptides in Silkworm. Life. 2023; 13(5):1161. https://doi.org/10.3390/life13051161
Chicago/Turabian StyleMakwana, Pooja, Kamidi Rahul, Katsuhiko Ito, and Bindu Subhadra. 2023. "Diversity of Antimicrobial Peptides in Silkworm" Life 13, no. 5: 1161. https://doi.org/10.3390/life13051161





