The Quantification of Bacterial Cell Size: Discrepancies Arise from Varied Quantification Methods
Abstract
1. Introduction
2. Quantification Methods Based on Microscopic Images
3. Quantification Methods Not Reliant on Microscopic Images
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Elledge, S.J. Cell cycle checkpoints: Preventing an identity crisis. Science 1996, 274, 1664–1672. [Google Scholar] [CrossRef] [PubMed]
- Cooper, S.; Helmstetter, C.E. Chromosome replication and the division cycle of Escherichia coli B/r. J. Mol. Biol. 1968, 31, 519–540. [Google Scholar] [CrossRef] [PubMed]
- Jun, S.; Si, F.; Pugatch, R.; Scott, M. Fundamental principles in bacterial physiology-history, recent progress, and the future with focus on cell size control: A review. Rep. Prog. Phys. 2018, 81, 056601. [Google Scholar] [CrossRef] [PubMed]
- Schaechter, M.; Maaløe, O.; Kjeldgaard, N.O. Dependency on medium and temperature of cell size and chemical composition during balanced growth of Salmonella typhimurium. Microbiology 1958, 19, 592–606. [Google Scholar] [CrossRef]
- Helmstetter, C.E.; Cummings, D.J. Bacterial Synchronization by Selection of Cells at Division. Proc. Natl. Acad. Sci. USA 1963, 50, 767–774. [Google Scholar] [CrossRef]
- Helmstetter, C.E.; Eenhuis, C.; Theisen, P.; Grimwade, J.; Leonard, A.C. Improved bacterial baby machine: Application to Escherichia coli K-12. J. Bacteriol. 1992, 174, 3445–3449. [Google Scholar] [CrossRef][Green Version]
- Bates, D.; Epstein, J.; Boye, E.; Fahrner, K.; Berg, H.; Kleckner, N. The Escherichia coli baby cell column: A novel cell synchronization method provides new insight into the bacterial cell cycle. Mol. Microbiol. 2005, 57, 380–391. [Google Scholar] [CrossRef]
- Helmstetter, C.E. Rate of DNA synthesis during the division cycle of Escherichia coli B/r. J. Mol. Biol. 1967, 24, 417–427. [Google Scholar] [CrossRef]
- Helmstetter, C.E.; Cooper, S. DNA synthesis during the division cycle of rapidly growing Escherichia coli B/r. J. Mol. Biol. 1968, 31, 507–518. [Google Scholar] [CrossRef]
- Donachie, W.D. Relationship between cell size and time of initiation of DNA replication. Nature 1968, 219, 1077–1079. [Google Scholar] [CrossRef]
- Koppes, L.; Meyer, M.; Oonk, H.; De Jong, M.; Nanninga, N. Correlation between size and age at different events in the cell division cycle of Escherichia coli. J. Bacteriol. 1980, 143, 1241–1252. [Google Scholar] [CrossRef] [PubMed]
- Boye, E.; Stokke, T.; Kleckner, N.; Skarstad, K. Coordinating DNA replication initiation with cell growth: Differential roles for DnaA and SeqA proteins. Proc. Natl. Acad. Sci. USA 1996, 93, 12206–12211. [Google Scholar] [CrossRef] [PubMed]
- Hill, N.S.; Kadoya, R.; Chattoraj, D.K.; Levin, P.A. Cell size and the initiation of DNA replication in bacteria. PLoS Genet. 2012, 8, e1002549. [Google Scholar] [CrossRef] [PubMed]
- Wallden, M.; Fange, D.; Lundius, E.G.; Baltekin, Ö.; Elf, J. The synchronization of replication and division cycles in individual E. coli cells. Cell 2016, 166, 729–739. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Ho, P.-Y.; Jiang, M.; Tang, B.; Liu, W.; Li, D.; Yu, X.; Kleckner, N.E.; Amir, A.; Liu, C. Interrogating the Escherichia coli cell cycle by cell dimension perturbations. Proc. Natl. Acad. Sci. USA 2016, 113, 15000–15005. [Google Scholar] [CrossRef] [PubMed]
- Si, F.; Li, D.; Cox, S.E.; Sauls, J.T.; Azizi, O.; Sou, C.; Schwartz, A.B.; Erickstad, M.J.; Jun, Y.; Li, X. Invariance of initiation mass and predictability of cell size in Escherichia coli. Curr. Biol. 2017, 27, 1278–1287. [Google Scholar] [CrossRef]
- Govers, S.K.; Campos, M.; Tyagi, B.; Laloux, G.; Jacobs-Wagner, C. Apparent simplicity and emergent robustness in bacterial cell cycle control. bioRxiv 2023. [Google Scholar] [CrossRef]
- Helmstetter, C.E. Initiation of chromosome replication in Escherichia coli: II. Analysis of the control mechanism. J. Mol. Biol. 1974, 84, 21–36. [Google Scholar] [CrossRef]
- Churchward, G.; Estiva, E.; Bremer, H. Growth rate-dependent control of chromosome replication initiation in Escherichia coli. J. Bacteriol. 1981, 145, 1232–1238. [Google Scholar] [CrossRef]
- Zaritsky, A.; Zabrovitz, S. DNA synthesis in Escherichia coli during a nutritional shift-up. Mol. Gen. Genet. 1981, 181, 564–566. [Google Scholar] [CrossRef]
- Wold, S.; Skarstad, K.; Steen, H.; Stokke, T.; Boye, E. The initiation mass for DNA replication in Escherichia coli K-12 is dependent on growth rate. EMBO J. 1994, 13, 2097–2102. [Google Scholar] [CrossRef] [PubMed]
- Bates, D.W.; Kleckner, N. Chromosome and replisome dynamics in E. coli: Loss of sister cohesion triggers global chromosome movement and mediates chromosome segregation. Cell 2005, 121, 899–911. [Google Scholar] [CrossRef]
- Zheng, H.; Bai, Y.; Jiang, M.; Tokuyasu, T.A.; Huang, X.; Zhong, F.; Wu, Y.; Fu, X.; Kleckner, N.; Hwa, T.; et al. General quantitative relations linking cell growth and the cell cycle in Escherichia coli. Nat. Microbiol. 2020, 5, 995–1001. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Marshall, D. Microfluidics for single cell analysis. Curr. Opin. Biotechnol. 2012, 23, 110–119. [Google Scholar] [CrossRef] [PubMed]
- Grünberger, A.; Wiechert, W.; Kohlheyer, D. Single-cell microfluidics: Opportunity for bioprocess development. Curr. Opin. Biotechnol. 2014, 29, 15–23. [Google Scholar] [CrossRef]
- Duncombe, T.A.; Tentori, A.M.; Herr, A.E. Microfluidics: Reframing biological enquiry. Nat. Rev. Mol. Cell Biol. 2015, 16, 554–567. [Google Scholar] [CrossRef]
- Potvin-Trottier, L.; Luro, S.; Paulsson, J. Microfluidics and single-cell microscopy to study stochastic processes in bacteria. Curr. Opin. Microbiol. 2018, 43, 186–192. [Google Scholar] [CrossRef]
- Gahlmann, A.; Moerner, W.E. Exploring bacterial cell biology with single-molecule tracking and super-resolution imaging. Nat. Rev. Microbiol. 2014, 12, 9–22. [Google Scholar] [CrossRef]
- Kuwada, N.J.; Traxler, B.; Wiggins, P.A. High-throughput cell-cycle imaging opens new doors for discovery. Curr. Genet. 2015, 61, 513–516. [Google Scholar] [CrossRef]
- Caicedo, J.C.; Cooper, S.J.; Heigwer, F.; Warchal, S.J.; Qiu, P.; Molnar, C.; Vasilevich, A.; Barry, J.D.; Bansal, H.S.; Kraus, O.Z. Data-analysis strategies for image-based cell profiling. Nat. Methods 2017, 14, 849–863. [Google Scholar] [CrossRef]
- Smith, K.; Piccinini, F.; Balassa, T.; Koos, K.; Danka, T.; Azizpour, H.; Horvath, P. Phenotypic Image Analysis Software Tools for Exploring and Understanding Big Image Data from Cell-Based Assays. Cell Syst. 2018, 6, 636–653. [Google Scholar] [CrossRef] [PubMed]
- Jeckel, H.; Drescher, K. Advances and opportunities in image analysis of bacterial cells and communities. FEMS Microbiol. Rev. 2021, 45, fuaa062. [Google Scholar] [CrossRef] [PubMed]
- Taheri-Araghi, S.; Brown, S.D.; Sauls, J.T.; McIntosh, D.B.; Jun, S. Single-Cell Physiology. Annu. Rev. Biophys. 2015, 44, 123–142. [Google Scholar] [CrossRef] [PubMed]
- Campos, M.; Surovtsev, I.V.; Kato, S.; Paintdakhi, A.; Beltran, B.; Ebmeier, S.E.; Jacobswagner, C. A Constant Size Extension Drives Bacterial Cell Size Homeostasis. Cell 2014, 159, 1433–1446. [Google Scholar] [CrossRef]
- Taheriaraghi, S.; Bradde, S.; Sauls, J.T.; Hill, N.S.; Levin, P.A.; Paulsson, J.; Vergassola, M.; Jun, S. Cell-Size Control and Homeostasis in Bacteria. Curr. Biol. 2015, 25, 385–391. [Google Scholar] [CrossRef]
- Jun, S.; Taheri-Araghi, S. Cell-size maintenance: Universal strategy revealed. Trends Microbiol. 2015, 23, 4–6. [Google Scholar] [CrossRef]
- Lau, I.; Filipe, S.R.; Soballe, B.; Okstad, O.; Barre, F.; Sherratt, D.J. Spatial and temporal organization of replicating Escherichia coli chromosomes. Mol. Microbiol. 2004, 49, 731–743. [Google Scholar] [CrossRef]
- Wang, X.; Possoz, C.; Sherratt, D.J. Dancing around the divisome: Asymmetric chromosome segregation in Escherichia coli. Genes Dev. 2005, 19, 2367–2377. [Google Scholar] [CrossRef][Green Version]
- Joshi, M.C.; Bourniquel, A.; Fisher, J.K.; Ho, B.T.; Magnan, D.; Kleckner, N.; Bates, D. Escherichia coli sister chromosome separation includes an abrupt global transition with concomitant release of late-splitting intersister snaps. Proc. Natl. Acad. Sci. USA 2011, 108, 2765–2770. [Google Scholar] [CrossRef]
- Santi, I.; Mckinney, J.D. Chromosome Organization and Replisome Dynamics in Mycobacterium smegmatis. Mbio 2015, 6, e01999-14. [Google Scholar] [CrossRef]
- Reyes-Lamothe, R.; Sherratt, D.J.; Leake, M.C. Stoichiometry and architecture of active DNA replication machinery in Escherichia coli. Science 2010, 328, 498–501. [Google Scholar] [CrossRef] [PubMed]
- Trojanowski, D.; Ginda, K.; Pioro, M.; Holowka, J.; Skut, P.; Jakimowicz, D.; Zakrzewskaczerwinska, J. Choreography of the Mycobacterium Replication Machinery during the Cell Cycle. mBio 2015, 6, e02125-14. [Google Scholar] [CrossRef] [PubMed]
- Mangiameli, S.M.; Veit, B.T.; Merrikh, H.; Wiggins, P.A. The Replisomes Remain Spatially Proximal throughout the Cell Cycle in Bacteria. PLoS Genet. 2017, 13, e1006582. [Google Scholar] [CrossRef] [PubMed]
- Iyer-Biswas, S.; Wright, C.S.; Henry, J.T.; Lo, K.; Burov, S.; Lin, Y.; Crooks, G.E.; Crosson, S.; Dinner, A.R.; Scherer, N.F. Scaling laws governing stochastic growth and division of single bacterial cells. Proc. Natl. Acad. Sci. USA 2014, 111, 15912–15917. [Google Scholar] [CrossRef]
- Tanouchi, Y.; Pai, A.; Park, H.; Huang, S.; Stamatov, R.; Buchler, N.E.; You, L. A noisy linear map underlies oscillations in cell size and gene expression in bacteria. Nature 2015, 523, 357–360. [Google Scholar] [CrossRef]
- Adiciptaningrum, A.; Osella, M.; Moolman, M.C.; Cosentino Lagomarsino, M.; Tans, S.J. Stochasticity and homeostasis in the E. coli replication and division cycle. Sci. Rep. 2015, 5, 18261. [Google Scholar] [CrossRef]
- Si, F.; Treut, G.L.; Sauls, J.T.; Vadia, S.; Levin, P.A.; Jun, S. Mechanistic Origin of Cell-Size Control and Homeostasis in Bacteria. Curr. Biol. 2019, 29, 1760. [Google Scholar] [CrossRef]
- Witz, G.; van Nimwegen, E.; Julou, T. Initiation of chromosome replication controls both division and replication cycles in E. coli through a double-adder mechanism. Elife 2019, 8, e48063. [Google Scholar] [CrossRef]
- Colin, A.; Micali, G.; Faure, L.; Cosentino Lagomarsino, M.; van Teeffelen, S. Two different cell-cycle processes determine the timing of cell division in Escherichia coli. Elife 2021, 10, e67495. [Google Scholar] [CrossRef]
- Ducret, A.; Quardokus, E.M.; Brun, Y.V. MicrobeJ, a tool for high throughput bacterial cell detection and quantitative analysis. Nat Microbiol 2016, 1, 16077. [Google Scholar] [CrossRef]
- Paintdakhi, A.; Parry, B.; Campos, M.; Irnov, I.; Elf, J.; Surovtsev, I.; Jacobs-Wagner, C. Oufti: An integrated software package for high-accuracy, high-throughput quantitative microscopy analysis. Mol. Microbiol. 2016, 99, 767–777. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, R.; van Teeseling, M.C.F.; Thanbichler, M.; Drescher, K. BacStalk: A comprehensive and interactive image analysis software tool for bacterial cell biology. Mol. Microbiol. 2020, 114, 140–150. [Google Scholar] [CrossRef]
- McQuin, C.; Goodman, A.; Chernyshev, V.; Kamentsky, L.; Cimini, B.A.; Karhohs, K.W.; Doan, M.; Ding, L.; Rafelski, S.M.; Thirstrup, D.; et al. CellProfiler 3.0: Next-generation image processing for biology. PLoS Biol. 2018, 16, e2005970. [Google Scholar] [CrossRef] [PubMed]
- Goñi-Moreno, Á.; Kim, J.; de Lorenzo, V. CellShape: A user-friendly image analysis tool for quantitative visualization of bacterial cell factories inside. Biotechnol. J. 2017, 12, 1600323. [Google Scholar] [CrossRef]
- Shal, K.; Choudhry, M.S. Evolution of Deep Learning Algorithms for MRI-Based Brain Tumor Image Segmentation. Crit. Rev. Biomed. Eng. 2021, 49, 77–94. [Google Scholar] [CrossRef] [PubMed]
- Cutler, K.J.; Stringer, C.; Lo, T.W.; Rappez, L.; Stroustrup, N.; Brook Peterson, S.; Wiggins, P.A.; Mougous, J.D. Omnipose: A high-precision morphology-independent solution for bacterial cell segmentation. Nat. Methods 2022, 19, 1438–1448. [Google Scholar] [CrossRef] [PubMed]
- Seo, H.; Badiei Khuzani, M.; Vasudevan, V.; Huang, C.; Ren, H.; Xiao, R.; Jia, X.; Xing, L. Machine learning techniques for biomedical image segmentation: An overview of technical aspects and introduction to state-of-art applications. Med. Phys. 2020, 47, e148–e167. [Google Scholar] [CrossRef] [PubMed]
- Malhotra, P.; Gupta, S.; Koundal, D.; Zaguia, A.; Enbeyle, W. Deep Neural Networks for Medical Image Segmentation. J. Health Eng. 2022, 2022, 9580991. [Google Scholar] [CrossRef]
- Spahn, C.; Gómez-de-Mariscal, E.; Laine, R.F.; Pereira, P.M.; von Chamier, L.; Conduit, M.; Pinho, M.G.; Jacquemet, G.; Holden, S.; Heilemann, M.; et al. DeepBacs for multi-task bacterial image analysis using open-source deep learning approaches. Commun. Biol. 2022, 5, 688. [Google Scholar] [CrossRef]
- Fishov, I.; Zaritsky, A.; Grover, N.B. On microbial states of growth. Mol. Microbiol. 1995, 15, 789–794. [Google Scholar] [CrossRef]
- Neidhardt, F.C. Bacterial Growth: Constant Obsession with dN/dt. J. Bacteriol. 1999, 181, 7405–7408. [Google Scholar] [CrossRef] [PubMed]
- Neidhardt, F.C. Apples, oranges and unknown fruit. Nat. Rev. Microbiol. 2006, 4, 876. [Google Scholar] [CrossRef] [PubMed]
- Falk, T.; Mai, D.; Bensch, R.; Çiçek, Ö.; Abdulkadir, A.; Marrakchi, Y.; Böhm, A.; Deubner, J.; Jäckel, Z.; Seiwald, K.; et al. U-Net: Deep learning for cell counting, detection, and morphometry. Nat. Methods 2019, 16, 67–70. [Google Scholar] [CrossRef] [PubMed]
- Lugagne, J.B.; Lin, H.; Dunlop, M.J. DeLTA: Automated cell segmentation, tracking, and lineage reconstruction using deep learning. PLoS Comput. Biol. 2020, 16, e1007673. [Google Scholar] [CrossRef]
- Bremer, H.; Churchward, G.; Young, R. Relation between growth and replication in bacteria. J. Theor. Biol. 1979, 81, 533–545. [Google Scholar] [CrossRef]
- Cooper, S. 1-Bacterial Growth. In Bacterial Growth and Division; Cooper, S., Ed.; Academic Press: San Diego, CA, USA, 1991; pp. 7–17. [Google Scholar]
- Sutton, S. Accuracy of Plate Counts. J. Valid. Technol. 2011, 17, 42–46. [Google Scholar]
- Ou, F.; Mcgoverin, C.; Swift, S.; Vanholsbeeck, F. Absolute bacterial cell enumeration using flow cytometry. J. Appl. Microbiol. 2017, 123, 464–477. [Google Scholar] [CrossRef]
- Von Freiesleben, U.; Rasmussen, K.V.; Atlung, T.; Hansen, F.G. Rifampicin-resistant initiation of chromosome replication from oriC in ihf mutants. Mol. Microbiol. 2000, 37, 1087–1093. [Google Scholar] [CrossRef]
- Morigen; Molina, F.; Skarstad, K. Deletion of the datA site does not affect once-per-cell-cycle timing but induces rifampin-resistant replication. J. Bacteriol. 2005, 187, 3913–3920. [Google Scholar] [CrossRef]
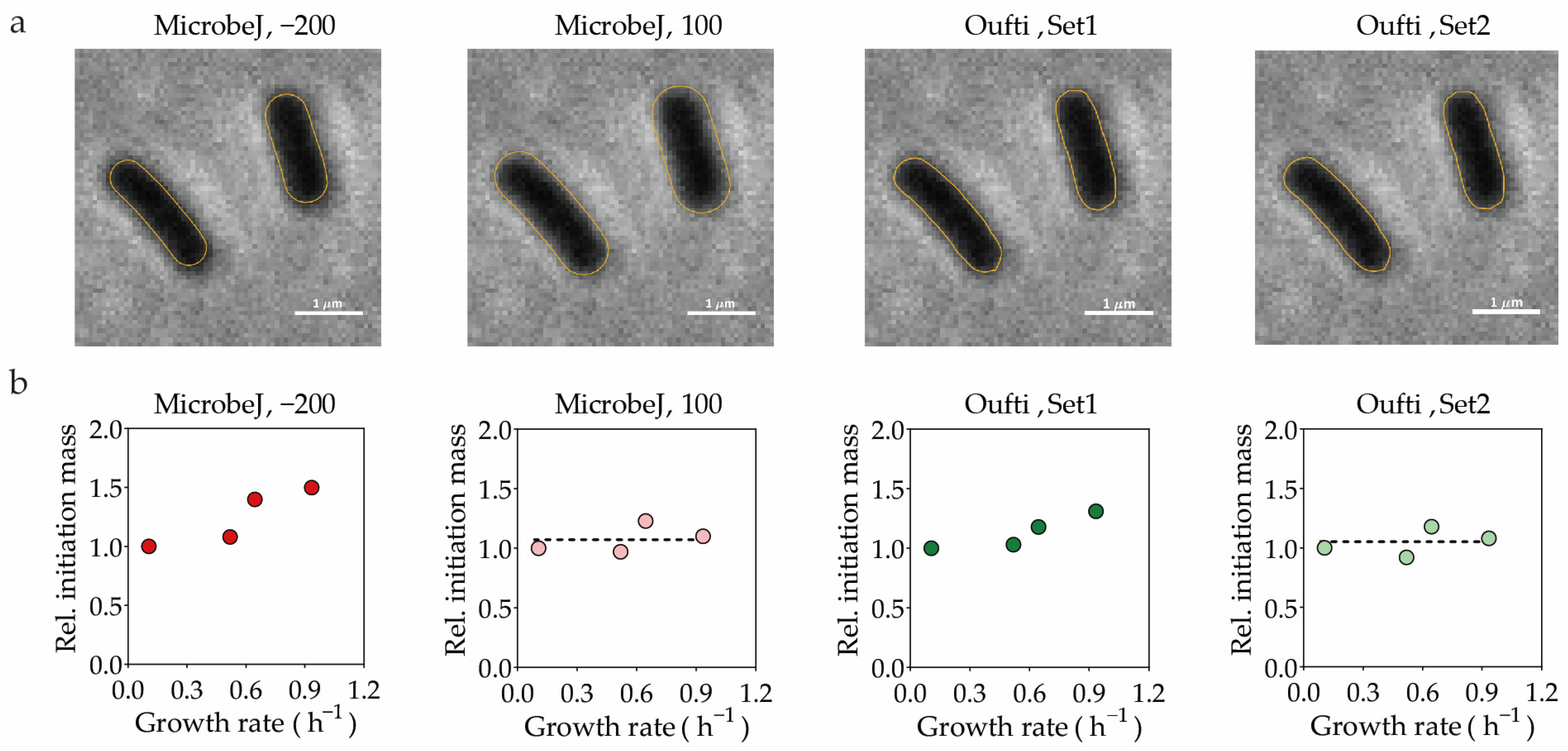

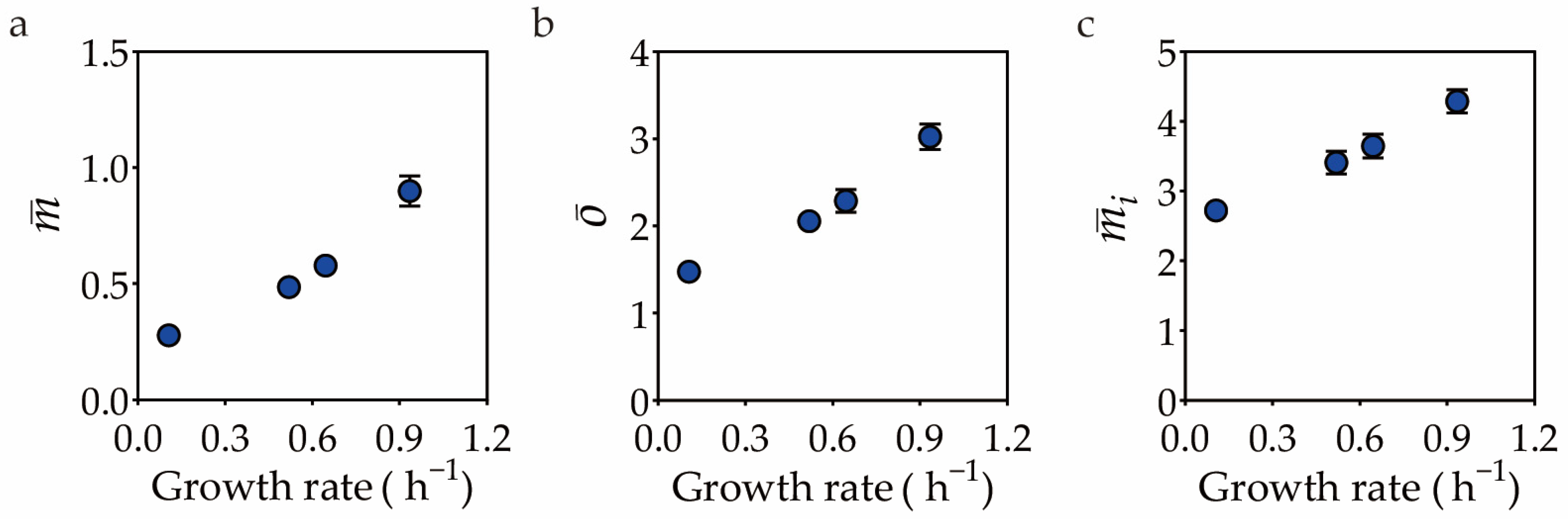
| Features 1 | Medium 2 | MicrobeJ | MicrobeJ | Oufti | Oufti | BacStalk | Custom |
|---|---|---|---|---|---|---|---|
| −200 | 100 | Set1 | Set2 | Scripts 3 | |||
| Length (µm) | Glutamine | 1.85 ± 0.44 | 2.16 ± 0.47 | 2.03 ± 0.47 | 2.04 ± 0.47 | 2.25 ± 0.48 | 2.11 ± 0.54 |
| Alanine | 2.22 ± 0.59 | 2.51 ± 0.67 | 2.38 ± 0.56 | 2.38 ± 0.56 | 2.63 ± 0.66 | 2.35 ± 0.45 | |
| Glycerol | 2.61 ± 0.67 | 2.94 ± 0.72 | 2.77 ± 0.65 | 2.78 ± 0.64 | 3.04 ± 0.69 | 2.79 ± 0.88 | |
| Glucose | 2.73 ± 0.65 | 2.97 ± 0.68 | 2.81 ± 0.67 | 2.80 ± 0.66 | 3.11 ± 0.67 | 2.88 ± 0.60 | |
| Width 4 (µm) | Glutamine | 0.55 ± 0.06 | 0.75 ± 0.06 | 0.52 ± 0.03 | 0.55 ± 0.03 | 0.79 ± 0.06 | 0.55 ± 0.05 |
| Alanine | 0.61 ± 0.06 | 0.81 ± 0.06 | 0.57 ± 0.02 | 0.57 ± 0.02 | 0.83 ± 0.06 | 0.60 ± 0.05 | |
| Glycerol | 0.67 ± 0.08 | 0.88 ± 0.08 | 0.60 ± 0.03 | 0.64 ± 0.03 | 0.90 ± 0.07 | 0.65 ± 0.05 | |
| Glucose | 0.79 ± 0.08 | 0.96 ± 0.08 | 0.72 ± 0.04 | 0.69 ± 0.03 | 1.00 ± 0.07 | 0.74 ± 0.06 | |
| Area (µm2) | Glutamine | 0.94 ± 0.26 | 1.51 ± 0.37 | 1.04 ± 0.25 | 1.10 ± 0.27 | 1.17 ± 0.26 | 1.02 ± 0.22 |
| Alanine | 1.28 ± 0.36 | 1.90 ± 0.54 | 1.35 ± 0.33 | 1.35 ± 0.33 | 1.47 ± 0.36 | 1.25 ± 0.24 | |
| Glycerol | 1.66 ± 0.48 | 2.46 ± 0.66 | 1.64 ± 0.42 | 1.74 ± 0.43 | 1.86 ± 0.44 | 1.60 ± 0.30 | |
| Glucose | 2.03 ± 0.52 | 2.70 ± 0.67 | 2.00 ± 0.51 | 1.92 ± 0.49 | 2.13 ± 0.47 | 1.90 ± 0.39 | |
| Volume 5 (µm3) | Glutamine | 0.39 ± 0.13 | 0.85 ± 0.24 | 0.44 ± 0.11 | 0.49 ± 0.13 | 0.96 ± 0.25 | 0.46 ± 0.13 |
| Alanine | 0.59 ± 0.19 | 1.15 ± 0.37 | 0.63 ± 0.16 | 0.63 ± 0.16 | 1.26 ± 0.36 | 0.59 ± 0.12 | |
| Glycerol | 0.85 ± 0.31 | 1.62 ± 0.51 | 0.80 ± 0.22 | 0.90 ± 0.23 | 1.74 ± 0.48 | 0.84 ± 0.26 | |
| Glucose | 1.21 ± 0.37 | 1.92 ± 0.56 | 1.18 ± 0.31 | 1.09 ± 0.29 | 2.18 ± 0.57 | 1.11 ± 0.25 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, Q.; Huang, W.; Zhang, Z.; Chu, P.; Wei, T.; Zheng, H.; Liu, C. The Quantification of Bacterial Cell Size: Discrepancies Arise from Varied Quantification Methods. Life 2023, 13, 1246. https://doi.org/10.3390/life13061246
Cao Q, Huang W, Zhang Z, Chu P, Wei T, Zheng H, Liu C. The Quantification of Bacterial Cell Size: Discrepancies Arise from Varied Quantification Methods. Life. 2023; 13(6):1246. https://doi.org/10.3390/life13061246
Chicago/Turabian StyleCao, Qian’andong, Wenqi Huang, Zheng Zhang, Pan Chu, Ting Wei, Hai Zheng, and Chenli Liu. 2023. "The Quantification of Bacterial Cell Size: Discrepancies Arise from Varied Quantification Methods" Life 13, no. 6: 1246. https://doi.org/10.3390/life13061246
APA StyleCao, Q., Huang, W., Zhang, Z., Chu, P., Wei, T., Zheng, H., & Liu, C. (2023). The Quantification of Bacterial Cell Size: Discrepancies Arise from Varied Quantification Methods. Life, 13(6), 1246. https://doi.org/10.3390/life13061246








