Mechanisms of Germline Stem Cell Competition across Species
Abstract
1. Introduction
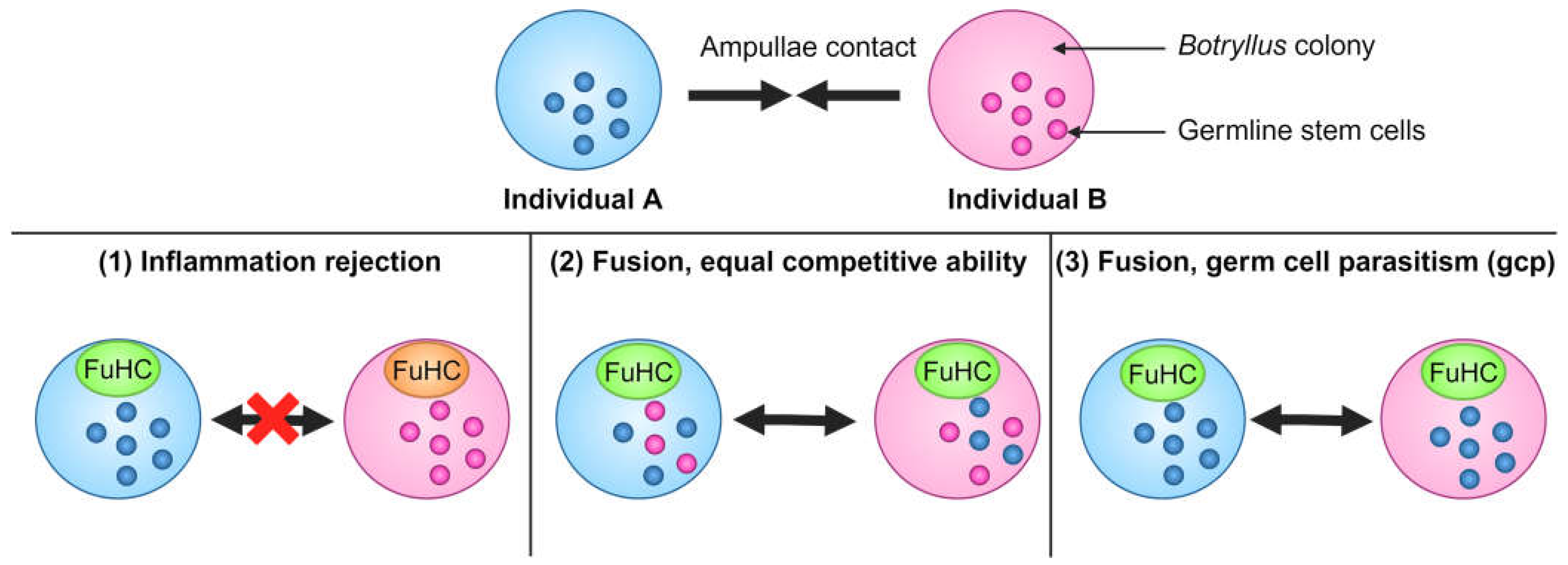
2. Early Studies of Germline Stem Cell Competition
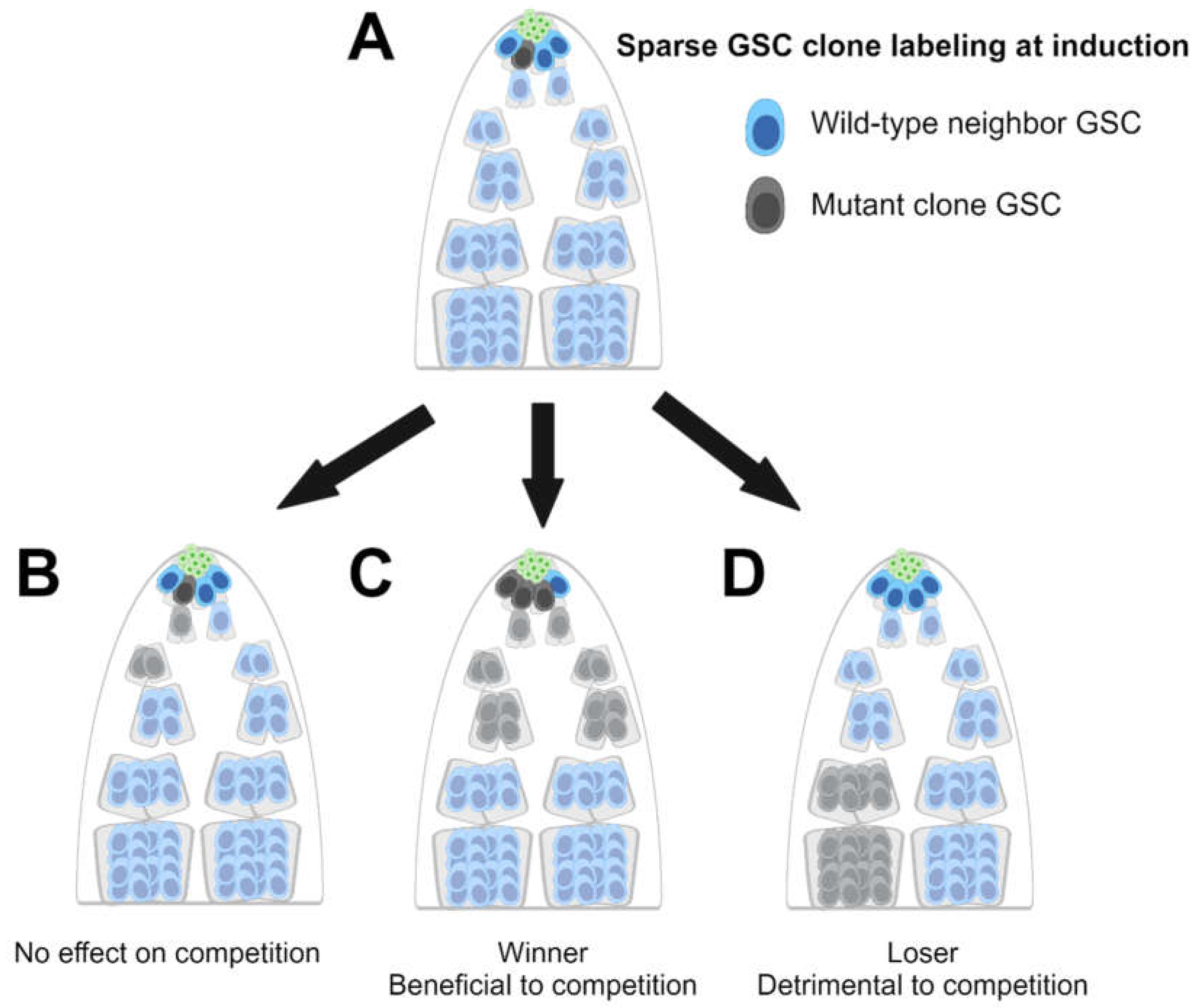
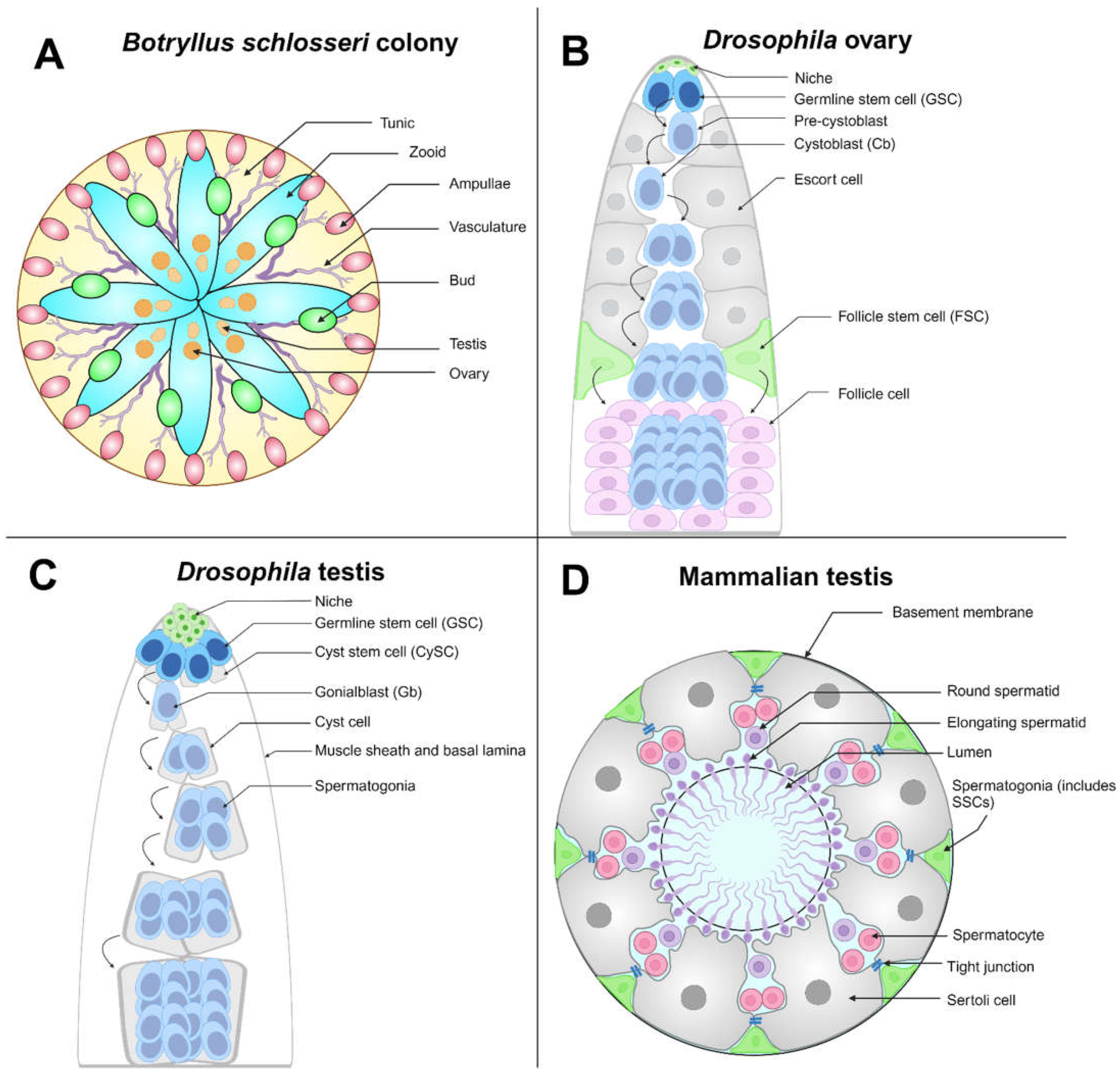
3. The Drosophila Germline Has Elucidated Cell Competition Mechanisms
4. Somatic Adult Stem Cells Compete for Niche Access in Gonads
5. Somatic Adult Stem Cells Compete for Niche Access in the Mammalian Intestine
6. Signaling Pathways in Somatic ASC Competition
7. Germline Stem Cell Competition Is Linked to Cancer
8. Adult Stem Cell Competition Causes Age-Related Disease
9. Paternal Age Effect (PAE) Disorders Are a Negative Outcome of Germline Competition
10. Discussion
| Gene | Organism/Tissue | Function | Role in Cell Competition | References |
|---|---|---|---|---|
| Germline stem cells | ||||
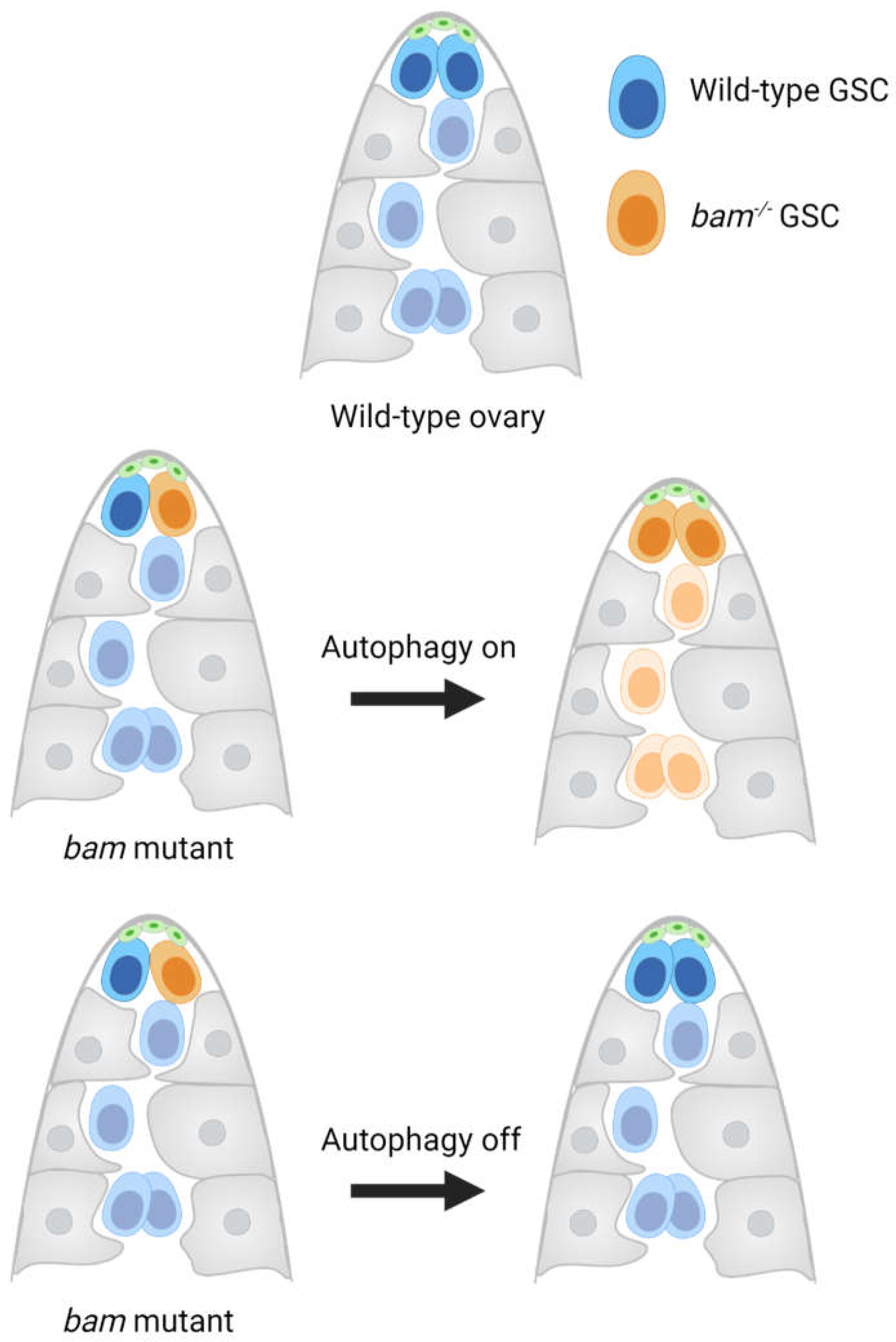
| bag of marbles (bam) | Drosophila ovary | Promotes differentiation of pre-cystoblast cells into cystoblasts | bam-mutant germline stem cell (GSC) clones upregulate autophagy leads and accumulate in the ovary stem cell niche. The role of E-Cadherin (E-Cad) in promoting the competitive abilities of bam-mutant GSCs is disputed. | [81,82,83] |
| benign gonial cell neoplasm (bgcn) | Drosophila ovary | Promotes differentiation of pre-cystoblast cells into cystoblasts | bgcn-mutant GSC clones upregulate E-Cad and force wild-type GSCs out of the ovarian stem cell niche. | [81] |
| Myc | Drosophila ovary | Growth-promoting pathway component | The role of Myc in competition is disputed. GSC clones with elevated Myc outcompete wild-type neighbors, suggesting that GSCs with lower Myc are replaced by those with higher Myc. However, Myc-null GSC clones are not outcompeted by wild-type GSCs. | [81,85] |
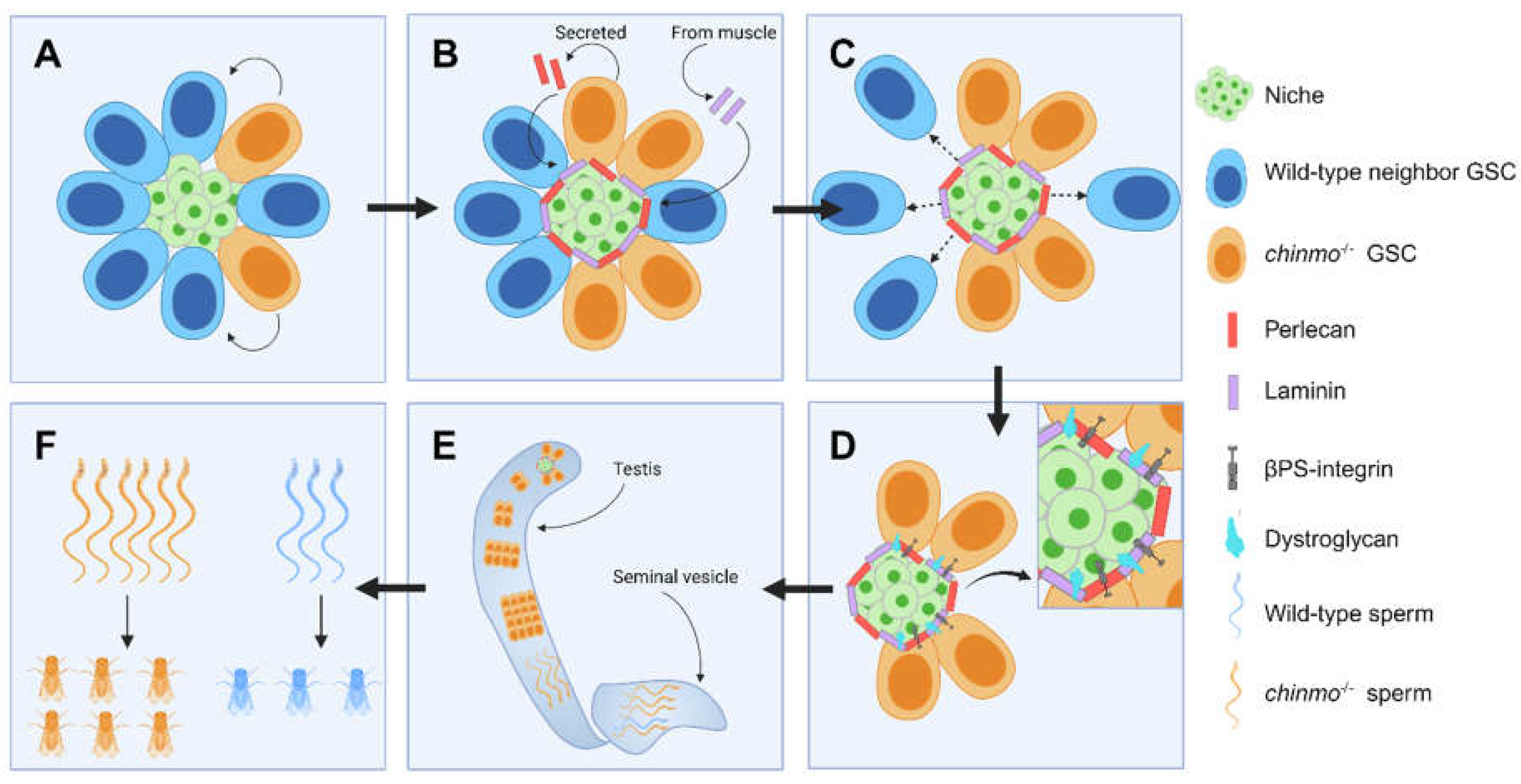
| chronologically inappropriate morphogenesis (chinmo) | Drosophila testis | Transcription factor which regulates neuronal temporal patterning; regulates eye development; maintains CySC sexual identity | chinmo-mutant GSC clones outcompete wild-type neighbors for niche access by forming an ECM ‘moat’ around the niche. | [92] |
| FGFR2 | Human testis | Growth-promoting pathway component | The FGFR2S252W gain-of-function allele is linked to the paternal age affect (PAE) disorder Apert syndrome. FGFR2 is presumed to have a role in SSC competition, but has not yet been tested in humans. | [179] |
| FGFR3 | Human testis | Growth-promoting pathway component | The FGFR3G380R gain-of-function allele is linked to the PAE disorder achondroplasia. FGFR3 is presumed to have a role in SSC competition, but has not yet been tested in humans. | [178] |
| HRAS | Human testis | RAS proteins downstream of several receptor tyrosine kinases (RTKs) | HRAS gain-of-function alleles are linked to the PAE disorder Costello syndrome. HRAS is presumed to have a role in SSC competition, but has not yet been tested in humans. | [182] |
| PTPN11 | Human testis | Protein tyrosine phosphatase downstream of several RTKs | PTPN11 gain-of-function alleles are linked to the PAE disorder Noonan syndrome. PTPN11 is presumed to have a role in SSC competition, but has not yet been tested in humans. | [181] |
| Fibroblast growth factor receptor (FGFR2) | Mouse testis | Growth-promoting pathway component | Murine spermatogonial stem cell (SSCs) expressing the Apert syndrome FGFR2S252W gain-of-function allele have increased competitiveness in in vitro and in vivo models. | [192] |
| Somatic adult stem cells | ||||
| hippo (hpo) | Drosophila ovary | Tumor suppressor which negatively regulates Yorkie, the Drosophila homolog of YAP | hpo-mutant follicle stem cell (FSC) clones outcompete wild-type neighbor FSCs. | [106] |
| hopscotch (hop) | Drosophila ovary | Janus tyrosine kinase, part of JAK/STAT pathway | hop-over-expressing FSC clones outcompete wild-type FSCs. | [105] |
| patched (ptc) | Drosophila ovary | Hedgehog pathway component; tumor suppressor | ptc-mutant FSC clones outcompete wild-type FSCs. | [105] |
| yorkie (yki) | Drosophila ovary | Growth-promoting pathway component | FSC clones over-expressing ykiS168A—a gain-of-function mutation—outcompete wild-type FSCs. | [106] |
| Abelson (Abl) | Drosophila testis | Kinase regulating growth, differentiation and adhesion | Abl-mutant cyst stem cell (CySC) clones outcompete wild-type neighbor CySCs. | [99] |
| hippo (hpo) | Drosophila testis | Tumor suppressor, negatively regulates Yorkie, the Drosophila homolog of YAP | hpo-mutant CySC clones outcompete wild-type neighbor CySCs. | [30] |
| patched (ptc) | Drosophila testis | Hedgehog pathway component; tumor suppressor | ptc-mutant CySC clones outcompete wild-type neighbor CySCs. | [30] |
| Ras | Drosophila testis | Kinase regulating growth | CySC clones over-expressing RasGV12—a gain-of-function mutation—outcompete wild-type neighbor CySCs. | [96] |
| Suppressor of cytokine signaling at 36E (Socs36E) | Drosophila testis | Negative regulator of JAK/STAT and EGFR | Socs36E-mutant CySC clones outcompete wild-type neighbor CySCs. | [96] |
| APC | Mammalian intestine | Tumor suppressor | APC-mutant intestinal stem cell (ISC) clones outcompete wild-type ISCs, creating a monoclonal crypt. | [29] |
| Kras | Mammalian intestine | Kinase regulating growth | ISC clones over-expressing KRASG12D—a gain-of-function mutation—outcompete wild-type ISCs, creating a monoclonal crypt. | [29] |
| DNMT3A | Human bone marrow | DNA methylation enzyme | Dnmt3A-mutant hematopoietic stem cells (HSCs) outcompete wild-type HSCs in competitive serial transplantation assays. | [144] |
| Tet2 | Human bone marrow | DNA methylcytosine dioxygenase | Tet2-mutant HSCs outcompete wild-type HSCs in a competitive transplantation assay. | [145] |
| Tp53 | Human bone marrow | Tumor suppressor | Tp53 status does not impact HSC competition during homeostasis, but after DNA damage, Tp53−/− HSCs have a competitive advantage over wild-type HSCs in mosaic animals. | [151] |
| Gene | Organism/Tissue | Function | Role in Cell Competition | References |
|---|---|---|---|---|
| Jak2 | Human bone marrow | Tyrosine kinase in the JAK/STAT pathway | A gain-of-function mutation in human Jak2V617F causes bone marrow cells to outcompete wild-type cells via increased cell cycling in a competitive transplantation assay. | [122] |
| RAS | Mammalian epithelia in vitro and in vivo | Kinase regulating growth | Epithelial cells over-expressing RASG12V—a gain-of-function mutation—are outcompeted by wild-type epithelia in EDAC. | [123] |
| Tp53 | Mammalian epithelia in vitro and in vivo | Tumor suppressor | Epithelial cells expressing a dominant-negative Tp53 are outcompeted by wild-type epithelia in a process termed epithelial defense against cancer (EDAC). | [125] |
| PIK3CA | Mammalian esophagus | Catalytic subunit of phosphoinositide 3-kinase (PI3K) | Esophageal epithelial cells heterozygous for the gain-of-function mutation Pik3CAH1047R/+ outcompete wild-type esophageal cells via biased cell fate toward proliferation. | [121] |
| APC | Mammalian intestine | Tumor suppressor | APC-mutant intestinal stem cell (ISC) clones cause wild-type ISCs to differentiate, thereby outcompeting them and leading to intestinal tumor initiation. | [44,45] |
| YAP | Mammalian intestine | Growth-promoting factor in the Hippo pathway | Epithelial cells over-expressing YAP5SA—a gain-of-function mutation—are outcompeted by wild-type epithelia in EDAC. | [124] |
| YAP | Mammalian liver | Growth-promoting factor in the Hippo pathway | Elevating YAP activity in hepatocytes through the 5SA mutation can drive liver tumor growth, if YAP activity is not higher in neighboring hepatocytes. | [126] |
| Glial cell derived neurotrophic factor (GDNF) | Mammalian testis | Secreted TGF-β ligand which is required for SSC niche maintenance | Misexpression in SSCs produces malignant tumors with germline markers. | [183] |
Funding
Conflicts of Interest
References
- Amoyel, M.; Bach, E.A. Cell competition: How to eliminate your neighbours. Development 2014, 141, 988–1000. [Google Scholar] [CrossRef] [PubMed]
- Bowling, S.; Lawlor, K.; Rodriguez, T.A. Cell competition: The winners and losers of fitness selection. Development 2019, 146, dev167486. [Google Scholar] [CrossRef]
- Moreno, E.; Basler, K.; Morata, G. Cells compete for decapentaplegic survival factor to prevent apoptosis in Drosophila wing development. Nature 2002, 416, 755–759. [Google Scholar] [CrossRef] [PubMed]
- Tyler, D.M.; Li, W.; Zhuo, N.; Pellock, B.; Baker, N.E. Genes affecting cell competition in Drosophila. Genetics 2007, 175, 643–657. [Google Scholar] [CrossRef] [PubMed]
- Baumgartner, M.E.; Dinan, M.P.; Langton, P.F.; Kucinski, I.; Piddini, E. Proteotoxic stress is a driver of the loser status and cell competition. Nat. Cell Biol. 2021, 23, 136–146. [Google Scholar] [CrossRef] [PubMed]
- Kucinski, I.; Dinan, M.; Kolahgar, G.; Piddini, E. Chronic activation of JNK JAK/STAT and oxidative stress signalling causes the loser cell status. Nat. Commun. 2017, 8, 136. [Google Scholar] [CrossRef]
- Kiparaki, M.; Khan, C.; Folgado-Marco, V.; Chuen, J.; Moulos, P.; Baker, N.E. The transcription factor Xrp1 orchestrates both reduced translation and cell competition upon defective ribosome assembly or function. eLife 2022, 11, e71705. [Google Scholar] [CrossRef]
- Blanco, J.; Cooper, J.C.; Baker, N.E. Roles of C/EBP class bZip proteins in the growth and cell competition of Rp (‘Minute’) mutants in Drosophila. eLife 2020, 9, e50535. [Google Scholar] [CrossRef]
- Ji, Z.; Kiparaki, M.; Folgado, V.; Kumar, A.; Blanco, J.; Rimesso, G.; Chuen, J.; Liu, Y.; Zheng, D.; Baker, N.E. Drosophila RpS12 controls translation, growth, and cell competition through Xrp1. PLoS Genet. 2019, 15, e1008513. [Google Scholar] [CrossRef]
- Lee, C.H.; Kiparaki, M.; Blanco, J.; Folgado, V.; Ji, Z.; Kumar, A.; Rimesso, G.; Baker, N.E. A Regulatory Response to Ribosomal Protein Mutations Controls Translation, Growth, and Cell Competition. Dev. Cell 2018, 46, 456–469.e4. [Google Scholar] [CrossRef]
- Kale, A.; Ji, Z.; Kiparaki, M.; Blanco, J.; Rimesso, G.; Flibotte, S.; Baker, N.E. Ribosomal Protein S12e Has a Distinct Function in Cell Competition. Dev. Cell 2018, 44, 42–55.e4. [Google Scholar] [CrossRef] [PubMed]
- de la Cova, C.; Senoo-Matsuda, N.; Ziosi, M.; Wu, D.C.; Bellosta, P.; Quinzii, C.M.; Johnston, L.A. Supercompetitor status of Drosophila Myc cells requires p53 as a fitness sensor to reprogram metabolism and promote viability. Cell Metab. 2014, 19, 470–483. [Google Scholar] [CrossRef] [PubMed]
- Moreno, E.; Basler, K. dMyc transforms cells into super-competitors. Cell 2004, 117, 117–129. [Google Scholar] [CrossRef] [PubMed]
- Vincent, J.P.; Kolahgar, G.; Gagliardi, M.; Piddini, E. Steep differences in wingless signaling trigger myc-independent competitive cell interactions. Dev. Cell 2011, 21, 366–374. [Google Scholar] [CrossRef]
- Rodrigues, A.B.; Zoranovic, T.; Ayala-Camargo, A.; Grewal, S.; Reyes-Robles, T.; Krasny, M.; Wu, D.C.; Johnston, L.A.; Bach, E.A. Activated STAT regulates growth and induces competitive interactions independently of Myc, Yorkie, Wingless and ribosome biogenesis. Development 2012, 139, 4051–4061. [Google Scholar] [CrossRef]
- Neto-Silva, R.M.; de Beco, S.; Johnston, L.A. Evidence for a Growth-Stabilizing Regulatory Feedback Mechanism between Myc and Yorkie, the Drosophila Homolog of Yap. Dev. Cell 2010, 19, 507–520. [Google Scholar] [CrossRef]
- Ziosi, M.; Baena-Lopez, L.A.; Grifoni, D.; Froldi, F.; Pession, A.; Garoia, F.; Trotta, V.; Bellosta, P.; Cavicchi, S.; Pession, A. dMyc Functions Downstream of Yorkie to Promote the Supercompetitive Behavior of Hippo Pathway Mutant Cells. PLoS Genet. 2010, 6, e1001140. [Google Scholar] [CrossRef]
- de la Cova, C.; Abril, M.; Bellosta, P.; Gallant, P.; Johnston, L.A. Drosophila myc regulates organ size by inducing cell competition. Cell 2004, 117, 107–116. [Google Scholar] [CrossRef]
- Morata, G.; Ripoll, P. Minutes: Mutants of drosophila autonomously affecting cell division rate. Dev. Biol. 1975, 42, 211–221. [Google Scholar] [CrossRef]
- Marygold, S.J.; Roote, J.; Reuter, G.; Lambertsson, A.; Ashburner, M.; Millburn, G.H.; Harrison, P.M.; Yu, Z.; Kenmochi, N.; Kaufman, T.C.; et al. The ribosomal protein genes and Minute loci of Drosophila melanogaster. Genome Biol. 2007, 8, R216. [Google Scholar] [CrossRef]
- Kongsuwan, K.; Yu, Q.; Vincent, A.; Frisardi, M.C.; Rosbash, M.; Lengyel, J.A.; Merriam, J. A Drosophila Minute gene encodes a ribosomal protein. Nature 1985, 317, 555–558. [Google Scholar] [CrossRef] [PubMed]
- Abrams, J.M. Competition and compensation: Coupled to death in development and cancer. Cell 2002, 110, 403–406. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Oliver, E.R.; Saunders, T.L.; Tarle, S.A.; Glaser, T. Ribosomal protein L24 defect in belly spot and tail (Bst), a mouse Minute. Development 2004, 131, 3907–3920. [Google Scholar] [CrossRef]
- Claveria, C.; Giovinazzo, G.; Sierra, R.; Torres, M. Myc-driven endogenous cell competition in the early mammalian embryo. Nature 2013, 500, 39–44. [Google Scholar] [CrossRef]
- Simons, B.D.; Clevers, H. Strategies for homeostatic stem cell self-renewal in adult tissues. Cell 2011, 145, 851–862. [Google Scholar] [CrossRef] [PubMed]
- Stine, R.R.; Matunis, E.L. Stem cell competition: Finding balance in the niche. Trends Cell Biol. 2013, 23, 357–364. [Google Scholar] [CrossRef]
- Snippert, H.J.; van der Flier, L.G.; Sato, T.; van Es, J.H.; van den Born, M.; Kroon-Veenboer, C.; Barker, N.; Klein, A.M.; van Rheenen, J.; Simons, B.D.; et al. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell 2010, 143, 134–144. [Google Scholar] [CrossRef]
- Issigonis, M.; Tulina, N.; de Cuevas, M.; Brawley, C.; Sandler, L.; Matunis, E. JAK-STAT signal inhibition regulates competition in the Drosophila testis stem cell niche. Science 2009, 326, 153–156. [Google Scholar] [CrossRef]
- Vermeulen, L.; Morrissey, E.; van der Heijden, M.; Nicholson, A.M.; Sottoriva, A.; Buczacki, S.; Kemp, R.; Tavare, S.; Winton, D.J. Defining stem cell dynamics in models of intestinal tumor initiation. Science 2013, 342, 995–998. [Google Scholar] [CrossRef]
- Amoyel, M.; Simons, B.D.; Bach, E.A. Neutral competition of stem cells is skewed by proliferative changes downstream of Hh and Hpo. EMBO J. 2014, 33, 2295–2313. [Google Scholar] [CrossRef]
- Baker, A.M.; Cereser, B.; Melton, S.; Fletcher, A.G.; Rodriguez-Justo, M.; Tadrous, P.J.; Humphries, A.; Elia, G.; McDonald, S.A.; Wright, N.A.; et al. Quantification of crypt and stem cell evolution in the normal and neoplastic human colon. Cell Rep. 2014, 8, 940–947. [Google Scholar] [CrossRef] [PubMed]
- Snippert, H.J.; Schepers, A.G.; van Es, J.H.; Simons, B.D.; Clevers, H. Biased competition between Lgr5 intestinal stem cells driven by oncogenic mutation induces clonal expansion. EMBO Rep. 2014, 15, 62–69. [Google Scholar] [CrossRef] [PubMed]
- Otto, S.P.; Hastings, I.M. Mutation and selection within the individual. Genetica 1998, 102–103, 507–524. [Google Scholar] [CrossRef]
- Courret, C.; Chang, C.H.; Wei, K.H.; Montchamp-Moreau, C.; Larracuente, A.M. Meiotic drive mechanisms: Lessons from Drosophila. Proc. Biol. Sci. 2019, 286, 20191430. [Google Scholar] [CrossRef]
- Lindholm, A.K.; Dyer, K.A.; Firman, R.C.; Fishman, L.; Forstmeier, W.; Holman, L.; Johannesson, H.; Knief, U.; Kokko, H.; Larracuente, A.M.; et al. The Ecology and Evolutionary Dynamics of Meiotic Drive. Trends Ecol. Evol. 2016, 31, 315–326. [Google Scholar] [CrossRef] [PubMed]
- McLaughlin, R.N., Jr.; Malik, H.S. Genetic conflicts: The usual suspects and beyond. J. Exp. Biol. 2017, 220, 6–17. [Google Scholar] [CrossRef]
- Bravo Nunez, M.A.; Lange, J.J.; Zanders, S.E. A suppressor of a wtf poison-antidote meiotic driver acts via mimicry of the driver’s antidote. PLoS Genet. 2018, 14, e1007836. [Google Scholar] [CrossRef]
- Saupe, S.J.; Johannesson, H. On the Mechanistic Basis of Killer Meiotic Drive in Fungi. Annu. Rev. Microbiol. 2022, 76, 305–323. [Google Scholar] [CrossRef]
- Lai, E.C.; Vogan, A.A. Proliferation and dissemination of killer meiotic drive loci. Curr. Opin. Genet. Dev. 2023, 82, 102100. [Google Scholar] [CrossRef]
- Dawe, R.K. The maize abnormal chromosome 10 meiotic drive haplotype: A review. Chromosome Res. 2022, 30, 205–216. [Google Scholar] [CrossRef]
- Wood, K.A.; Goriely, A. The impact of paternal age on new mutations and disease in the next generation. Fertil. Steril. 2022, 118, 1001–1012. [Google Scholar] [CrossRef] [PubMed]
- Almoguera, C.; Shibata, D.; Forrester, K.; Martin, J.; Arnheim, N.; Perucho, M. Most human carcinomas of the exocrine pancreas contain mutant c-K-ras genes. Cell 1988, 53, 549–554. [Google Scholar] [CrossRef]
- Goriely, A.; Wilkie, A.O. Paternal age effect mutations and selfish spermatogonial selection: Causes and consequences for human disease. Am. J. Hum. Genet. 2012, 90, 175–200. [Google Scholar] [CrossRef]
- Flanagan, D.J.; Pentinmikko, N.; Luopajarvi, K.; Willis, N.J.; Gilroy, K.; Raven, A.P.; McGarry, L.; Englund, J.I.; Webb, A.T.; Scharaw, S.; et al. NOTUM from Apc-mutant cells biases clonal competition to initiate cancer. Nature 2021, 594, 430–435. [Google Scholar] [CrossRef] [PubMed]
- van Neerven, S.M.; de Groot, N.E.; Nijman, L.E.; Scicluna, B.P.; van Driel, M.S.; Lecca, M.C.; Warmerdam, D.O.; Kakkar, V.; Moreno, L.F.; Vieira Braga, F.A.; et al. Apc-mutant cells act as supercompetitors in intestinal tumour initiation. Nature 2021, 594, 436–441. [Google Scholar] [CrossRef]
- Madan, E.; Pelham, C.J.; Nagane, M.; Parker, T.M.; Canas-Marques, R.; Fazio, K.; Shaik, K.; Yuan, Y.; Henriques, V.; Galzerano, A.; et al. Flower isoforms promote competitive growth in cancer. Nature 2019, 572, 260–264. [Google Scholar] [CrossRef]
- Harris, B.B. The effects of the aging of X-rayed males upon mutation frequency in Drosophila. J. Hered. 1929, 10, 299–302. [Google Scholar] [CrossRef]
- Pontecorvo, L. Synchronous mitoses and differentiation, sheltering the germ track. Drosoph. Inf. Serv. 1944, 18, 54–55. [Google Scholar]
- Chandley, A.; Bateman, A.J. Mutagenic sensitivity of sperm, spermatids, spermatocytes and spermatogonia in Drosophila Melanogaster. Heredity 1960, 14, 363–375. [Google Scholar] [CrossRef][Green Version]
- Alexander, M.L. Mutation Rates at Specific Autosomal Loci in the Mature and Immature Germ Cells of Drosophila Melanogaster. Genetics 1954, 39, 409–428. [Google Scholar] [CrossRef]
- Abrahamson, S.; Meyer, H.U.; Himoe, E.; Daniel, G. Further evidence demonstrating germinal selection in early premeiotic germ cells of Drosophila males. Genetics 1966, 54, 687–696. [Google Scholar] [CrossRef] [PubMed]
- Wieschaus, E.; Szabad, J. The development and function of the female germ line in Drosophila Melanogaster: A cell lineage study. Dev. Biol. 1979, 68, 29–46. [Google Scholar] [CrossRef]
- Germani, F.; Bergantinos, C.; Johnston, L.A. Mosaic Analysis in Drosophila. Genetics 2018, 208, 473–490. [Google Scholar] [CrossRef]
- Perrimon, N. Clonal Analysis of Dominant Female-Sterile, Germline-Dependent Mutations in Drosophila melanogaster. Genetics 1984, 108, 927–939. [Google Scholar] [CrossRef]
- Extavour, C.; Garcia-Bellido, A. Germ cell selection in genetic mosaics in Drosophila Melanogaster. Proc. Natl. Acad. Sci. USA 2001, 98, 11341–11346. [Google Scholar] [CrossRef] [PubMed]
- Stoner, D.S.; Rinkevich, B.; Weissman, I.L. Heritable germ and somatic cell lineage competitions in chimeric colonial protochordates. Proc. Natl. Acad. Sci. USA 1999, 96, 9148–9153. [Google Scholar] [CrossRef] [PubMed]
- Stoner, D.S.; Weissman, I.L. Somatic and germ cell parasitism in a colonial ascidian: Possible role for a highly polymorphic allorecognition system. Proc. Natl. Acad. Sci. USA 1996, 93, 15254–15259. [Google Scholar] [CrossRef]
- Rodriguez, D.; Kassmer, S.H.; De Tomaso, A.W. Gonad development and hermaphroditism in the ascidian Botryllus schlosseri. Mol. Reprod. Dev. 2017, 84, 158–170. [Google Scholar] [CrossRef]
- Brown, F.D.; Tiozzo, S.; Roux, M.M.; Ishizuka, K.; Swalla, B.J.; De Tomaso, A.W. Early lineage specification of long-lived germline precursors in the colonial ascidian Botryllus schlosseri. Development 2009, 136, 3485–3494. [Google Scholar] [CrossRef]
- Langenbacher, A.D.; De Tomaso, A.W. Temporally and spatially dynamic germ cell niches in Botryllus schlosseri revealed by expression of a TGF-beta family ligand and vasa. Evodevo 2016, 7, 9. [Google Scholar] [CrossRef]
- Scofield, V.L.; Schlumpberger, J.M.; West, L.A.; Weissman, I.L. Protochordate allorecognition is controlled by a MHC-like gene system. Nature 1982, 295, 499–502. [Google Scholar] [CrossRef]
- Weissman, I.L.; Saito, Y.; Rinkevich, B. Allorecognition histocompatibility in a protochordate species: Is the relationship to MHC somatic or structural? Immunol. Rev. 1990, 113, 227–241. [Google Scholar] [CrossRef] [PubMed]
- Laird, D.J.; De Tomaso, A.W.; Weissman, I.L. Stem cells are units of natural selection in a colonial ascidian. Cell 2005, 123, 1351–1360. [Google Scholar] [CrossRef] [PubMed]
- Weissman, I.L. Stem cells are units of natural selection for tissue formation, for germline development, and in cancer development. Proc. Natl. Acad. Sci. USA 2015, 112, 8922–8928. [Google Scholar] [CrossRef]
- Fentress, M.K.; De Tomaso, A.W. Increased collective migration correlates with germline stem cell competition in a basal chordate. PLoS ONE 2023, 18, e0291104. [Google Scholar] [CrossRef]
- King, R.C. Ovarian Development in Drosophila melanogaster; Academic Press: New York, NY, USA, 1970; p. 227. [Google Scholar]
- Spradling, A.C. Drosophila genetics of oogenesis. In The Development of Drosophila melanogaster; Bate, A.M., Martinez Arias, A., Eds.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1993; pp. 1–70. [Google Scholar]
- McKearin, D.M.; Spradling, A.C. bag-of-marbles: A Drosophila gene required to initiate both male and female gametogenesis. Genes. Dev. 1990, 4, 2242–2251. [Google Scholar] [CrossRef]
- Kirilly, D.; Xie, T. The Drosophila ovary: An active stem cell community. Cell Res. 2007, 17, 15–25. [Google Scholar] [CrossRef]
- Xu, T.; Rubin, G.M. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development 1993, 117, 1223–1237. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.; Luo, L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron 1999, 22, 451–461. [Google Scholar] [CrossRef]
- Brand, A.H.; Perrimon, N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 1993, 118, 401–415. [Google Scholar] [CrossRef]
- Potter, C.J.; Tasic, B.; Russler, E.V.; Liang, L.; Luo, L. The Q system: A repressible binary system for transgene expression, lineage tracing, and mosaic analysis. Cell 2010, 141, 536–548. [Google Scholar] [CrossRef] [PubMed]
- Evans, C.J.; Olson, J.M.; Ngo, K.T.; Kim, E.; Lee, N.E.; Kuoy, E.; Patananan, A.N.; Sitz, D.; Tran, P.; Do, M.T.; et al. G-TRACE: Rapid Gal4-based cell lineage analysis in Drosophila. Nat. Methods 2009, 6, 603–605. [Google Scholar] [CrossRef] [PubMed]
- Ni, J.Q.; Markstein, M.; Binari, R.; Pfeiffer, B.; Liu, L.P.; Villalta, C.; Booker, M.; Perkins, L.; Perrimon, N. Vector and parameters for targeted transgenic RNA interference in Drosophila Melanogaster. Nat. Methods 2008, 5, 49–51. [Google Scholar] [CrossRef] [PubMed]
- Ni, J.Q.; Zhou, R.; Czech, B.; Liu, L.P.; Holderbaum, L.; Yang-Zhou, D.; Shim, H.S.; Tao, R.; Handler, D.; Karpowicz, P.; et al. A genome-scale shRNA resource for transgenic RNAi in Drosophila. Nat. Methods 2011, 8, 405–407. [Google Scholar] [CrossRef]
- Adams, M.D.; Celniker, S.E.; Holt, R.A.; Evans, C.A.; Gocayne, J.D.; Amanatides, P.G.; Scherer, S.E.; Li, P.W.; Hoskins, R.A.; Galle, R.F.; et al. The genome sequence of Drosophila Melanogaster. Science 2000, 287, 2185–2195. [Google Scholar] [CrossRef]
- Song, X.; Zhu, C.H.; Doan, C.; Xie, T. Germline stem cells anchored by adherens junctions in the Drosophila ovary niches. Science 2002, 296, 1855–1857. [Google Scholar] [CrossRef]
- McKearin, D.; Ohlstein, B. A role for the Drosophila bag-of-marbles protein in the differentiation of cystoblasts from germline stem cells. Development 1995, 121, 2937–2947. [Google Scholar] [CrossRef]
- Ohlstein, B.; Lavoie, C.A.; Vef, O.; Gateff, E.; McKearin, D.M. The Drosophila cystoblast differentiation factor, benign gonial cell neoplasm, is related to DExH-box proteins and interacts genetically with bag-of-marbles. Genetics 2000, 155, 1809–1819. [Google Scholar] [CrossRef]
- Jin, Z.; Kirilly, D.; Weng, C.; Kawase, E.; Song, X.; Smith, S.; Schwartz, J.; Xie, T. Differentiation-defective stem cells outcompete normal stem cells for niche occupancy in the Drosophila ovary. Cell Stem Cell 2008, 2, 39–49. [Google Scholar] [CrossRef]
- Zhao, S.; Fortier, T.M.; Baehrecke, E.H. Autophagy Promotes Tumor-like Stem Cell Niche Occupancy. Curr. Biol. 2018, 28, 3056–3064.e3053. [Google Scholar] [CrossRef]
- Zhang, Q.; Zhang, Y.; Zhang, Q.; Li, L.; Zhao, S. Division promotes adult stem cells to perform active niche competition. Genetics 2023, 224, iyad035. [Google Scholar] [CrossRef] [PubMed]
- Barth, J.M.; Szabad, J.; Hafen, E.; Kohler, K. Autophagy in Drosophila ovaries is induced by starvation and is required for oogenesis. Cell Death Differ. 2011, 18, 915–924. [Google Scholar] [CrossRef] [PubMed]
- Rhiner, C.; Diaz, B.; Portela, M.; Poyatos, J.F.; Fernandez-Ruiz, I.; Lopez-Gay, J.M.; Gerlitz, O.; Moreno, E. Persistent competition among stem cells and their daughters in the Drosophila ovary germline niche. Development 2009, 136, 995–1006. [Google Scholar] [CrossRef]
- Greenspan, L.J.; de Cuevas, M.; Matunis, E. Genetics of gonadal stem cell renewal. Annu. Rev. Cell Dev. Biol. 2015, 31, 291–315. [Google Scholar] [CrossRef]
- Wang, H.; Singh, S.R.; Zheng, Z.; Oh, S.W.; Chen, X.; Edwards, K.; Hou, S.X. Rap-GEF signaling controls stem cell anchoring to their niche through regulating DE-cadherin-mediated cell adhesion in the Drosophila testis. Dev. Cell 2006, 10, 117–126. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, Y.M.; Jones, D.L.; Fuller, M.T. Orientation of asymmetric stem cell division by the APC tumor suppressor and centrosome. Science 2003, 301, 1547–1550. [Google Scholar] [CrossRef]
- Voog, J.; D’Alterio, C.; Jones, D.L. Multipotent somatic stem cells contribute to the stem cell niche in the Drosophila testis. Nature 2008, 454, 1132–1136. [Google Scholar] [CrossRef]
- Wallenfang, M.R.; Nayak, R.; DiNardo, S. Dynamics of the male germline stem cell population during aging of Drosophila melanogaster. Aging Cell 2006, 5, 297–304. [Google Scholar] [CrossRef]
- Sheng, X.R.; Matunis, E. Live imaging of the Drosophila spermatogonial stem cell niche reveals novel mechanisms regulating germline stem cell output. Development 2011, 138, 3367–3376. [Google Scholar] [CrossRef]
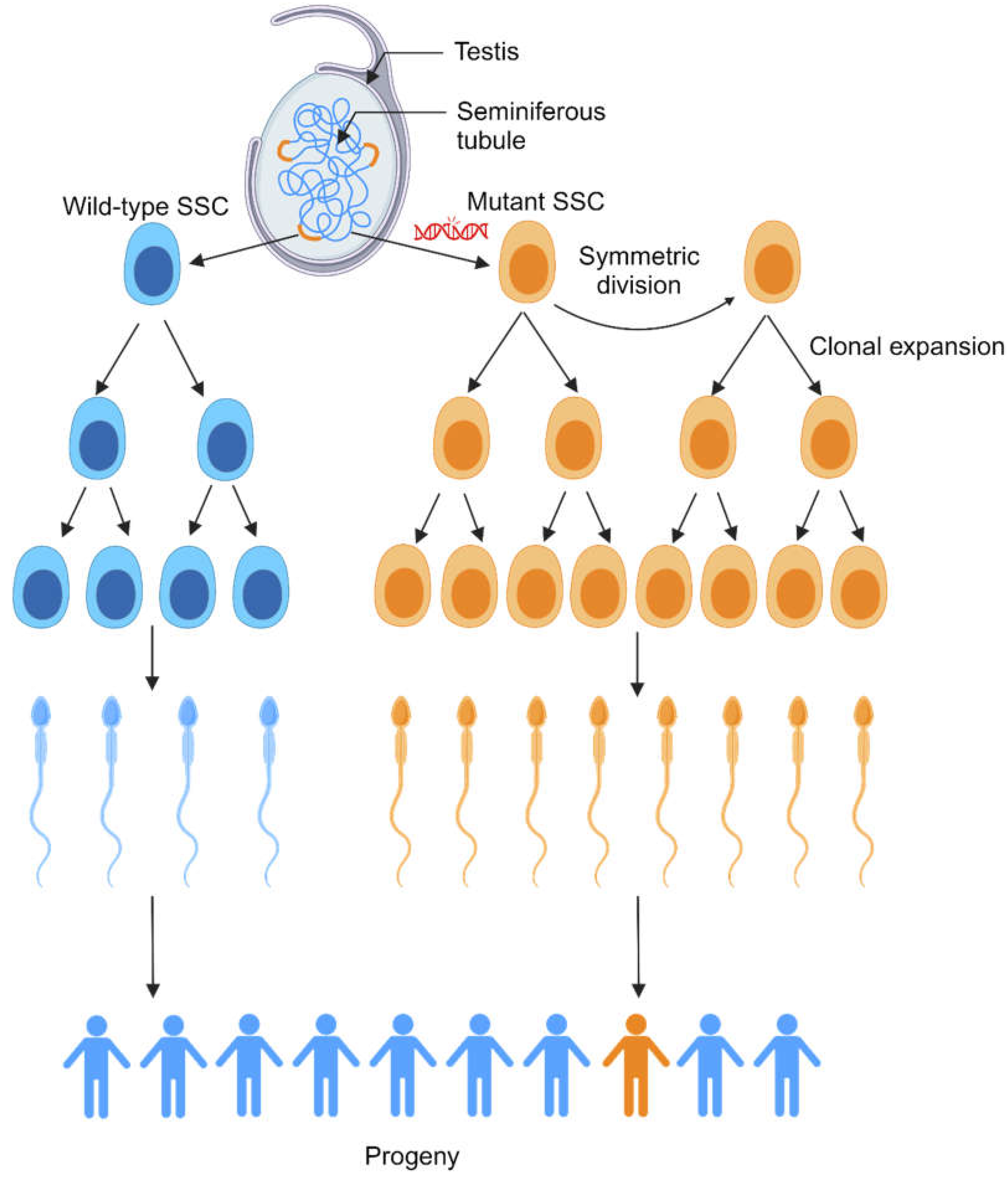
- Tseng, C.Y.; Burel, M.; Cammer, M.; Harsh, S.; Flaherty, M.S.; Baumgartner, S.; Bach, E.A. chinmo-mutant spermatogonial stem cells cause mitotic drive by evicting non-mutant neighbors from the niche. Dev. Cell 2022, 57, 80–94.e87. [Google Scholar] [CrossRef]
- Zanetti, C.; Krause, D.S. “Caught in the net”: The extracellular matrix of the bone marrow in normal hematopoiesis and leukemia. Exp. Hematol. 2020, 89, 13–25. [Google Scholar] [CrossRef] [PubMed]
- Lu, P.; Weaver, V.M.; Werb, Z. The extracellular matrix: A dynamic niche in cancer progression. J. Cell Biol. 2012, 196, 395–406. [Google Scholar] [CrossRef] [PubMed]
- Giussani, M.; Triulzi, T.; Sozzi, G.; Tagliabue, E. Tumor Extracellular Matrix Remodeling: New Perspectives as a Circulating Tool in the Diagnosis and Prognosis of Solid Tumors. Cells 2019, 8, 81. [Google Scholar] [CrossRef] [PubMed]
- Amoyel, M.; Anderson, J.; Suisse, A.; Glasner, J.; Bach, E.A. Socs36E Controls Niche Competition by Repressing MAPK Signaling in the Drosophila Testis. PLoS Genet. 2016, 12, e1005815. [Google Scholar] [CrossRef]
- Singh, S.R.; Zheng, Z.; Wang, H.; Oh, S.W.; Chen, X.; Hou, S.X. Competitiveness for the niche and mutual dependence of the germline and somatic stem cells in the Drosophila testis are regulated by the JAK/STAT signaling. J. Cell. Physiol. 2010, 223, 500–510. [Google Scholar] [CrossRef]
- Singh, S.R.; Liu, Y.; Zhao, J.; Zeng, X.; Hou, S.X. The novel tumour suppressor Madm regulates stem cell competition in the Drosophila testis. Nat. Commun. 2016, 7, 10473. [Google Scholar] [CrossRef]
- Stine, R.R.; Greenspan, L.J.; Ramachandran, K.V.; Matunis, E.L. Coordinate regulation of stem cell competition by Slit-Robo and JAK-STAT signaling in the Drosophila testis. PLoS Genet. 2014, 10, e1004713. [Google Scholar] [CrossRef] [PubMed]
- Cook, M.S.; Cazin, C.; Amoyel, M.; Yamamoto, S.; Bach, E.; Nystul, T. Neutral Competition for Drosophila Follicle and Cyst Stem Cell Niches Requires Vesicle Trafficking Genes. Genetics 2017, 206, 1417–1428. [Google Scholar] [CrossRef]
- Herranz, H.; Hong, X.; Hung, N.T.; Voorhoeve, P.M.; Cohen, S.M. Oncogenic cooperation between SOCS family proteins and EGFR identified using a Drosophila epithelial transformation model. Genes. Dev. 2012, 26, 1602–1611. [Google Scholar] [CrossRef]
- Nystul, T.; Spradling, A. An epithelial niche in the Drosophila ovary undergoes long-range stem cell replacement. Cell Stem Cell 2007, 1, 277–285. [Google Scholar] [CrossRef]
- Nystul, T.; Spradling, A. Regulation of epithelial stem cell replacement and follicle formation in the Drosophila ovary. Genetics 2010, 184, 503–515. [Google Scholar] [CrossRef] [PubMed]
- Kronen, M.R.; Schoenfelder, K.P.; Klein, A.M.; Nystul, T.G. Basolateral junction proteins regulate competition for the follicle stem cell niche in the Drosophila ovary. PLoS ONE 2014, 9, e101085. [Google Scholar] [CrossRef] [PubMed]
- Vied, C.; Reilein, A.; Field, N.S.; Kalderon, D. Regulation of stem cells by intersecting gradients of long-range niche signals. Dev. Cell 2012, 23, 836–848. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Kalderon, D. Coupling of Hedgehog and Hippo pathways promotes stem cell maintenance by stimulating proliferation. J. Cell Biol. 2014, 205, 325–338. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.A.; Kalderon, D. Cyclin E-dependent protein kinase activity regulates niche retention of Drosophila ovarian follicle stem cells. Proc. Natl. Acad. Sci. USA 2009, 106, 21701–21706. [Google Scholar] [CrossRef]
- Wang, Z.A.; Huang, J.; Kalderon, D. Drosophila follicle stem cells are regulated by proliferation and niche adhesion as well as mitochondria and ROS. Nat. Commun. 2012, 3, 769. [Google Scholar] [CrossRef]
- Barker, N.; van Es, J.H.; Kuipers, J.; Kujala, P.; van den Born, M.; Cozijnsen, M.; Haegebarth, A.; Korving, J.; Begthel, H.; Peters, P.J.; et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 2007, 449, 1003–1007. [Google Scholar] [CrossRef]
- Gehart, H.; Clevers, H. Tales from the crypt: New insights into intestinal stem cells. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 19–34. [Google Scholar] [CrossRef]
- Griffiths, D.F.; Davies, S.J.; Williams, D.; Williams, G.T.; Williams, E.D. Demonstration of somatic mutation and colonic crypt clonality by X-linked enzyme histochemistry. Nature 1988, 333, 461–463. [Google Scholar] [CrossRef]
- Lopez-Garcia, C.; Klein, A.M.; Simons, B.D.; Winton, D.J. Intestinal stem cell replacement follows a pattern of neutral drift. Science 2010, 330, 822–825. [Google Scholar] [CrossRef]
- de Navascues, J.; Perdigoto, C.N.; Bian, Y.; Schneider, M.H.; Bardin, A.J.; Martinez-Arias, A.; Simons, B.D. Drosophila midgut homeostasis involves neutral competition between symmetrically dividing intestinal stem cells. EMBO J. 2012, 31, 2473–2485. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, V.H.; Nadarajan, P.; Graham, T.A.; Pipinikas, C.P.; Brown, J.M.; Falzon, M.; Nye, E.; Poulsom, R.; Lawrence, D.; Wright, N.A.; et al. Stochastic homeostasis in human airway epithelium is achieved by neutral competition of basal cell progenitors. eLife 2013, 2, e00966. [Google Scholar] [CrossRef] [PubMed]
- Ritsma, L.; Ellenbroek, S.I.; Zomer, A.; Snippert, H.J.; de Sauvage, F.J.; Simons, B.D.; Clevers, H.; van Rheenen, J. Intestinal crypt homeostasis revealed at single-stem-cell level by in vivo live imaging. Nature 2014, 507, 362–365. [Google Scholar] [CrossRef]
- Baker, N.E. Emerging mechanisms of cell competition. Nat. Rev. Genet. 2020, 21, 683–697. [Google Scholar] [CrossRef] [PubMed]
- Vishwakarma, M.; Piddini, E. Outcompeting cancer. Nat. Rev. Cancer 2020, 20, 187–198. [Google Scholar] [CrossRef]
- Vogelstein, B.; Papadopoulos, N.; Velculescu, V.E.; Zhou, S.; Diaz, L.A., Jr.; Kinzler, K.W. Cancer genome landscapes. Science 2013, 339, 1546–1558. [Google Scholar] [CrossRef]
- Parker, T.M.; Gupta, K.; Palma, A.M.; Yekelchyk, M.; Fisher, P.B.; Grossman, S.R.; Won, K.J.; Madan, E.; Moreno, E.; Gogna, R. Cell competition in intratumoral and tumor microenvironment interactions. EMBO J. 2021, 40, e107271. [Google Scholar] [CrossRef]
- Yum, M.K.; Han, S.; Fink, J.; Wu, S.S.; Dabrowska, C.; Trendafilova, T.; Mustata, R.; Chatzeli, L.; Azzarelli, R.; Pshenichnaya, I.; et al. Tracing oncogene-driven remodelling of the intestinal stem cell niche. Nature 2021, 594, 442–447. [Google Scholar] [CrossRef]
- Herms, A.; Colom, B.; Piedrafita, G.; Kalogeropoulou, A.; Banerjee, U.; King, C.; Abby, E.; Murai, K.; Caseda, I.; Fernandez-Antoran, D.; et al. Organismal metabolism regulates the expansion of oncogenic PIK3CA mutant clones in normal esophagus. Nat. Genet. 2024. [Google Scholar] [CrossRef]
- Lundberg, P.; Takizawa, H.; Kubovcakova, L.; Guo, G.; Hao-Shen, H.; Dirnhofer, S.; Orkin, S.H.; Manz, M.G.; Skoda, R.C. Myeloproliferative neoplasms can be initiated from a single hematopoietic stem cell expressing JAK2-V617F. J. Exp. Med. 2014, 211, 2213–2230. [Google Scholar] [CrossRef]
- Hogan, C.; Dupre-Crochet, S.; Norman, M.; Kajita, M.; Zimmermann, C.; Pelling, A.E.; Piddini, E.; Baena-Lopez, L.A.; Vincent, J.P.; Itoh, Y.; et al. Characterization of the interface between normal and transformed epithelial cells. Nat. Cell Biol. 2009, 11, 460–467. [Google Scholar] [CrossRef] [PubMed]
- Chiba, T.; Ishihara, E.; Miyamura, N.; Narumi, R.; Kajita, M.; Fujita, Y.; Suzuki, A.; Ogawa, Y.; Nishina, H. MDCK cells expressing constitutively active Yes-associated protein (YAP) undergo apical extrusion depending on neighboring cell status. Sci. Rep. 2016, 6, 28383. [Google Scholar] [CrossRef]
- Watanabe, H.; Ishibashi, K.; Mano, H.; Kitamoto, S.; Sato, N.; Hoshiba, K.; Kato, M.; Matsuzawa, F.; Takeuchi, Y.; Shirai, T.; et al. Mutant p53-Expressing Cells Undergo Necroptosis via Cell Competition with the Neighboring Normal Epithelial Cells. Cell Rep. 2018, 23, 3721–3729. [Google Scholar] [CrossRef]
- Moya, I.M.; Castaldo, S.A.; Van den Mooter, L.; Soheily, S.; Sansores-Garcia, L.; Jacobs, J.; Mannaerts, I.; Xie, J.; Verboven, E.; Hillen, H.; et al. Peritumoral activation of the Hippo pathway effectors YAP and TAZ suppresses liver cancer in mice. Science 2019, 366, 1029–1034. [Google Scholar] [CrossRef] [PubMed]
- Martins, V.C.; Busch, K.; Juraeva, D.; Blum, C.; Ludwig, C.; Rasche, V.; Lasitschka, F.; Mastitsky, S.E.; Brors, B.; Hielscher, T.; et al. Cell competition is a tumour suppressor mechanism in the thymus. Nature 2014, 509, 465–470. [Google Scholar] [CrossRef] [PubMed]
- Carriere, P.; Baade, P.; Fritschi, L. Population based incidence and age distribution of spermatocytic seminoma. J. Urol. 2007, 178, 125–128. [Google Scholar] [CrossRef]
- Goriely, A.; Hansen, R.M.; Taylor, I.B.; Olesen, I.A.; Jacobsen, G.K.; McGowan, S.J.; Pfeifer, S.P.; McVean, G.A.; Rajpert-De Meyts, E.; Wilkie, A.O. Activating mutations in FGFR3 and HRAS reveal a shared genetic origin for congenital disorders and testicular tumors. Nat. Genet. 2009, 41, 1247–1252. [Google Scholar] [CrossRef] [PubMed]
- Eble, J.N. Spermatocytic seminoma. Hum. Pathol. 1994, 25, 1035–1042. [Google Scholar] [CrossRef]
- Rajpert-De Meyts, E.; Jacobsen, G.K.; Bartkova, J.; Aubry, F.; Samson, M.; Bartek, J.; Skakkebaek, N.E. The immunohistochemical expression pattern of Chk2, p53, p19INK4d, MAGE-A4 and other selected antigens provides new evidence for the premeiotic origin of spermatocytic seminoma. Histopathology 2003, 42, 217–226. [Google Scholar] [CrossRef]
- Giannoulatou, E.; Maher, G.J.; Ding, Z.; Gillis, A.J.M.; Dorssers, L.C.J.; Hoischen, A.; Rajpert-De Meyts, E.; Consortium, W.G.S.; McVean, G.; Wilkie, A.O.M.; et al. Whole-genome sequencing of spermatocytic tumors provides insights into the mutational processes operating in the male germline. PLoS ONE 2017, 12, e0178169. [Google Scholar] [CrossRef]
- Nagano, M.C. Homing efficiency and proliferation kinetics of male germ line stem cells following transplantation in mice. Biol. Reprod. 2003, 69, 701–707. [Google Scholar] [CrossRef] [PubMed]
- Nagano, M.C.; Yeh, J.R. The identity and fate decision control of spermatogonial stem cells: Where is the point of no return? Curr. Top. Dev. Biol. 2013, 102, 61–95. [Google Scholar] [CrossRef] [PubMed]
- Oatley, J.M.; Brinster, R.L. The germline stem cell niche unit in mammalian testes. Physiol. Rev. 2012, 92, 577–595. [Google Scholar] [CrossRef]
- Reuter, V.E. Origins and molecular biology of testicular germ cell tumors. Mod. Pathol. 2005, 18 (Suppl. 2), S51–S60. [Google Scholar] [CrossRef]
- Lim, J.; Goriely, A.; Turner, G.D.; Ewen, K.A.; Jacobsen, G.K.; Graem, N.; Wilkie, A.O.; Rajpert-De Meyts, E. OCT2, SSX and SAGE1 reveal the phenotypic heterogeneity of spermatocytic seminoma reflecting distinct subpopulations of spermatogonia. J. Pathol. 2011, 224, 473–483. [Google Scholar] [CrossRef]
- Maher, G.J.; Goriely, A.; Wilkie, A.O. Cellular evidence for selfish spermatogonial selection in aged human testes. Andrology 2014, 2, 304–314. [Google Scholar] [CrossRef]
- Wetherell, D.; Lawrentschuk, N.; Gyomber, D. Spermatocytic seminoma with sarcoma: An indication for adjuvant chemotherapy in localized disease. Korean J. Urol. 2013, 54, 884–887. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Genovese, G.; Kahler, A.K.; Handsaker, R.E.; Lindberg, J.; Rose, S.A.; Bakhoum, S.F.; Chambert, K.; Mick, E.; Neale, B.M.; Fromer, M.; et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N. Engl. J. Med. 2014, 371, 2477–2487. [Google Scholar] [CrossRef]
- Jaiswal, S.; Fontanillas, P.; Flannick, J.; Manning, A.; Grauman, P.V.; Mar, B.G.; Lindsley, R.C.; Mermel, C.H.; Burtt, N.; Chavez, A.; et al. Age-related clonal hematopoiesis associated with adverse outcomes. N. Engl. J. Med. 2014, 371, 2488–2498. [Google Scholar] [CrossRef]
- Watson, C.J.; Papula, A.L.; Poon, G.Y.P.; Wong, W.H.; Young, A.L.; Druley, T.E.; Fisher, D.S.; Blundell, J.R. The evolutionary dynamics and fitness landscape of clonal hematopoiesis. Science 2020, 367, 1449–1454. [Google Scholar] [CrossRef]
- Jaiswal, S.; Ebert, B.L. Clonal hematopoiesis in human aging and disease. Science 2019, 366, eaan4673. [Google Scholar] [CrossRef] [PubMed]
- Challen, G.A.; Sun, D.; Jeong, M.; Luo, M.; Jelinek, J.; Berg, J.S.; Bock, C.; Vasanthakumar, A.; Gu, H.; Xi, Y.; et al. Dnmt3a is essential for hematopoietic stem cell differentiation. Nat. Genet. 2011, 44, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Ko, M.; Bandukwala, H.S.; An, J.; Lamperti, E.D.; Thompson, E.C.; Hastie, R.; Tsangaratou, A.; Rajewsky, K.; Koralov, S.B.; Rao, A. Ten-Eleven-Translocation 2 (TET2) negatively regulates homeostasis and differentiation of hematopoietic stem cells in mice. Proc. Natl. Acad. Sci. USA 2011, 108, 14566–14571. [Google Scholar] [CrossRef] [PubMed]
- Hormaechea-Agulla, D.; Matatall, K.A.; Le, D.T.; Kain, B.; Long, X.; Kus, P.; Jaksik, R.; Challen, G.A.; Kimmel, M.; King, K.Y. Chronic infection drives Dnmt3a-loss-of-function clonal hematopoiesis via IFNgamma signaling. Cell Stem Cell 2021, 28, 1428–1442.e1426. [Google Scholar] [CrossRef] [PubMed]
- Zioni, N.; Bercovich, A.A.; Chapal-Ilani, N.; Bacharach, T.; Rappoport, N.; Solomon, A.; Avraham, R.; Kopitman, E.; Porat, Z.; Sacma, M.; et al. Inflammatory signals from fatty bone marrow support DNMT3A driven clonal hematopoiesis. Nat. Commun. 2023, 14, 2070. [Google Scholar] [CrossRef]
- Caiado, F.; Kovtonyuk, L.V.; Gonullu, N.G.; Fullin, J.; Boettcher, S.; Manz, M.G. Aging drives Tet2+/- clonal hematopoiesis via IL-1 signaling. Blood 2023, 141, 886–903. [Google Scholar] [CrossRef]
- Jakobsen, N.A.; Turkalj, S.; Zeng, A.G.X.; Stoilova, B.; Metzner, M.; Rahmig, S.; Nagree, M.S.; Shah, S.; Moore, R.; Usukhbayar, B.; et al. Selective advantage of mutant stem cells in human clonal hematopoiesis is associated with attenuated response to inflammation and aging. Cell Stem Cell 2024, 31, 1127–1144.e1117. [Google Scholar] [CrossRef]
- Nam, A.S.; Dusaj, N.; Izzo, F.; Murali, R.; Myers, R.M.; Mouhieddine, T.H.; Sotelo, J.; Benbarche, S.; Waarts, M.; Gaiti, F.; et al. Single-cell multi-omics of human clonal hematopoiesis reveals that DNMT3A R882 mutations perturb early progenitor states through selective hypomethylation. Nat. Genet. 2022, 54, 1514–1526. [Google Scholar] [CrossRef]
- Bondar, T.; Medzhitov, R. p53-mediated hematopoietic stem and progenitor cell competition. Cell Stem Cell 2010, 6, 309–322. [Google Scholar] [CrossRef]
- Kahn, J.D.; Miller, P.G.; Silver, A.J.; Sellar, R.S.; Bhatt, S.; Gibson, C.; McConkey, M.; Adams, D.; Mar, B.; Mertins, P.; et al. PPM1D-truncating mutations confer resistance to chemotherapy and sensitivity to PPM1D inhibition in hematopoietic cells. Blood 2018, 132, 1095–1105. [Google Scholar] [CrossRef]
- Hsu, J.I.; Dayaram, T.; Tovy, A.; De Braekeleer, E.; Jeong, M.; Wang, F.; Zhang, J.; Heffernan, T.P.; Gera, S.; Kovacs, J.J.; et al. PPM1D Mutations Drive Clonal Hematopoiesis in Response to Cytotoxic Chemotherapy. Cell Stem Cell 2018, 23, 700–713.e706. [Google Scholar] [CrossRef] [PubMed]
- Kralovics, R.; Passamonti, F.; Buser, A.S.; Teo, S.S.; Tiedt, R.; Passweg, J.R.; Tichelli, A.; Cazzola, M.; Skoda, R.C. A gain-of-function mutation of JAK2 in myeloproliferative disorders. New Engl. J. Med. 2005, 352, 1779–1790. [Google Scholar] [CrossRef] [PubMed]
- Margolis, J.; Borrello, I.; Flinn, I.W. New approaches to treating malignances with stem cell transplantation. Semin. Oncol. 2000, 27, 524–530. [Google Scholar] [PubMed]
- Cuckle, H.; Morris, J. Maternal age in the epidemiology of common autosomal trisomies. Prenat. Diagn. 2021, 41, 573–583. [Google Scholar] [CrossRef]
- Frederiksen, L.E.; Ernst, A.; Brix, N.; Braskhoj Lauridsen, L.L.; Roos, L.; Ramlau-Hansen, C.H.; Ekelund, C.K. Risk of Adverse Pregnancy Outcomes at Advanced Maternal Age. Obs. Gynecol. 2018, 131, 457–463. [Google Scholar] [CrossRef]
- Pinheiro, R.L.; Areia, A.L.; Mota Pinto, A.; Donato, H. Advanced Maternal Age: Adverse Outcomes of Pregnancy, A Meta-Analysis. Acta Med. Port. 2019, 32, 219–226. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.K.; Wen, S.W.; Krewski, D.; Fleming, N.; Yang, Q.; Walker, M.C. Paternal age and adverse birth outcomes: Teenager or 40+, who is at risk? Hum. Reprod. 2008, 23, 1290–1296. [Google Scholar] [CrossRef]
- Khandwala, Y.S.; Baker, V.L.; Shaw, G.M.; Stevenson, D.K.; Lu, Y.; Eisenberg, M.L. Association of paternal age with perinatal outcomes between 2007 and 2016 in the United States: Population based cohort study. BMJ 2018, 363, k4372. [Google Scholar] [CrossRef]
- Astolfi, P.; De Pasquale, A.; Zonta, L.A. Paternal age and preterm birth in Italy, 1990 to 1998. Epidemiology 2006, 17, 218–221. [Google Scholar] [CrossRef]
- Urhoj, S.K.; Andersen, P.K.; Mortensen, L.H.; Davey Smith, G.; Nybo Andersen, A.M. Advanced paternal age and stillbirth rate: A nationwide register-based cohort study of 944,031 pregnancies in Denmark. Eur. J. Epidemiol. 2017, 32, 227–234. [Google Scholar] [CrossRef]
- Michaelson, J.J.; Shi, Y.; Gujral, M.; Zheng, H.; Malhotra, D.; Jin, X.; Jian, M.; Liu, G.; Greer, D.; Bhandari, A.; et al. Whole-genome sequencing in autism identifies hot spots for de novo germline mutation. Cell 2012, 151, 1431–1442. [Google Scholar] [CrossRef] [PubMed]
- Paavilainen, M.; Bloigu, A.; Hemminki, E.; Gissler, M.; Klemetti, R. Aging fatherhood in Finland—First-time fathers in Finland from 1987 to 2009. Scand. J. Public Health 2016, 44, 423–430. [Google Scholar] [CrossRef]
- Khandwala, Y.S.; Zhang, C.A.; Lu, Y.; Eisenberg, M.L. The age of fathers in the USA is rising: An analysis of 168 867 480 births from 1972 to 2015. Hum. Reprod. 2017, 32, 2110–2116. [Google Scholar] [CrossRef]
- Chatsirisupachai, K.; de Magalhaes, J.P. Somatic mutations in human ageing: New insights from DNA sequencing and inherited mutations. Ageing Res. Rev. 2024, 96, 102268. [Google Scholar] [CrossRef] [PubMed]
- Jonsson, H.; Sulem, P.; Kehr, B.; Kristmundsdottir, S.; Zink, F.; Hjartarson, E.; Hardarson, M.T.; Hjorleifsson, K.E.; Eggertsson, H.P.; Gudjonsson, S.A.; et al. Parental influence on human germline de novo mutations in 1,548 trios from Iceland. Nature 2017, 549, 519–522. [Google Scholar] [CrossRef]
- Rahbari, R.; Wuster, A.; Lindsay, S.J.; Hardwick, R.J.; Alexandrov, L.B.; Turki, S.A.; Dominiczak, A.; Morris, A.; Porteous, D.; Smith, B.; et al. Timing, rates and spectra of human germline mutation. Nat. Genet. 2016, 48, 126–133. [Google Scholar] [CrossRef] [PubMed]
- Goldmann, J.M.; Wong, W.S.; Pinelli, M.; Farrah, T.; Bodian, D.; Stittrich, A.B.; Glusman, G.; Vissers, L.E.; Hoischen, A.; Roach, J.C.; et al. Parent-of-origin-specific signatures of de novo mutations. Nat. Genet. 2016, 48, 935–939. [Google Scholar] [CrossRef]
- Goldmann, J.M.; Veltman, J.A.; Gilissen, C. De Novo Mutations Reflect Development and Aging of the Human Germline. Trends Genet. TIG 2019, 35, 828–839. [Google Scholar] [CrossRef]
- Wang, X.; Pepling, M.E. Regulation of Meiotic Prophase One in Mammalian Oocytes. Front. Cell Dev. Biol. 2021, 9, 667306. [Google Scholar] [CrossRef]
- Solc, P.; Schultz, R.M.; Motlik, J. Prophase I arrest and progression to metaphase I in mouse oocytes: Comparison of resumption of meiosis and recovery from G2-arrest in somatic cells. Mol. Hum. Reprod. 2010, 16, 654–664. [Google Scholar] [CrossRef]
- Crow, J.F. The origins, patterns and implications of human spontaneous mutation. Nat. Rev. Genet. 2000, 1, 40–47. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, M.E.; Noyes, M.D.; Eichler, E.E.; Quinlan, A.R.; Harris, K. Effects of parental age and polymer composition on short tandem repeat de novo mutation rates. Genetics 2024, 226, iyae013. [Google Scholar] [CrossRef] [PubMed]
- Kong, A.; Frigge, M.L.; Masson, G.; Besenbacher, S.; Sulem, P.; Magnusson, G.; Gudjonsson, S.A.; Sigurdsson, A.; Jonasdottir, A.; Jonasdottir, A.; et al. Rate of de novo mutations and the importance of father’s age to disease risk. Nature 2012, 488, 471–475. [Google Scholar] [CrossRef] [PubMed]
- Hebron, K.E.; Hernandez, E.R.; Yohe, M.E. The RASopathies: From pathogenetics to therapeutics. Dis. Models Mech. 2022, 15, dmm049107. [Google Scholar] [CrossRef]
- Maher, G.J.; Ralph, H.K.; Ding, Z.; Koelling, N.; Mlcochova, H.; Giannoulatou, E.; Dhami, P.; Paul, D.S.; Stricker, S.H.; Beck, S.; et al. Selfish mutations dysregulating RAS-MAPK signaling are pervasive in aged human testes. Genome Res. 2018, 28, 1779–1790. [Google Scholar] [CrossRef]
- Wilkin, D.J.; Szabo, J.K.; Cameron, R.; Henderson, S.; Bellus, G.A.; Mack, M.L.; Kaitila, I.; Loughlin, J.; Munnich, A.; Sykes, B.; et al. Mutations in fibroblast growth-factor receptor 3 in sporadic cases of achondroplasia occur exclusively on the paternally derived chromosome. Am. J. Hum. Genet. 1998, 63, 711–716. [Google Scholar] [CrossRef]
- Wilkie, A.O.; Slaney, S.F.; Oldridge, M.; Poole, M.D.; Ashworth, G.J.; Hockley, A.D.; Hayward, R.D.; David, D.J.; Pulleyn, L.J.; Rutland, P.; et al. Apert syndrome results from localized mutations of FGFR2 and is allelic with Crouzon syndrome. Nat. Genet. 1995, 9, 165–172. [Google Scholar] [CrossRef]
- Rutland, P.; Pulleyn, L.J.; Reardon, W.; Baraitser, M.; Hayward, R.; Jones, B.; Malcolm, S.; Winter, R.M.; Oldridge, M.; Slaney, S.F.; et al. Identical mutations in the FGFR2 gene cause both Pfeiffer and Crouzon syndrome phenotypes. Nat. Genet. 1995, 9, 173–176. [Google Scholar] [CrossRef]
- Tartaglia, M.; Mehler, E.L.; Goldberg, R.; Zampino, G.; Brunner, H.G.; Kremer, H.; van der Burgt, I.; Crosby, A.H.; Ion, A.; Jeffery, S.; et al. Mutations in PTPN11, encoding the protein tyrosine phosphatase SHP-2, cause Noonan syndrome. Nat. Genet. 2001, 29, 465–468. [Google Scholar] [CrossRef]
- Aoki, Y.; Niihori, T.; Kawame, H.; Kurosawa, K.; Ohashi, H.; Tanaka, Y.; Filocamo, M.; Kato, K.; Suzuki, Y.; Kure, S.; et al. Germline mutations in HRAS proto-oncogene cause Costello syndrome. Nat. Genet. 2005, 37, 1038–1040. [Google Scholar] [CrossRef]
- Meng, X.; de Rooij, D.G.; Westerdahl, K.; Saarma, M.; Sariola, H. Promotion of seminomatous tumors by targeted overexpression of glial cell line-derived neurotrophic factor in mouse testis. Cancer Res. 2001, 61, 3267–3271. [Google Scholar] [PubMed]
- Puri, P.; Phillips, B.T.; Suzuki, H.; Orwig, K.E.; Rajkovic, A.; Lapinski, P.E.; King, P.D.; Feng, G.S.; Walker, W.H. The transition from stem cell to progenitor spermatogonia and male fertility requires the SHP2 protein tyrosine phosphatase. Stem Cells 2014, 32, 741–753. [Google Scholar] [CrossRef]
- Risch, N.; Reich, E.W.; Wishnick, M.M.; McCarthy, J.G. Spontaneous mutation and parental age in humans. Am. J. Hum. Genet. 1987, 41, 218–248. [Google Scholar]
- Goriely, A.; McVean, G.A.; Rojmyr, M.; Ingemarsson, B.; Wilkie, A.O. Evidence for selective advantage of pathogenic FGFR2 mutations in the male germ line. Science 2003, 301, 643–646. [Google Scholar] [CrossRef]
- Prior, I.A.; Hood, F.E.; Hartley, J.L. The Frequency of Ras Mutations in Cancer. Cancer Res. 2020, 80, 2969–2974. [Google Scholar] [CrossRef] [PubMed]
- Yamada, M.; De Chiara, L.; Seandel, M. Spermatogonial Stem Cells: Implications for Genetic Disorders and Prevention. Stem Cells Dev. 2016, 25, 1483–1494. [Google Scholar] [CrossRef] [PubMed]
- Shinohara, T.; Orwig, K.E.; Avarbock, M.R.; Brinster, R.L. Germ line stem cell competition in postnatal mouse testes. Biol. Reprod. 2002, 66, 1491–1497. [Google Scholar] [CrossRef][Green Version]
- Kitadate, Y.; Jorg, D.J.; Tokue, M.; Maruyama, A.; Ichikawa, R.; Tsuchiya, S.; Segi-Nishida, E.; Nakagawa, T.; Uchida, A.; Kimura-Yoshida, C.; et al. Competition for Mitogens Regulates Spermatogenic Stem Cell Homeostasis in an Open Niche. Cell Stem Cell 2019, 24, 79–92.e6. [Google Scholar] [CrossRef]
- Ryu, B.Y.; Orwig, K.E.; Oatley, J.M.; Avarbock, M.R.; Brinster, R.L. Effects of aging and niche microenvironment on spermatogonial stem cell self-renewal. Stem Cells 2006, 24, 1505–1511. [Google Scholar] [CrossRef]
- Martin, L.A.; Assif, N.; Gilbert, M.; Wijewarnasuriya, D.; Seandel, M. Enhanced fitness of adult spermatogonial stem cells bearing a paternal age-associated FGFR2 mutation. Stem Cell Rep. 2014, 3, 219–226. [Google Scholar] [CrossRef]
- Yi, H.; Talmon, G.; Wang, J. Glutamate in cancers: From metabolism to signaling. J. Biomed. Res. 2019, 34, 260–270. [Google Scholar] [CrossRef] [PubMed]
- Soares, C.C.; Rizzo, A.; Maresma, M.F.; Meier, P. Autocrine glutamate signaling drives cell competition in Drosophila. Dev. Cell in press. 2024. [Google Scholar] [CrossRef] [PubMed]
- Lenhart, K.F.; DiNardo, S. Somatic cell encystment promotes abscission in germline stem cells following a regulated block in cytokinesis. Dev. Cell 2015, 34, 192–205. [Google Scholar] [CrossRef] [PubMed]
- Vollset, S.E.; Ababneh, H.S.; Abate, Y.H.; Abbafati, C.; Abbasgholizadeh, R.; Abbasian, M.; Abbastabar, H.; Magied, A.H.A.A.A.; ElHafeez, S.A.; Abdelkader, A.; et al. Burden of disease scenarios for 204 countries and territories, 2022–2050: A forecasting analysis for the Global Burden of Disease Study 2021. Lancet 2024, 403, 2204–2256. [Google Scholar] [CrossRef] [PubMed]
- Cleary, A.S.; Leonard, T.L.; Gestl, S.A.; Gunther, E.J. Tumour cell heterogeneity maintained by cooperating subclones in Wnt-driven mammary cancers. Nature 2014, 508, 113–117. [Google Scholar] [CrossRef]
- Janiszewska, M.; Tabassum, D.P.; Castano, Z.; Cristea, S.; Yamamoto, K.N.; Kingston, N.L.; Murphy, K.C.; Shu, S.; Harper, N.W.; Del Alcazar, C.G.; et al. Subclonal cooperation drives metastasis by modulating local and systemic immune microenvironments. Nat. Cell Biol. 2019, 21, 879–888. [Google Scholar] [CrossRef]
- Dong, Y.L.; Vadla, G.P.; Lu, J.J.; Ahmad, V.; Klein, T.J.; Liu, L.F.; Glazer, P.M.; Xu, T.; Chabu, C.Y. Cooperation between oncogenic Ras and wild-type p53 stimulates STAT non-cell autonomously to promote tumor radioresistance. Commun. Biol. 2021, 4, 374. [Google Scholar] [CrossRef]
- Sheng, X.R.; Brawley, C.M.; Matunis, E.L. Dedifferentiating spermatogonia outcompete somatic stem cells for niche occupancy in the Drosophila testis. Cell Stem Cell 2009, 5, 191–203. [Google Scholar] [CrossRef]
- Flaherty, M.S.; Salis, P.; Evans, C.J.; Ekas, L.A.; Marouf, A.; Zavadil, J.; Banerjee, U.; Bach, E.A. chinmo is a functional effector of the JAK/STAT pathway that regulates eye development, tumor formation, and stem cell self-renewal in Drosophila. Dev. Cell 2010, 18, 556–568. [Google Scholar] [CrossRef]
- Chao, C.F.; Pesch, Y.Y.; Yu, H.; Wang, C.; Aristizabal, M.J.; Huan, T.; Tanentzapf, G.; Rideout, E. An important role for triglyceride in regulating spermatogenesis. eLife 2024, 12, RP87523. [Google Scholar] [CrossRef]
- Moore, L.; Cagan, A.; Coorens, T.H.H.; Neville, M.D.C.; Sanghvi, R.; Sanders, M.A.; Oliver, T.R.W.; Leongamornlert, D.; Ellis, P.; Noorani, A.; et al. The mutational landscape of human somatic and germline cells. Nature 2021, 597, 381–386. [Google Scholar] [CrossRef] [PubMed]
- Franco, I.; Johansson, A.; Olsson, K.; Vrtacnik, P.; Lundin, P.; Helgadottir, H.T.; Larsson, M.; Revechon, G.; Bosia, C.; Pagnani, A.; et al. Somatic mutagenesis in satellite cells associates with human skeletal muscle aging. Nat. Commun. 2018, 9, 800. [Google Scholar] [CrossRef] [PubMed]
- Kaplanis, J.; Ide, B.; Sanghvi, R.; Neville, M.; Danecek, P.; Coorens, T.; Prigmore, E.; Short, P.; Gallone, G.; McRae, J.; et al. Genetic and chemotherapeutic influences on germline hypermutation. Nature 2022, 605, 503–508. [Google Scholar] [CrossRef] [PubMed]
- Xia, B.; Yan, Y.; Baron, M.; Wagner, F.; Barkley, D.; Chiodin, M.; Kim, S.Y.; Keefe, D.L.; Alukal, J.P.; Boeke, J.D.; et al. Widespread Transcriptional Scanning in the Testis Modulates Gene Evolution Rates. Cell 2020, 180, 248–262.e21. [Google Scholar] [CrossRef]
- Baert, Y.; De Kock, J.; Alves-Lopes, J.P.; Soder, O.; Stukenborg, J.B.; Goossens, E. Primary Human Testicular Cells Self-Organize into Organoids with Testicular Properties. Stem Cell Rep. 2017, 8, 30–38. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hodge, R.A.; Bach, E.A. Mechanisms of Germline Stem Cell Competition across Species. Life 2024, 14, 1251. https://doi.org/10.3390/life14101251
Hodge RA, Bach EA. Mechanisms of Germline Stem Cell Competition across Species. Life. 2024; 14(10):1251. https://doi.org/10.3390/life14101251
Chicago/Turabian StyleHodge, Rachel A., and Erika A. Bach. 2024. "Mechanisms of Germline Stem Cell Competition across Species" Life 14, no. 10: 1251. https://doi.org/10.3390/life14101251
APA StyleHodge, R. A., & Bach, E. A. (2024). Mechanisms of Germline Stem Cell Competition across Species. Life, 14(10), 1251. https://doi.org/10.3390/life14101251









